Abstract
MRP1 (multidrug-resistance-associated protein 1; also known as ABCC1) is a member of the human ABC (ATP-binding cassette) transporter superfamily that confers cell resistance to chemotherapeutic agents. Considering the structural and functional similarities to the other ABC proteins, the interaction between its two NBDs (nucleotide-binding domains), NBD1 (N-terminal NBD) and NBD2 (C-terminal NBD), is proposed to be essential for the regulation of the ATP-binding/ATP-hydrolysis cycle of MRP1. We were interested in the ability of recombinant NBD1 and NBD2 to interact with each other and to influence ATPase activity. We purified NBD1 (Asn642–Ser871) and NBD2 (Ser1286–Val1531) as soluble monomers under native conditions. We measured extremely low intrinsic ATPase activity of NBD1 (10−5 s−1) and NBD2 (6×10−6 s−1) and no increase in the ATP-hydrolysis rate could be detected in an NBD1+NBD2 mixture, with concentrations up to 200 μM. Despite the fact that both monomers bind ATP, no stable NBD1·NBD2 heterodimer could be isolated by gel-filtration chromatography or native-PAGE, but we observed some significant modifications of the heteronuclear single-quantum correlation NMR spectrum of 15N-NBD1 in the presence of NBD2. This apparent NBD1·NBD2 interaction only occurred in the presence of Mg2+ and ATP. Partial sequential assignment of the NBD1 backbone resonances shows that residue Gly771 of the LSGGQ sequence is involved in NBD1·NBD2 complex formation. This is the first NMR observation of a direct interaction between the ABC signature and the opposite NBD. Our study also reveals that the NBD1·NBD2 heterodimer of MRP1 is a transient complex. This labile interaction is not sufficient to induce an ATPase co-operativity of the NBDs and suggests that other structures are required for the ATPase activation mechanism.
Keywords: ABC signature, ABC transporter, ABCC1, biochemical and NMR studies, MRP1, nucleotide-binding domain (NBD)
Abbreviations: ABC, ATP-binding cassette; CFTR, cystic fibrosis transmembrane conductance regulator; HSQC, heteronuclear single-quantum correlation; IPTG, isopropyl β-D-thiogalactoside; ME, 2-mercaptoethanol; MRP1, multidrug-resistance-associated protein 1; NBD, nucleotide-binding domain; NBD1, N-terminal NBD; NBD2, C-terminal NBD; P-gp, P-glycoprotein; RLI, RNase-L inhibitor; RT, reverse transcriptase; RTS, rapid translation system
INTRODUCTION
ABC (ATP-binding cassette) transporters couple ATP hydrolysis with the transport of endogenous and exogenous molecules across biological membranes. The canonical functional form of ABC proteins consists of two hydrophilic NBDs (nucleotide-binding domains) located at the cytoplasmic surface of the membrane and hydrophobic transmembrane spanning domains that are thought to form the translocation pathway [1].
MRP1 (multidrug-resistance-associated protein 1; also known as ABCC1) is a 1531 amino acid membrane protein belonging to the ‘C’ subfamily of the ABC superfamily (see [2]). MRP1 was discovered by virtue of its ability to cause multidrug resistance in human cell lines [3] and has been detected in tumours. This ABC protein confers resistance to diverse chemotherapeutic agents and is able to transport anionic conjugate compounds out of cells [4,5]. MRP1 is an energy-dependent efflux pump under the control of ATP binding/hydrolysis and ATPase activity takes place within the NBDs. Although ABC transporters have been studied for several years, little is known about the mechanism by which the energy of ATP hydrolysis is used to transport substrate across the membrane. The NBDs of MRP1 contain conserved motifs that are common to all ABC family members: the Walker A and B motifs required for nucleotide binding and hydrolysis and the ABC signature sequence (LSGGQ) or C-motif.
Dimer formation between NBDs of ABC transporters, either as homo- or hetero-dimers, appears as a general rule of their structural organization and is required for the functional activation of transport. Homodimers are commonly proposed for bacterial ABC proteins, which contain only one NBD. The structures of several isolated NBD domains have been solved by X-ray crystallography and different organization of homodimers has been proposed. However, the head-to-tail model revealed by the crystal structures of Rad50 [6], MutS [7,8], MJ0796-E171Q [9], BtuCD [10] and MalK [11] is now considered to reflect the physiological NBD dimer. In the dimer model, the two bound nucleotides are at the interface between the subunits and are co-ordinated by amino acids from both NBDs. The residues of the ABC signature sequence of one subunit complete the nucleotide-binding site of the opposite monomer. The very recent X-ray crystallographic structure of RLI (RNase-L inhibitor) [12] characterizes the first bacterial heterodimer of a twin cassette ATPase. However, there are no three-dimensional structural data available yet describing the NBD1–NBD2 (where NBD1 stands for N-terminal NBD and NBD2 stands for C-terminal NBD) heterodimer interface of human multidrug transporters.
In the eukaryotic ABC transporters of the C-subfamily, a single polypeptide chain spans two non-identical NBDs, NBD1 and NBD2. The inactivation of one of the two NBDs abolishes the transport activity of the protein. In MRP1, both NBDs contain the conserved sequences Walker A and B motifs, H-switch and Q-loop. The ABC signature sequence LSGGQ is replaced by LSVGQ in NBD2 and the catalytic glutamate is substituted by an aspartate residue in NBD1. Moreover, the NBD1 is characterized by the 13 amino acid deletion preceding the Q-loop as compared with NBD2. Furthermore, it has been described that the two NBDs of MRP1 have different properties and functions based on mutations, ATP binding and ATP-dependent LTC4 (leukotriene C4) transport [13–15]. Mutagenesis studies of the ABC signature sequence in MRP1-NBDs were recently carried out to determine its role in the communication between NBDs [16–18]. The loss of ATPase and transport activity of mutated MRP1 highlights the importance of these residues of the LSGGQ motif in MRP1 function.
In a previous study, we showed that isolated MRP1-NBD1 has a very low intrinsic ATPase activity [19]. We proposed that this could be due to a lack of additional interactions localized at the NBD1·NBD2 heterodimer interface in MRP1, suggesting that the interaction with NBD2 is required to enhance this activity. The objective of the present study was to examine how NBD1 and NBD2 interact. We produced highly purified NBD2 in monomeric form, as well as high amounts of isotopically labelled NBD1. Using biochemical and NMR techniques, we compared the ATPase activity of both domains and we looked for a possible interaction and stimulation of the ATPase activity induced by the heterodimer formation. We illustrate here the first NMR study of the interaction in solution between the isolated NBD1 and NBD2 of an ABC transporter.
MATERIALS AND METHODS
Construction of the recombinant NBD expression vectors
Total RNAs were purified from the human cell line HL60/ADR (Oncodesign, Dijon, France), using the High Pure RNA Isolation kit (Roche Diagnostics, Meylan, France). RT (reverse transcriptase)–PCR amplification of the MRP1-NBD2 cDNA fragment was performed with the Titan one-tube RT–PCR system (Roche Diagnostics). The forward primer 5′-acatatgagctggccccagg-3′ and the reverse primer 5′-agcggccgctcacaccaagccg-3′ were used, where underlined bases correspond to the NdeI and NotI restriction sites respectively. The MRP1-NBD2 PCR fragment was cloned into T-overhanging pBlueScript and subcloned into pET28a (Novagen, Madison, WI, U.S.A.), using appropriate enzymes and was then sequenced. The construction of the expression vector used for the production of the MRP1-NBD1 domain has been described previously [19]. The pET vectors allow the expression of N-terminal His-tagged NBD fusion protein, encompassing residues Asn642–Ser871 and Ser1286–Val1531 for the NBD1 and NBD2 domains respectively.
Production of recombinant NBDs in Escherichia coli
NBD2 production
The E. coli BL21(DE3)pLysS (Novagen) was transformed with recombinant pET28a-MRP1-NBD2 vector and grown at 37 °C in 2YT (yeast extract tryptone; 1.6%, w/v, tryptone, 1%, w/v, yeast extract and 0.5%, w/v, NaCl) medium (Difco, West Molesey, Surrey, U.K.) containing kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml). Gene expression was induced (A600 0.6) with 0.5 mM IPTG (isopropyl β-D-thiogalactoside; Promega, Charbonnières, France) for 20 h at 16 °C (A600 6–7). Then, cells were harvested by centrifugation (10000 g for 15 min) at 4 °C, frozen in liquid nitrogen and stored at −80 °C until use.
NBD1 production and 15N13C2H isotopic labelling
The overexpression of the unlabelled NBD1 and the production of the 15N-labelled domain, using the pET28a-MRP1-NBD1 vector in E. coli, have been described previously [19]. In order to obtain 15N13C2H-labelled NBD1 domain for the NMR studies, cultures were grown in 3 litres of M9 minimal medium containing 0.1% [15N]ammonium chloride (Cambridge Isotope Laboratories, Andover, MA, U.S.A.), 0.33% [13C]glucose (Spectra Stable Isotopes, Columbia, MD, U.S.A.) and 99% 2H2O (Spectra Stable Isotopes). The production of 15N13C2H-labelled NBD1 domain was induced with 0.5 mM IPTG at a cell density of 0.5 unit at 600 nm and continued overnight at 28 °C.
Protein purification
NBD2 purification
All purification steps were performed at 4 °C. The cell pellet from 3 litres of culture was resuspended in 80 ml of buffer A (25 mM Tris/HCl, pH 8.0, 500 mM KCl and 10%, v/v, glycerol) with 1 mM Pefabloc-SC (Roche Diagnostics). Cells were lysed by sonication (5 s on/5 s off, 36 cycles) in a Vibro-cell sonicator (Fisher Bioblock Scientific, Illkirch, France) and ultracentrifuged at 200000 g for 120 min. The supernatant was applied on to a 5 ml Talon-Superflow cobalt-affinity resin column (BD Biosciences Clontech, Mountain View, CA, U.S.A.) connected to an AKTAexplorer system (Amersham Biosciences, Saclay, France). Proteins were eluted from the column with a 10–200 mM imidazole gradient in buffer A at a flow rate of 2 ml/min. Fractions were analysed by SDS/PAGE and NBD2 (eluted between 70 and 100 mM imidazole) was collected. After concentration to a final volume of 10 ml, NBD2 was loaded on to a 50 ml HiPrep 26/10 desalting column (Amersham Biosciences) at a flow rate of 2 ml/min in buffer A. NBD2 fractions were collected and purified on to the 5 ml Talon-Superflow cobalt-affinity resin column for a second time, with a 30–150 mM imidazole gradient. NBD2 fractions were then concentrated up to 4 ml and loaded on to a 180 ml Superdex 75 PrepGrade column (Amersham Biosciences) in buffer B [25 mM Tris/HCl, pH 8.0, 7 mM ME (2-mercaptoethanol) and 10% glycerol] with 150 mM KCl, using a flow rate of 1 ml/min. The NBD2 fractions, containing only the monomeric form of the protein, were collected and applied on to the HiPrep 26/10 desalting column in buffer B. The last purification step was performed on a 6 ml Resource Q column (Amersham Biosciences), using a linear KCl gradient in buffer B. After SDS/PAGE analysis of the fractions and the control of the ATPase activity over the gradient, NBD2 was stored at −80 °C at a concentration of 300 μM in 25 mM Tris/HCl (pH 8.0) and 150 mM KCl buffer. For the NMR experiments, NBD2 was stored in buffer C (200 mM Na/K phosphate, pH 7.0, 10 mM NaCl and 7 mM ME).
NBD1, [15N]NBD1 and [15N13C2H]NBD1 purifications
The NBD1 preparations used for the biochemical experiments and the [15N]NBD1 or [15N13C2H]NBD1 used for the NMR studies were purified as described previously [19]. The proteins were stored at −80 °C in buffer C at an NBD1 concentration of approx. 500 μM.
Amino acid-specific labelling of NBD1 in vitro
To obtain a selective amino acid labelling of NBD1 for structural studies, a cell-free protein synthesis technique was used. Protein production was performed by using the RTS 500 (rapid translation system 500) instrument (Roche Diagnostics). The RTS 500 E. coli HY kit and the pET28a-MRP1-NBD1 as template vector were used. For the specific incorporation of the selected amino acids, [15N]Leu, [15N]Ser or [15N]Val (Spectra Stable Isotopes) were combined with unlabelled amino acid solutions of the RTS amino acid sampler as described by the manufacturer's instructions. Protein synthesis reaction was continued for 16 h at 30 °C. Reaction products were filtered and purified on to a 1 ml HisTrap HP column (Amersham Biosciences) with a linear imidazole gradient. Proteins were stored in buffer C and the yield of purified [15N]Leu–NBD1, [15N]Ser–NBD1 and [15N]Val–NBD1 was approx. 1–1.5 mg.
ATPase activity
The NBD ATPase activity was measured as the production of [33P]Pi from [γ-33P]ATP (PerkinElmer, Applied Biosystems, Warrrington, Cheshire, U.K.) using the charcoal method [19]. Briefly, the assay was performed at 30 °C in a reaction mixture (70 μl) containing 30–100 μM of NBDs in a buffer containing 50 mM Tris/HCl (pH 8.0), 100 mM KCl and [γ-33P]ATP (0.5–1.5 mM, 10–30 Bq/pmol). MgCl2 was added as indicated in the Figure legends. Aliquots (5 μl) of the reaction mixture were withdrawn at the indicated time to be treated with activated charcoal. [33P]Pi was counted in a Wallac1414 liquid-scintillation counter.
NMR experiments
Samples for NMR were prepared in buffer C. Samples with both NBD1 and NBD2 were prepared on Microcon YM-10 centrifugal filter devices (Millipore, Watford, Herts., U.K.), with final concentrations of 220 μM NBD1, 1 mM Mg-ATP and 75–350 μM NBD2, or by 1:1 (v/v) dilution of 450 μM NBD1 with 340 μM NBD2 without subsequent concentration with 4 mM ATP and 8 mM MgCl2. 15N HSQC (heteronuclear single-quantum correlation) spectra of uniformly or selectively 15N-labelled NBD1 were recorded at 298 K on a Bruker DRX600 spectrometer equipped with a triple resonance 1H13C15N 5 mm gradient cryoprobe. HNCA, HNCO, HNCACB, HNCOCA and HNCOCACB triple resonance experiments were performed on [15N13C2H]NBD1 at 295 K. The NMR signal was locked with 5% 2H2O. For each HSQC spectrum, 120 complex t1 (15N) and 512 complex t2 (1H) points were acquired with 48 scans/increment. The spectral widths were 28 (15N) and 5 p.p.m. (1H), the frequency offsets at 118.5 (15N) and 8 p.p.m. (1H). The spectra were apodized with shifted sine-bell functions before Fourier transformation and zero-filled to obtain 256×512 real matrixes using XWINNMR. Small zero-order phase adjustments in the t2 dimension were necessary. A polynomial automatic baseline correction was also performed in this dimension. The spectra were analysed with SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco, CA, U.S.A.). Peak line widths and heights were determined by a Gaussian fit for the 15N and 1H dimensions using the data above the lowest contour.
Other methods
Protein concentrations were determined with the Bio-Rad protein assay using BSA as a standard. SDS/PAGE was performed using a 15:0.37 acrylamide/bisacrylamide gel and stained with Coomassie Blue. SDS/PAGE molecular mass standards were from Amersham Biosciences (LMW-SDS marker kit). Native-gel electrophoresis (6.6%, w/v, acrylamide in Tris/glycine buffer, pH 8.9) was run for 4 h at 10 mA and 10 °C.
RESULTS
We have previously reported the monomeric state and the extremely low ATPase activity of isolated NBD1 of MRP1 with biochemical and NMR data [19]. In order to determine the contribution of NBD2 in the ATPase activity of MRP1 and the role of the NBD1·NBD2 heterodimer in the enhancement of the catalytic activity, we produced homogeneous NBD2 to check its influence on the biochemical and structural properties of NBD1.
Purification of recombinant NBD2
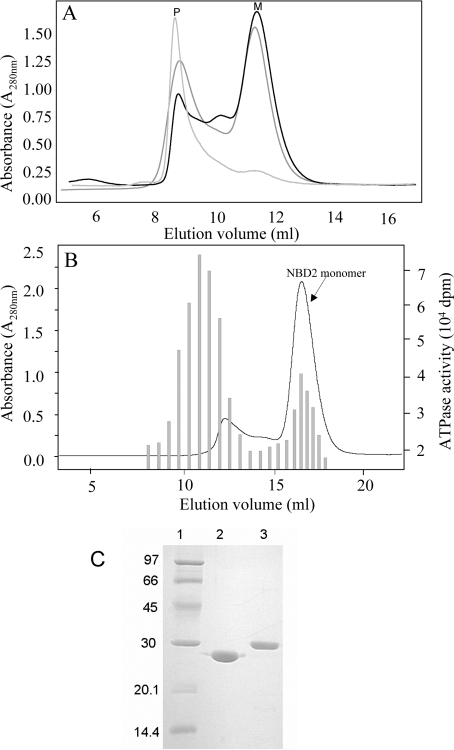
The N-terminal boundary of the MRP1-NBD2 fragment was defined with the help of sequence alignments with NBDs of known structures. This domain was extended from Ser1286 to Val1531, the C-terminus of the full-length protein. NBD2 was produced as an N-terminal His6-tag fusion protein in E. coli (theoretical molecular mass of 29.7 kDa). The optimal yield of soluble NBD2 that can be extracted from cells without detergent was obtained from cultures induced for 20 h at 16 °C. During the purification procedure, particular care was taken to control the monomeric or oligomeric state of NBD2. Figure 1(A) shows the chromatographic profiles from a gel-filtration column of three different NBD2 preparations. The apparent molecular mass of the NBD2 domain varies from 30 kDa (Figure 1A, peak M) to more than 150 kDa (Figure 1A, peak P). NBD2 was eluted as an oligomer in peak P, migrated as a single band on SDS/PAGE, but appeared as a smear on native-PAGE (results not shown). Thus, to ensure that the interaction experiments are performed only with NBD2 monomers, the first steps of purification were performed very rapidly and at 4 °C. Excessive concentration and extensive dialysis of NBD2 between different chromatographic separations were avoided. A rapid desalting of the fractions by passage through a HiPrep™ 26/10 gel-filtration column maintained most of the NBD2 as monomers.
Figure 1. Purification of MRP1-NBD2.
(A) Gel-filtration chromatographic profiles of NBD2 preparations. NBD2 samples of three different preparations were applied to a gel-filtration Superdex 75 HR 10/30 column. Protein elution was followed by absorption at A280. Monomeric NBD2 was eluted in the fractions corresponding to peak M, whereas oligomers of NBD2 were recovered in the fractions corresponding to peak P. The Superdex 75 column was calibrated using the low molecular mass gel-filtration kit (Amersham Biosciences, 13.7, 25, 43, 67 and 2000 kDa) and alcohol dehydrogenase (Sigma–Aldrich, Poole, Dorset, U.K., 150 kDa). (B) ATPase activity in Superdex 200 elution fractions. An NBD2 sample purified on cobalt-affinity resin was applied to a gel-filtration Superdex 200HR 10/30 column. Protein elution was followed by A280 (line curve). ATPase activity was followed in 25 μl of aliquots of each fraction by adding 5 μl of [γ-33P]ATP (final concentration 0.3 mM, 22 Bq/pmol) to start the reaction. ATPase activity (shaded bars) was monitored by measuring the amount of [33P]Pi produced during 4 h at 30 °C. (C) SDS/PAGE of NBD1 and NBD2. Purified NBDs were analysed by SDS/PAGE (15% polyacrylamide) and revealed by Coomassie Blue staining. Lane 1, molecular mass standards; lane 2, 5 μg of NBD1; lane 3, 5 μg of NBD2.
We paid special attention to avoid nucleotidase contaminations in the NBD2 preparations, a recurrent difficulty observed during the purification of NBD1. This is important considering the particularly low ATPase activity of these domains (see below), as compared with the ATPase from E. coli present in the cell extract. Figure 1(B) shows the importance of such contaminants in partially purified NBD2. These ATPase activities are not dependent on NBD2, since their maximum did not overlap with fractions containing the NBD2 domain and were removed by gel filtration and/or anionic chromatography. The yield of NBD2 purified to homogeneity was rather low (3–5 mg/l of culture), but the protein can be stored at −80 °C as a monomer protein until used. Figure 1(C) presents an SDS/PAGE analysis after loading with the NBD1 and NBD2 purified domains produced from E. coli.
ATPase activity of the isolated and combined NBD1/NBD2
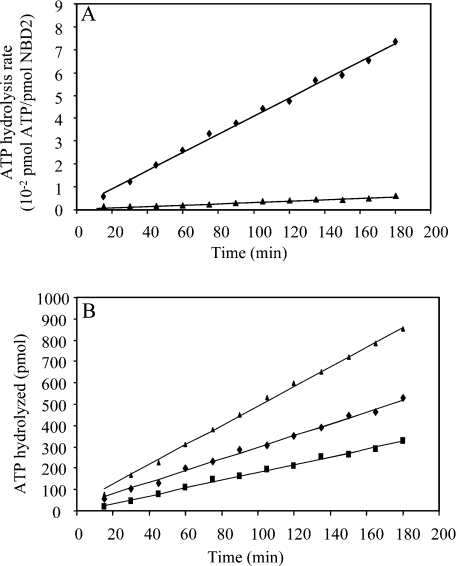
Biochemical studies of the full-length MRP1 have provided evidence that the two MRP1-NBDs are not equivalent in their ability to hydrolyse ATP [13–15]. The characterization of the isolated domains could help us to understand whether this difference is due to an intrinsic property of the NBD. The homogeneity of the NBDs preparations and the correct folding of NBD2 were controlled by the binding of the nucleotide to the isolated domains. The binding efficiency of TNP-ATP [2′,3′-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate] was measured for both domains and gave similar dissociation constant values in the μM range (results not shown). To determine the ATPase activity of isolated NBD2, we quantified the [33P]Pi produced during the hydrolysis of [γ-33P]ATP mediated by NBD2. The rate of ATP hydrolysis by the isolated domain was found to be very low. In the experiment described in Figure 2(A), the ATP hydrolysis rate of NBD2 was 6×10−6 s−1 (0.01 nmol of Pi·mg−1·min−1). This ATPase activity is fully dependent on the presence of Mg2+, since no activity could be detected in the presence of EDTA. The low value of NBD2 ATPase activity was highly reproducible over several protein preparations.
Figure 2. NBD2 and NBD1+NBD2 ATPase activity.
(A) NBD2 ATPase activity was determined using [γ-33P]ATP as substrate. The reaction mixtures contained NBD2 (66 μM) and ATP (1 mM) in 70 μl of buffer (50 mM Tris/HCl, pH 8 and 100 mM KCl) in the presence of 3 mM MgCl2 (◆) or 1 mM EDTA (▲). Aliquots (5 μl) were withdrawn at indicated times and [33P]Pi produced was counted. (B) ATPase activity of the NBDs was followed in standard buffer (50 mM Tris/HCl, pH 8, 100 mM KCl and 3 mM MgCl2) at 30 °C. The ATPase activity was measured as the amount of [γ-33P]ATP hydrolysed in aliquots (5 μl) at the indicated times and in the presence of 66 μM NBD2 (■); 66 μM NBD1 (◆); 66 μM NBD1 and 66 μM NBD2 (▲).
In our previous study, we characterized the ATPase activity of isolated NBD1 of MRP1 [19]. In the present study, we compare it with that of NBD2 under the same conditions. Moreover, we mixed the NBDs to investigate the potential co-operativity of the NBDs in the ATP hydrolysis reaction. As shown in Figure 2(B), NBD2 ATPase activity (6×10−6 s−1) is somewhat lower than that of NBD1 (10×10−6 s−1). The equimolar mixture of NBD1 and NBD2 has an ATPase activity that corresponds strictly to the sum of the ATPase of the isolated domains. Similar experiments with different NBD concentrations and a variety of buffer conditions were also performed. The results did not show any significant increase in the ATPase activity and led us to conclude that there was no detectable co-operative effect on the ATPase, induced by the presence of both domains.
No evidence for stable NBD1·NBD2 heterodimer formation
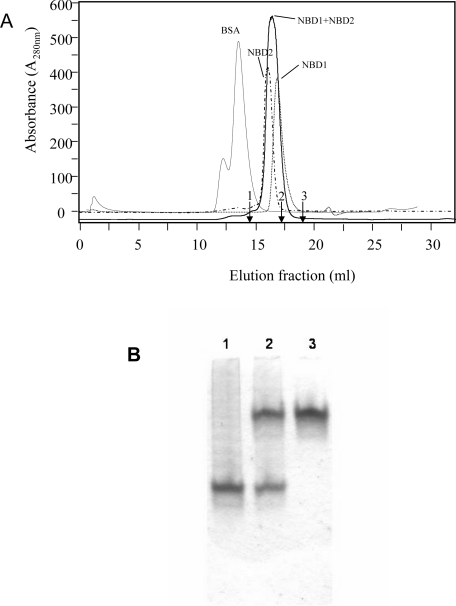
With the purified NBD1 and NBD2, it became possible to address the question of whether a stable NBD1·NBD2 heterodimer formation occurs. If such an interaction takes place, it should be detectable by the techniques of gel-filtration chromatography or native PAGE. NBD1 or NBD2 proteins were subjected to gel filtration on a Superdex 200 column that should efficiently resolve the NBD1 and NBD2 monomers from a stable NBD1·NBD2 heterodimer. The isolated domains are eluted as single peaks (Figure 3A) with apparent molecular masses close to those predicted (∼27–30 kDa). A mixture containing an equimolar amount of both domains was analysed under the same conditions. The mixture of NBD1+NBD2 proteins was eluted with an apparent molecular mass similar to that of the monomers. Several experiments were reproduced in the presence of nucleotides and/or magnesium, but these two components did not favour the formation of the expected NBD1·NBD2 complex. Figure 3(B) presents an attempt to visualize the heterodimer by native PAGE. The single bands observed with NBD1 or NBD2 confirm the homogeneity of the protein preparations. The higher negative charge of NBD2 results in faster migration of this domain as compared with NBD1. We did not detect any NBD1·NBD2 complex by this method, since no additional bands appeared. The finding that the heterodimer could not be isolated by gel filtration or native PAGE suggests that the isolated NBDs do not interact or, if they do, they form a very transient complex that cannot be visualized using these techniques.
Figure 3. Gel-filtration chromatography and native PAGE of NBD1+NBD2 mixture.
(A) Chromatographic profiles of Superdex 200 column fractions showing the elution of NBD1, 27 kDa; NBD2, 30 kDa; an NBD1+NBD2 mixture; and BSA, 67 kDa. The Superdex 200 column was calibrated with other proteins (indicated by arrows): 1, 43 kDa ovalbumin; 2, 25 kDa chymotrypsinogen A; 3, 13.7 kDa RNase A. Protein elution was recorded at A280. (B) Native PAGE was loaded with NBD2 (3 μg, lane 1), a mixture of NBD1 and NBD2 (3 μg for each NBD, lane 2) and NBD1 (3 μg, lane 3).
NBD1·NBD2 association detected by NMR line broadening
NMR perturbation mapping is an easily applicable NMR technique to identify protein–protein complexes of weak affinity and has been previously used to identify several protein–protein-binding sites [20]. It is based on the measurement of two-dimensional 1H-15N HSQC spectra, which correlate the 1H and 15N chemical shifts of the protein backbone amide of each residue as well as those of the side-chain amine groups of Gln and Asn. The 1H-15N HSQC spectrum of a protein is considered as a fingerprint of its backbone conformation. Changes in chemical shifts, resonance intensities or line widths in the HSQC spectrum of a protein after the addition of its partner are indicative of dynamic processes, such as chemical exchange between the free and complexed forms. In order to detect an association between isolated NBD1 and NBD2, we recorded 1H-15N HSQC spectra of 15N-labelled NBD1 in the absence and in the presence of unlabelled NBD2, with various ATP and Mg2+ concentrations.
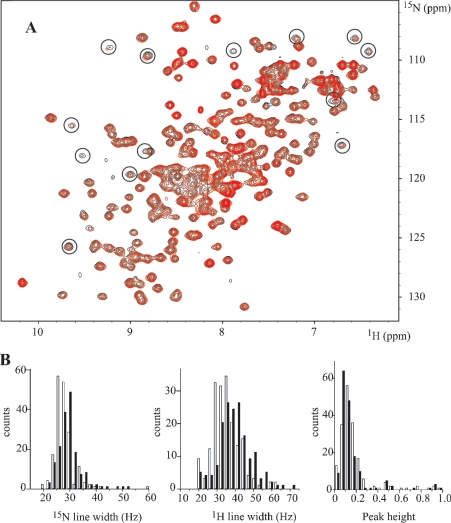
As we have previously reported [19], binding of ATP to NBD1 is revealed by shifts of several resonances in the HSQC spectrum of NBD1. We have also observed no line broadening of NBD1 in the ATP-bound conformation. When we added NBD2, ATP and Mg2+ were kept in excess compared with NBDs to avoid the ATP-free conformation of NBD1. Under these experimental conditions, NBD2 does not induce any major variation in the chemical shifts of NBD1 (Figure 4A): no peak is shifted by more than half its line width, which indicates that no conformational changes take place. However, line broadening and concomitant decrease in intensities are detected for NBD1 when NBD2 is added. The most striking of these are highlighted by circles on the contour plot in Figure 4(A). Moreover, these changes are ATP-dependent. They are not observed in an NBD1+NBD2 mixture with only Mg2+. A more detailed analysis of the distributions of 1H/15N line widths and peak heights was performed for isolated peaks or peaks that can be easily convoluted to a Gaussian form (Figure 4B). The intensities were normalized relative to the intensity of peaks that do not broaden on addition of NBD2. The maximum of the line width distribution of NBD1 is shifted by approx. 5 and 10 Hz for 15N and 1H respectively. We exclude auto-association of NBD1 and an effect of viscosity due to higher protein concentrations, on the basis that these changes are observed neither with more concentrated NBD1 samples nor in an NBD1+NBD2 mixture in the absence of ATP (results not shown). Line broadening is more likely to originate in slow or intermediate exchange between monomer and NBD2-complexed NBD1 or in an increase in effective molecular mass, which is approx. 57 kDa for the NBD1·NBD2 dimer as compared with 27 kDa for monomer NBD1. The concomitant increase in the global reorientational correlation time of NBD1, when associated with NBD2, is consistent with a more efficient transversal relaxation.
Figure 4. 1H-15N HSQC spectra of NBD1 with and without NBD2: distribution of line widths and intensities.
(A) Overlaid contour plots of the HSQC spectra of NBD1 (220 μM) at 298 K in the presence of 0:1 (black) and 0.75:1 (red) molar equivalents of NBD2. The solutions contained 4 mM ATP and 8 mM MgCl2. (B) Distribution of 15N and 1H line widths and normalized peak heights in the 15N HSQC spectra without (white bars) and with (black bars) NBD2.
Assignment of the NBD1 backbone resonances shows that Gly771 is involved in a transient NBD1·NBD2 complex
Line broadening is not homogeneous. Several peaks, which are broadened, sometimes beyond the detection limit, are probably associated with residues directly involved in the NBD1·NBD2 interaction. To identify these residues, we have undertaken the sequential assignment of NBD1, based on triple resonance NMR experiments. We have achieved so far the assignment of 80% of the backbone. In order to validate these assignments, we compared them with the 1H-15N HSQC spectra of selectively 15N-labelled NBD1, produced by the RTS technique (see the Materials and methods section). Good quality spectra were obtained by using a cryoprobe with protein concentrations of 50–100 μM. Contrary with in vivo production of NBD1 with selected 15N-labelled amino acids, the in vitro RTS method allows very selective labelling (Figure 5). Cross-labelling, especially of Ile and Val in the case of Leu, is avoided with RTS. Moreover, by overlaying the spectra of in vivo and in vitro produced proteins, we conclude that the RTS produced proteins fold normally (Figure 5). [15N]Leu and [15N]Ser labellings were crucial to assign the residues of the ABC signature LSGGQ with a high degree of confidence. Combined with the partial sequential assignment, it allowed us to discriminate between M841SGGK845 and the ABC signature residues L768SGGQ772 (assignments shown in Figure 5).
Figure 5. 1H-15N HSQC spectra of NBD1 with different 15N-labelling methods.
Overlaid contour plots of the HSQC spectra at 298 K of NBD1 obtained with uniform labelling in E. coli (black, 220 μM), in vivo selective labelling with 15N-Leu in E. coli (blue, 220 μM) and in vitro selective labelling with 15N-Leu by cell-free expression (red, 80 μM). The last HSQC spectrum shows the 23 Leu signals from NBD1 and that of the His tag.
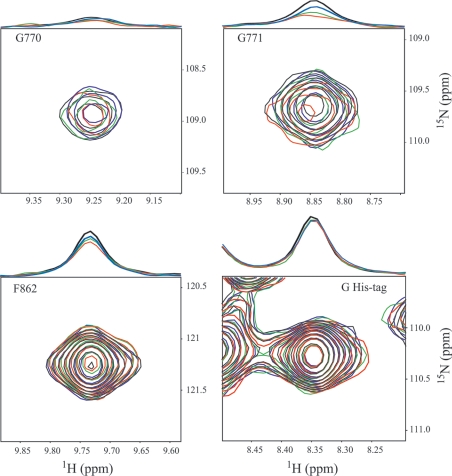
Titration experiments with increasing amounts of NBD2 showed that a relatively high NBD2/NBD1 ratio is required to detect significant modifications in the NBD1 HSQC spectrum (Figure 6). Not all signals are affected in the same way, due to differences in local internal motions. In the general case, the intensity decreases of approx. 20%, while the peaks broaden by a few Hz, e.g. Phe862. Side chains and the residues belonging to the His tag, which are subject to fast local motions, remain unchanged (see Figure 6 for Gly of His-tag). For a few residues, such as Gly771 and Gly770 to a lesser extent, the loss of intensity and line broadening are more significant, especially when more than 1 molar equivalent of NBD2 is added (Figure 6). This result can be explained by the formation of a transient NBD1·NBD2 complex and shows that Gly771 is located at the interface, as the interaction of Gly771 with NBD2 would result in a local decrease in flexibility. The special case of Gly771 shows that the ABC signature motif of MRP1 is directly involved in ATP-dependent NBD1·NBD2 complex formation.
Figure 6. NMR titration experiments of NBD1 by NBD2.
The HSQC spectra of NBD1 (220 μM) were titrated in the presence of 1 mM Mg-ATP by addition of 0:1 (black), 0.35:1 (blue), 1:1 (green) and 1.6:1 (red) molar equivalents of NBD2. The selected regions are centred on the backbone correlations of Gly770, Gly771, Phe862 and one Gly of His tag. On the top of each two-dimensional spectrum, the extracted 1H spectrum at the 15N chemical shift of these residues is drawn, which has been normalized relative to the Gly His-tag signal.
DISCUSSION
In the present study, we investigated the biochemical and structural consequences of heterodimer formation between the two NBDs, NBD1 and NBD2, of MRP1. For this purpose, we purified labelled NBD1 to develop NMR studies and we isolated NBD2 in a monomeric form to examine heterodimer formation. Both NBD domains are folded, monomeric and can bind nucleotide, despite a very low intrinsic ATPase activity. Our NMR data indicate that isolated NBD1 interacts with NBD2, showing that a complex involving Gly771 of the signature sequence can be formed. Thus complex formation between the isolated NBDs of MRP1 is shown here for the first time. The interaction of two NBDs in homo- or hetero-dimers is assumed to be a key element of the three-dimensional structural arrangement of the ABC transporters (for reviews, see [21–25]). This NBD dimerization process is thought to be essential for the activation of the transport mechanism for this family of proteins, as it has been recently shown for CFTR (cystic fibrosis transmembrane conductance regulator; also known as ABCC7) [26].
A number of three-dimensional crystal structures of bacterial ABC transporter NBDs have been reported and several homodimer interfaces are described. The first NBD heterodimer structure [12], which has been published very recently, is the MgADP bound structure of the Pyrococcus furiosus twin cassette ATPase RLI. Nevertheless, there are only three mammalian ABC transporters for which high-resolution three-dimensional NBD structures are available: NBD of TAP1 (transporter associated with antigen processing 1) [27], NBD1 of mouse CFTR [28,29] and NBD1 of human CFTR [30]. No structure of the NBD2 of any ABC B- or C-subfamily protein has been solved so far. Structural information about NBD heterodimers could only be derived by molecular modelling, as described for the full-length ABC transporters P-gp (P-glycoprotein; ABCB1) [31] or SUR1/ABCC8 [32], or with the first two-dimensional crystal structure of MRP1 [33] and P-gp [34]. Recently, a three-dimensional model of the CFTR heterodimer was built [35]. The authors argued, based on this model, that the buried interface between CFTR-NBD1 and CFTR-NBD2 is rather small for a specific dimer contact. This can be compared with the extensive buried surface (>2000 Å2; where 1 Å=10−10 m) of the ATP-bound homodimer of bacterial MalK, whose additional C-terminal regulatory domain contributes substantially to dimer formation both in the presence and absence of nucleotide.
NMR allows characterization of weak affinity protein–protein complexes and is complementary to structure determination by X-ray crystallography. To our knowledge, only one other NBD of an ABC transporter protein, MJ1267 (LivG) of Methanococcus jannaschii, has been studied by NMR [36,37]. But no information about a putative homodimer formation in solution was reported. We could not obtain any evidence for an NBD1·NBD2 complex of MRP1 by gel filtration or native PAGE. However, relatively modest but significant changes in the 15N HSQC NMR spectrum of 15N-labelled NBD1 were observed on the addition of NBD2. The results suggest that the presence of NBD2 does not induce a structural rearrangement of NBD1, but demonstrate a weak but specific interaction of NBD1 in its Mg-ATP-bound conformation with NBD2. This interaction is ATP-dependent, since it is not detected in the absence of the nucleotide or in the presence of Mg2+ alone. The combined use of [15N13C2H]NBD1 for triple resonance experiments and specifically amino acid-labelled NBD1 enabled us to assign one of the major changes in the HSQC spectrum induced by the presence of NBD2 to residue Gly771 of the LSGGQ sequence. Thus our NMR results show that the labile NBD1·NBD2 association involves the ABC sequence signature.
The ABC signature region LSGGQ is the most characteristic motif of the whole ABC transporter superfamily and is found between the two Walker motifs. The prokaryotic ‘head-to-tail’ crystallographic structure, where the two monomers complement each other's active site, is now commonly accepted as the physiological NBD dimer form. The ABC signature motif of one subunit interacts with the ATP bound at the Walker motifs of the opposite subunit. Influence of the nucleotide on the homodimer conformation of the NBDs was revealed by comparison of the nucleotide-free and ATP-bound structures of bacterial Rad50 [6,38]. The work of Hopfner et al. [6,38] shows that the second Gly residue of the Rad50 LSGGQ motif interacts with the oxygen of the γ-phosphate of the ATP bound at the Walker motif of the opposite subunit. This residue in Rad50 is analogous to Gly771 affected in the MRP1-NBD1 spectrum. Similar arrangements have been found in the MsbA lipid transporter of Vibrio cholerae [39] and of E. coli [40], the vitamin B12 transporter BtuCD [10] and the maltose transporter MalK [11] of E. coli.
Several recent biochemical studies also described the close proximity of the two NBDs in the homodimeric MalFGK ABC protein by vanadate-induced cleavage experiments [41] and for the human multidrug transporter MDR1 using Walker A and signature sequence mutants in cross-linking experiments [42,43]. In the absence of any heterodimer structure and to more fully understand the role of the ABC signature sequence in MRP1, mutation experiments have been recently reported. In these studies, Gly771 of MRP1 was substituted for Asp [16,17]: Ren et al. [16] suggested that both signature sequences of MRP1 are involved in the ATP hydrolysis and transport activity of MRP1; Szentpetery et al. [17] proposed that the conserved Gly residue is part of the network responsible for the accelerated hydrolytic activity upon interaction of the protein with its transported substrate. In agreement with these recent biochemical studies, our NMR data reinforce the essential role of Gly771 in the direct interaction of the two subunits in the absence of other intramolecular constraints and support the similarities to the three-dimensional structures of known homodimers.
However, our findings underline some specific differences as compared with other studies. We were unable to isolate a stable NBD1·NBD2 complex, although a stable dimer could be expected in the absence of ATPase activity. Indeed, the absence of a stable heterodimer cannot be explained by the dissociation of the NBD upon ATP hydrolysis, because the estimated time to hydrolyse one ATP per NBD is much longer than the duration of the interaction experiments. The fast equilibrium between NBD monomers and dimers was reported [44,45]. Mutants of the ABC-ATPase GlcV from Sulfolobus solfataricus [45], defective in ATP hydrolysis, were used to reveal the homodimer in gel-filtration experiments. The authors further mutated the GlcV-Gly144 residue (homologous with MRP1-Gly771) and showed that this mutant failed to dimerize, indicating also an essential role for this residue in stabilizing the productive dimeric state of the protein. In MRP1, the weak NBD1·NBD2 affinity observed may be due to the fact that the NBDs are isolated from the other domains of the full-length molecule. There is very little information characterizing recombinant heterodimers of this ABC family because of the difficulties in isolating recombinant NBDs and probably because of the weakness of the dimer interaction.
An additional unexpected result of our study was the complete absence of stimulation of the ATPase activity measured in the presence of both MRP1-NBDs. This observation differs from the recently published results about the NBDs of CFTR [46], showing that the co-assembly of purified and renatured NBD1 and NBD2 exhibited a 2–3-fold enhancement in the catalytic activity relative to the isolated domains. Kidd et al. [46] concluded that the heterodimerization of the two NBD domains of CFTR induces co-operativity of the ATP hydrolysis reaction. The CFTR-NBD boundaries used in that study are quite similar to those we defined for the MRP1-NBDs. CFTR is another member of the C-subfamily of human ABC transporters and its NBDs, like those of MRP1, are also functionally asymmetric [47,48]. The ABC signature motif LSGGQ is substituted for LSHGH in CFTR-NBD2 and the mutation of Gly551, analogous to MRP1-Gly771, in the LSGGQ of CFTR-NBD1 is associated with a severe form of cystic fibrosis. As compared with our results, the NBD-dimer formation of these two structurally close ABC-family members must induce different conformational modifications at the ATPase catalytic centre. Moreover, we cannot rule out the possibility that the precise molecular mechanism required to generate an optimal catalytic activity is different for these two ABC transporters, which have highly divergent physiological functions. Other intramolecular contacts of the NBDs with the intracytoplasmic domains should be necessary in MRP1 to reveal the NBD1·NBD2 ATPase co-operativity. Our knowledge of the architecture of the ABC transporters shows that the dimerization of the NBDs is essential, but their interfaces seem often limited and without strong interaction. Additional structural elements, which are different according to the particular ABC enzyme (see [49]), are required to stabilize the NBD-dimer conformation. In MRP1, these domains still have to be identified to explain the drug-stimulated ATPase activity of the full-length protein.
Acknowledgments
We are grateful to F. Bontems (ICSN, Ecole Polytechnique, Palaiseau, France) for help in NMR measurements and valuable suggestions. We acknowledge F. Duffieux (Aventis Pharma, Romainville, France) and V. Stoven (Institut Pasteur, Paris, France) for initial studies on NBD1 and stimulating discussion. We thank Dr P. Genne (Oncodesign, Dijon, France) for the gift of HL60/ADR cells and J. Bignon (ICSN, CNRS, Gif-Sur-Yvette Cedex, France) for a critical reading of this paper.
References
- 1.Holland B., Cole S. P., Kuchler K., Higgins C. F., editors. London: Academic Press; 2003. ABC Proteins from Bacteria to M. [Google Scholar]
- 2.Hipfner D. R., Deeley R. G., Cole S. P. Structural, mechanistic and clinical aspects of MRP1. Biochim. Biophys. Acta. 1999;1461:359–376. doi: 10.1016/s0005-2736(99)00168-6. [DOI] [PubMed] [Google Scholar]
- 3.Cole S. P., Bhardwaj G., Gerlach J. H., Mackie J. E., Grant C. E., Almquist K. C., Stewart A. J., Kurz E. U., Duncan A. M., Deeley R. G. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 4.Cole S. P., Deeley R. G. Multidrug resistance mediated by the ATP-binding cassette transporter protein MRP. BioEssays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Leslie E. M., Deeley R. G., Cole S. P. Toxicological relevance of the multidrug resistance protein 1, MRP1 (ABCC1) and related transporters. Toxicology. 2001;167:3–23. doi: 10.1016/s0300-483x(01)00454-1. [DOI] [PubMed] [Google Scholar]
- 6.Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell (Cambridge, Mass.) 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 7.Lamers M. H., Perrakis A., Enzlin J. H., Winterwerp H. H., de Wind N., Sixma T. K. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature (London) 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 8.Obmolova G., Ban C., Hsieh P., Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature (London) 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 9.Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locher K. P., Lee A. T., Rees D. C. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 11.Chen J., Lu G., Lin J., Davidson A. L., Quiocho F. A. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Karcher A., Buttner K., Martens B., Jansen R. P., Hopfner K. P. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure (Camb.) 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Gao M., Cui H. R., Loe D. W., Grant C. E., Almquist K. C., Cole S. P., Deeley R. G. Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J. Biol. Chem. 2000;275:13098–13108. doi: 10.1074/jbc.275.17.13098. [DOI] [PubMed] [Google Scholar]
- 14.Hou Y., Cui L., Riordan J. R., Chang X. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J. Biol. Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- 15.Nagata K., Nishitani M., Matsuo M., Kioka N., Amachi T., Ueda K. Nonequivalent nucleotide trapping in the two nucleotide binding folds of the human multidrug resistance protein MRP1. J. Biol. Chem. 2000;275:17626–17630. doi: 10.1074/jbc.M000792200. [DOI] [PubMed] [Google Scholar]
- 16.Ren X. Q., Furukawa T., Haraguchi M., Sumizawa T., Aoki S., Kobayashi M., Akiyama S. Function of the ABC signature sequences in the human multidrug resistance protein 1. Mol. Pharmacol. 2004;65:1536–1542. doi: 10.1124/mol.65.6.1536. [DOI] [PubMed] [Google Scholar]
- 17.Szentpetery Z., Kern A., Liliom K., Sarkadi B., Varadi A., Bakos E. The role of the conserved glycines of ABC signature motifs of MRP1 in the communications between the substrate binding site and the catalytic centers. J. Biol. Chem. 2004;279:41670–41678. doi: 10.1074/jbc.M406484200. [DOI] [PubMed] [Google Scholar]
- 18.Szentpetery Z., Sarkadi B., Bakos E., Varadi A. Functional studies on the MRP1 multidrug transporter: characterization of ABC-signature mutant variants. Anticancer Res. 2004;24:449–455. [PubMed] [Google Scholar]
- 19.Ramaen O., Masscheleyn S., Duffieux F., Pamlard O., Oberkampf M., Lallemand J. Y., Stoven V., Jacquet E. Biochemical characterization and NMR studies of the nucleotide-binding domain 1 of multidrug-resistance-associated protein 1: evidence for interaction between ATP and Trp653. Biochem. J. 2003;376:749–756. doi: 10.1042/BJ20030998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuiderweg E. R. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 21.Kerr I. D. Structure and association of ATP-binding cassette transporter nucleotide-binding domains. Biochim. Biophys. Acta. 2002;1561:47–64. doi: 10.1016/s0304-4157(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt L., Tampe R. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 2002;12:754–760. doi: 10.1016/s0959-440x(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 23.Locher K. P. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 2004;14:426–431. doi: 10.1016/j.sbi.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Higgins C. F., Linton K. J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 25.Jones P. M., George A. M. The ABC transporter structure and mechanism: perspectives on recent research. Cell. Mol. Life Sci. 2004;61:682–699. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vergani P., Lockless S. W., Nairn A. C., Gadsby D. C. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature (London) 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaudet R., Wiley D. C. Structure of the ABC ATPase domain of human TAP1, the transporter associated with antigen processing. EMBO J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thibodeau P. H., Brautigam C. A., Machius M., Thomas P. J. Side chain and backbone contribution of Phe508 to CFTR folding. Nat. Struct. Mol. Biol. 2004;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis H. A., Buchanan S. G., Burley S. K., Conners K., Dickey M., Dorwart M., Fowler R., Gao X., Guggino W. B., Hendrickson W. A., et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis H. A., Zhao X., Wang C., Sauder J. M., Rooney I., Noland B. W., Lorimer D., Kearins M. C., Conners K., Condon B., et al. Impact of the DeltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J. Biol. Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 31.Stenham D. R., Campbell J. D., Sansom M. S. P., Higgins C. F., Kerr I. D., Linton K. J. An atomic detail model for the human ATP binding cassette transporter P-glycoprotein derived from disulfide cross-linking and homology modelling. FASEB J. 2003;17:2287–2289. doi: 10.1096/fj.03-0107fje. [DOI] [PubMed] [Google Scholar]
- 32.Campbell J. D., Sansom M. S. P., Ashcroft F. M. Potassium channel regulation. EMBO Rep. 2003;4:1038–1042. doi: 10.1038/sj.embor.7400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg M. F., Mao Q., Holzenburg A., Ford R. C., Deeley R. G., Cole S. P. The structure of the multidrug resistance protein 1 (MRP1/ABCC1). Crystallization and single-particle analysis. J. Biol. Chem. 2001;276:16076–16082. doi: 10.1074/jbc.M100176200. [DOI] [PubMed] [Google Scholar]
- 34.Rosenberg M. F., Callaghan R., Modok S., Higgins C. F., Ford R. C. Three-dimensional structure of P-glycoprotein: the transmembrane regions adopt an asymmetric configuration in the nucleotide-bound state. J. Biol. Chem. 2005;280:2857–2862. doi: 10.1074/jbc.M410296200. [DOI] [PubMed] [Google Scholar]
- 35.Callebaut I., Eudes R., Mornon J. P., Lehn P. Nucleotide-binding domains of human cystic fibrosis transmembrane conductance regulator: detailed sequence analysis and three-dimensional modeling of the heterodimer. Cell. Mol. Life Sci. 2004;61:230–242. doi: 10.1007/s00018-003-3386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C., Hunt J. F., Rance M., Palmer A. G., III 1H, 15N and 13C backbone assignment of MJ1267, an ATP-binding cassette. J. Biomol. NMR. 2002;24:167–168. doi: 10.1023/a:1020959907518. [DOI] [PubMed] [Google Scholar]
- 37.Wang C., Karpowich N., Hunt J. F., Rance M., Palmer A. G., III Dynamics of ATP-binding cassette contribute to allosteric control, nucleotide binding and energy transduction in ABC transporters. J. Mol. Biol. 2004;342:525–537. doi: 10.1016/j.jmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Hopfner K. P., Karcher A., Craig L., Woo T. T., Carney J. P., Tainer J. A. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell (Cambridge, Mass.) 2001;105:473–485. doi: 10.1016/s0092-8674(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 39.Chang G. Structure of MsbA from Vibrio cholera: a multidrug resistance ABC transporter homolog in a closed conformation. J. Mol. Biol. 2003;330:419–430. doi: 10.1016/s0022-2836(03)00587-4. [DOI] [PubMed] [Google Scholar]
- 40.Chang G., Roth C. B. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science. 2001;293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 41.Fetsch E. E., Davidson A. L. Vanadate-catalysed photocleavage of the signature motif of an ATP-binding cassette (ABC) transporter. Proc. Natl. Acad. Sci. U.S.A. 2002;99:9685–9690. doi: 10.1073/pnas.152204499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loo T. W., Bartlett M. C., Clarke D. M. The LSGGQ motif in each nucleotide-binding domain of human P-glycoprotein is adjacent to the opposing walker A sequence. J. Biol. Chem. 2002;277:41303–41306. doi: 10.1074/jbc.C200484200. [DOI] [PubMed] [Google Scholar]
- 43.Loo T. W., Bartlett M. C., Clarke D. M. Drug binding in human P-glycoprotein causes conformational changes in both nucleotide-binding domains. J. Biol. Chem. 2003;278:1575–1578. doi: 10.1074/jbc.M211307200. [DOI] [PubMed] [Google Scholar]
- 44.Benabdelhak H., Schmitt L., Horn C., Jumel K., Blight M. A., Holland I. B. Positive co-operative activity and dimerization of the isolated ABC ATPase domain of HlyB from Escherichia coli. Biochem. J. 2005;386:489–495. doi: 10.1042/BJ20041282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdon G., Albers S. V., van Oosterwijk N., Dijkstra B. W., Driessen A. J., Thunnissen A. M. Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus. J. Mol. Biol. 2003;334:255–267. doi: 10.1016/j.jmb.2003.08.065. [DOI] [PubMed] [Google Scholar]
- 46.Kidd J. F., Ramjeesingh M., Stratford F., Huan L. J., Bear C. E. A heteromeric complex of the two nucleotide binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) mediates ATPase activity. J. Biol. Chem. 2004;279:41664–41669. doi: 10.1074/jbc.M407666200. [DOI] [PubMed] [Google Scholar]
- 47.Szabo K., Szakacs G., Hegeds T., Sarkadi B. Nucleotide occlusion in the human cystic fibrosis transmembrane conductance regulator. Different patterns in the two nucleotide binding domains. J. Biol. Chem. 1999;274:12209–12212. doi: 10.1074/jbc.274.18.12209. [DOI] [PubMed] [Google Scholar]
- 48.Nagel G. Differential function of the two nucleotide binding domains on cystic fibrosis transmembrane conductance regulator. Biochim. Biophys. Acta. 1999;1461:263–274. doi: 10.1016/s0005-2736(99)00162-5. [DOI] [PubMed] [Google Scholar]
- 49.Lebbink J. H., Sixma T. K. Variations on the ABC. Structure (Camb.) 2005;13:498–500. doi: 10.1016/j.str.2005.03.003. [DOI] [PubMed] [Google Scholar]