Abstract
Nontypeable Haemophilus influenzae (NTHi) causes a wide variety of respiratory tract infections in humans. It is capable of invading and surviving in epithelial cells and has also been shown to persist in macrophage-like cell line J774A.1. To determine the molecular mechanisms which enable NTHi to survive in an intracellular environment, differential display reverse transcriptase PCR was used to identify genes which were either induced or upregulated by NTHi residing in macrophages. Using this approach, we identified one transcript which was consistently amplified from intracellular NTHi cDNA. Nucleotide sequence analysis of this product revealed that it spanned the 3′ and 5′ ends of rpoE and rseB, respectively, which form part of the extracytoplasmic stress operon that encodes and regulates expression of alternate sigma factor sigma E (ςE). To confirm that expression of rpoE was upregulated following uptake of NTHi by macrophages, an rpoE-lacZ transcriptional fusion was constructed, and expression of β-galactosidase activity in broth-grown NTHi was compared with expression of β-galactosidase activity in intracellular NTHi. The level of β-galactosidase activity in NTHi 4 h after phagocytosis by macrophages was found to be 100-fold higher than that of broth-grown organisms, suggesting that genes of the ςE regulon may be important for persistence of NTHi in mammalian cells. The hypothesis that ςE plays a role in the intracellular survival of NTHi was subsequently confirmed by the decreased ability of an rpoE insertion mutant to survive in macrophages.
Nontypeable Haemophilus influenzae (NTHi) is a nonencapsulated bacterium which forms part of the commensal flora of the human upper respiratory tract. This organism causes a variety of infections, including sinusitis, bronchitis, otitis media, and pneumonia (29). One problem associated with disease caused by NTHi is the ability of this organism to persist in the respiratory tract and cause recurrent infections once antibiotic therapy has ceased (2, 23). In the case of otitis media, recrudescent infections occurring as much as 5 months after the original onset of symptoms have been described. Such persistent reinfection is common in preschool children and can result in serious sequelae, including perforation of the eardrums and hearing impairment in adulthood (28).
Although NTHi is not generally considered an invasive microorganism, increasing evidence suggests that this bacterium is able to survive in host cells. It has been demonstrated that NTHi can penetrate and survive in Chang and human bronchial epithelial cell monolayers (22, 35). In the case of bronchial epithelial cells adhesion and invasion of NTHi are mediated by the interaction of lipooligosaccharide with the platelet-activating factor receptor (36). Intracellular organisms have also been found in biopsy samples from patients with chronic bronchitis and in NTHi-infected adenoid organ cultures, in which the organism either is clustered between adjacent epithelial cells or occurs intracellularly in mononuclear cells (11). NTHi has also been identified in macrophage-like cells in adenoid tissue obtained from children persistently infected with NTHi. These organisms were shown to be viable, suggesting that NTHi could survive macrophage-mediated killing (15). More recently, we demonstrated that some NTHi isolates are capable of surviving in mouse macrophage cell line J774A.1 for at least 72 h (6). This apparent ability of NTHi to sequester itself within macrophages and other host cells, thereby avoiding circulating antibodies and antibiotics, may contribute to its ability to cause endogenous reinfection.
The ability to survive in macrophages is a potent strategy for preventing clearance from the host and has been adopted by other intracellular pathogens, such as Salmonella enterica serovar Typhimurium and Legionella pneumophila. One feature of these organisms is that following phagocytosis they either upregulate or induce expression of a specific set of stress proteins essential for intracellular survival (13). In S. enterica serovar Typhimurium phagocytosis by macrophages is accompanied by de novo synthesis of more than 30 proteins (4). This adaptive response provides protection from a diverse range of antimicrobial mechanisms inherent in macrophages, including oxidative free radicals, lysosomal enzymes, and transient acidification of the phagosome. In order for NTHi to survive in macrophages, it too must be able to protect itself from the environmental stresses encountered in an intracellular environment. Identifying the genes involved in this process is therefore critical to our understanding of the host-pathogen interaction.
In this study we sought to identify genes which were either induced or upregulated by NTHi in response to the intracellular environment of macrophages. To compare the pattern of genes expressed by intracellular NTHi with the pattern of genes expressed by extracellular organisms, we used the ubiquitous H. influenzae uptake signal sequence (USS) as a primer in a differential display reverse transcriptase PCR (dd-RT-PCR). There are approximately 1,500 USSs distributed largely at random in the genome of H. influenzae. Each comprises a 29-bp consensus sequence containing the core sequence 5′-AAAGTGCGGT-3′ at its 5′ end (34). When used as a primer in dd-rtPCR, this consensus sequence allows expression of a comprehensive number of genes to be monitored simultaneously. Using this approach, we determined that following phagocytosis by macrophages, the profile of genes expressed by NTHi changes, consistent with an adaptive response. We found that one of the genes upregulated during this adaptive response encodes the sigma E (ςE) subunit of RNA polymerase (rpoE). In Escherichia coli RpoE is a prerequisite for the extracytoplasmic stress response and directs expression of a specific regulon following detection of environmental stresses or protein misfolding in the periplasm (8, 9, 24). ςE has also recently been shown to be critically important for the virulence of S. enterica serovar Typhimurium. The hypothesis that ςE plays a role in the survival of NTHi in macrophages was confirmed by the observation that the intracellular survival of an NTHi rpoE mutant was impaired compared to the intracellular survival of its wild-type parent (21). Genes of the ςE regulon are therefore implicated in promotion of survival of NTHi in macrophages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used or constructed in this study are listed in Table 1. All H. influenzae strains were grown in brain heart infusion (BHI) broth supplemented with hemin (10 μg ml−1) and NAD (20 μg ml−1). BHI agar plates were prepared by adding Bacto Agar (1.5%, wt/vol; Difco) and Levinthal’s base (10%, vol/vol) (1). When required, ampicillin was used at a concentration of 50 μg ml−1 and kanamycin was used at a concentration of 50 μg ml−1 for growth of E. coli, and kanamycin was used at a concentration of 10 μg ml−1 for growth of H. influenzae.
TABLE 1.
Bacterial strains and plasmids used or constructed in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| H. influenzae strains | ||
| NT1008 | NTHi clinical isolate | |
| Rd | Nonencapsulated variant of a sero- type d strain | 5 |
| NT1008E | NT1008 ςE::Kan | This study |
| E. coli DH5α | 17 | |
| Plasmids | ||
| pCRII | PCR cloning vector | Invitrogen |
| pBluescript SK | Apr cloning vector | Stratagene |
| pUC4K | Kmr cassette | 30 |
| pJEC62 | rpoE promoter region in pBluescript | This study |
| pJEC65 | rpoE in pBluescript | This study |
| pJEC67 | XbaI fragment from pLic2A in pBluescript | This study |
| pJEC69 | rpoE::kan in pBluescript | This study |
| pLK | Promoterless lacZ gene | |
| pJEC72 | rpoE′-′lacZ in pBluescript | This study |
| pLic2A | lic2 locus | 19 |
| pJEC73 | rpoE-lacZ in ksgA gene in pJEC67 | This study |
DNA manipulation.
Restriction and modifying enzymes were obtained from Boehringer Mannheim. Standard methods were used for restriction enzyme digestion, ligation, transformation, and preparation of plasmid DNA from E. coli. Transformation of H. influenzae was performed as described elsewhere (18). Double-stranded sequencing of cloned reverse transcriptase PCR bands was carried out as described in the Sequenase handbook (U.S. Biochemicals).
Cell lines.
A phagocytic cell line, BALB/c mouse macrophage-like cell line J774A.1 (ATCC 91051511), and a nonphagocytic cell line, human epithelial cell line Hep2 (ATCC 86030501), were used. J774A.1 cells were grown in Dulbecco’s modified Eagle medium (DMEM), and Hep2 cells were grown in minimal essential medium containing Earle’s balanced salts solution. Both media were supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Gibco BRL). The cells were maintained at 37°C in the presence of 5% CO2 in a moist chamber and were subcultured every 3 to 5 days.
Invasion assays.
For invasion assays (and RNA isolation), cells were seeded into six-well tissue culture plates (Costar) at a concentration of approximately 2 × 106 cells/well and incubated overnight at 37°C in the presence of 5% CO2, and the invasion assay was carried out as previously described (6). Prior to infection, the monolayers were washed three times with phosphate-buffered saline (PBS) and then overlaid on fresh tissue culture medium without serum. Exponentially growing bacteria were then added to the monolayers at a multiplicity of infection of 100:1. Infection was allowed to proceed for 1 h, and then the monolayers were washed three times with PBS and overlaid with fresh media containing serum and 50 μg of polymyxin B sulfate per ml to kill any residual external bacteria. The efficiency of killing of extracellular NTHi by polymyxin B sulfate was determined by Craig et al. (6), who showed that polymyxin B sulfate at a concentration of 10 mg/ml can kill a suspension containing 5 × 108 CFU of NTHi per ml in 1 h. At various times after polymyxin B sulfate was added, the monolayers were again washed three times with PBS and then lysed, and serial dilutions of each lysate were plated onto BHI agar to determine cell numbers; alternatively, RNA was extracted from the intracellular bacteria. At each time a viable count analysis was also performed with the tissue culture supernatant to ensure that no viable extracellular bacteria were present.
RNA isolation from broth-grown bacteria and from intracellular bacteria.
Total RNA was isolated from BHI broth-grown NTHi by the method of Figueroa et al. (12). To isolate RNA from intracellular bacteria, infected monolayers that had been incubated for 4 h in the presence of polymyxin B sulfate were washed thoroughly with PBS to remove nonviable extracellular bacteria (6). Each monolayer was then lysed with water, and the bacteria were pelleted by centrifugation at 5,000 × g for 10 min. RNA was then isolated immediately and stored at −80°C before it was used.
dd-RT-PCR.
A random primer (5′-WWWWWYNACCGCACTTT-3′) based on the ubiquitous H. influenzae DNA USS was used for dd-RT-PCR (33). RNAs isolated from broth-grown and intracellular bacteria were initially subjected to reverse transcription. Each reaction mixture contained 2 μg of RNA, each deoxynucleoside triphosphate at a concentration of 20 nM, 50 ng of uptake sequence primer, 25 U of RNase inhibitor (Boehringer Mannheim), and 40 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). The reaction mixtures were incubated at 25°C for 10 min and then at 60°C for 1 h. They were then heated to 99°C for 5 min to denature the avian myeloblastosis virus reverse transcriptase. The resulting cDNA was amplified by PCR by using the uptake sequence primer. Each 50-μl PCR mixture contained 5 μl of cDNA, 100 pmol of uptake sequence primer, each deoxynucleoside triphosphate at a concentration of 200 μM, 5 μCi of [32P]dCTP, and 1 U of Taq polymerase (Boehringer Mannheim) in buffer supplied by the manufacturer. PCR amplification was performed by using a denaturation step consisting of 95°C for 5 min before the Taq polymerase was added, followed by 30 cycles of 95°C for 1 min, 42°C for 1 min, and 72°C for 2 min and a final extension step consisting of 72°C for 10 min.
To visualize the PCR products, equal volumes were separately mixed with Sequenase termination solution (U.S. Biochemicals) and denatured at 90°C for 5 min before electrophoresis on a 6% sequencing gel. The gel was dried and then subjected to autoradiography with Kodak Biomax film.
Following autoradiography dd-RT-PCR bands of interest were cut from the polyacrylamide gel, and the DNA was eluted by boiling in a 0.5 M ammonium acetate-10 mM magnesium acetate-1 mM EDTA (pH 8.0)-0.1% sodium dodecyl sulfate solution for 30 min. The DNA was then ethanol precipitated and subsequently reamplified by using the conditions described above without the radioisotope. PCR products were then cloned in pCR 2.1-TOPO (Invitrogen) according to the manufacturer’s instructions.
Construction of an NTHi rpoE mutant.
The H. influenzae rpoE gene was isolated by PCR from an NT1008 colony template by using primers JEC11 (5′-CTTGCTCGGAAGATTCGG-3′) and JEC12 (5′-GTTAGATAAAATACTAGTGCC-3′). The amplified DNA was cloned in pCR 2.1-TOPO (Invitrogen) and then subcloned in pBluescript SK(+) by using EcoRI sites to create plasmid pJEC65. The rpoE gene fragment was mutated by inserting a kanamycin antibiotic resistance cassette isolated from plasmid pUC4K. pUC4K was digested with BamHI to isolate the kanamycin resistance cassette, which was then inserted into a unique MfeI site in the rpoE fragment in pJEC65 following Klenow treatment of all sticky ends. The resulting plasmid (pJEC69) containing the mutated rpoE gene fragment (rpoE::Kmr) was linearized with SacI and used to transform NT1008, selecting for kanamycin resistance. Mutation of the chromosomal rpoE gene was confirmed by PCR performed with primers JEC11 and JEC12, which revealed that in the mutant the size of the PCR product had increased by approximately 1.2 kb as a result of insertion of the Kmr cassette (data not shown).
Construction and assay of an rpoE′-′lacZ reporter fusion.
To construct an rpoE′-′lacZ fusion, a PCR-amplified (using primers 5′-GGCTTTTGGATCCCCTTGCTG-3′ and 5′-GTTAGATAAAATACTAGTGCC-3′) SpeI-BamHI fragment containing 310 bp upstream of the rpoE gene and 54 bp of the coding region was cloned in pBluescript SK(+) to obtain plasmid pJEC62. The 4-kb BamHI fragment carrying a promoterless lacZ fragment from plasmid pLK was inserted into pJEC62 digested with BamHI to obtain plasmid pJEC72 containing rpoE′-′lacZ. The fusion was inserted into the H. influenzae chromosome at the lic2 locus in the nonessential ksgA gene (19). This was accomplished by digesting pJEC72 with SmaI and SpeI to liberate the ςE′-lacZ fragment. This fragment was then inserted into pJEC67 at a unique HindIII site following end repair with the Klenow fragment. pJEC67 is pBluescript SK(+) containing a 1.7-kb XbaI fragment from pLic2A and has a unique HindIII site in the ksgA gene. The resulting plasmid (pJEC73) contained rpoE′-′lacZ cloned in the orientation opposite that of ksgA to prevent transcriptional readthrough (this was confirmed by restriction digestion). pJEC73 was linearized with SmaI and used to transform NT1008, selecting for Kmr colonies. Insertion of the rpoE′-′lacZ fusion into the ksgA gene was confirmed by Southern blot analysis (data not shown).
β-Galactosidase activities produced by intracellular and broth-grown bacteria were quantified with a β-galactosidase reporter gene assay kit (Boehringer Mannheim). Briefly, monolayers were infected as described above, and at zero time and 2 and 4 h after polymyxin B sulfate treatment the monolayers were washed three times with PBS and lysed, and the β-galactosidase assay was carried out according to the manufacturer’s instructions. One aliquot of cells was used to determine the number of CFU of intracellular bacteria at each time, and the amount of β-galactosidase present in each sample (in femtograms per CFU) was also determined. Zero time was the time when polymyxin B sulfate was added to the infected monolayers. The number of CFU at zero time therefore represented the total cell-associated NTHi, including both adherent and intracellular organisms.
RESULTS
Comparison of the patterns of genes expressed by in vitro broth-grown and cell-associated NTHi.
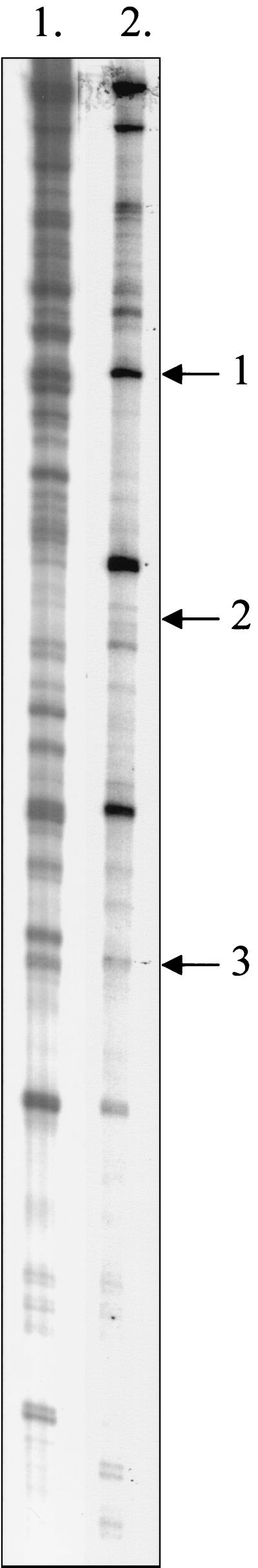
To establish whether NTHi elicits an adaptive response following uptake by macrophages, the pattern of genes expressed by intracellular NTHi was determined and compared to the pattern of genes expressed by broth-grown organisms. Gene expression was monitored by dd-RT-PCR by using the H. influenzae USS as a primer. cDNAs were synthesized from total RNAs isolated from broth-grown organisms resuspended in DMEM and organisms isolated from macrophages 4 h after phagocytosis had taken place. The cDNAs were then amplified by PCR by using the USS as a primer. The resultant PCR products were then separated by polyacrylamide gel electrophoresis. To ensure that the resultant PCR products were derived from cDNAs and were not due to contaminating genomic DNAs, control experiments were performed with purified RNAs as the templates. Using this procedure, we generated expression profiles, comprised of ladders of bands at different molecular weights, for broth-grown NTHi and intracellular NTHi (Fig. 1). A comparison of these expression profiles appeared to indicate that intracellular NTHi expressed a different repertoire of genes than organisms grown in BHI broth expressed. We identified transcripts which appeared to be either induced or upregulated in response to the intracellular environment of macrophages. Transcripts which were not expressed by NTHi or had been down regulated were also apparent. Taken as a whole, the observed differences in the expression profiles of the in vitro broth-grown organisms and the intracellular organisms suggest that NTHi does elicit an adaptive response following uptake by macrophages.
FIG. 1.
32P-labeled dd-RT-PCR products derived from BHI medium-grown H. influenzae (lane 1) and from intracellular bacteria recovered 4 h after infection of mouse J774A.1 macrophages (lane 2). The positions of the two 23S rRNA fragments are indicated by arrows 1 and 2. The position of the rpoE fragment is indicated by arrow 3.
Identification of NTHi rpoE locus by dd-RT-PCR technique.
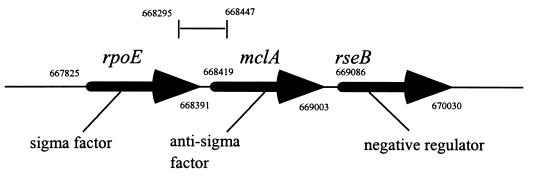
To identify potential genes that were upregulated by NTHi following phagocytosis by macrophages, dd-RT-PCR products unique to intracellular microorganisms were excised from the polyacrylamide gel and reamplified by PCR by using the USS primer. The amplified products were then cloned into the pCR 2.1-TOPO vector, and their nucleotide sequences were determined. At this point it became apparent that many of the bands excised from the polyacrylamide gel represented a heterogeneous mixture of PCR products since the nucleotide sequences of individual clones were different. To identify the most abundant PCR product, the cloned DNAs of at least 10 clones derived from each band were T-track sequenced. The most common clone, which was presumed to represent the most abundant PCR product, was then analyzed further. Using this approach, we found that the compositions of many of the major bands in Fig. 1 were so heterogeneous that it was impossible to proceed any further with them. However, three bands (Fig. 1) were found to comprise predominantly one DNA species. The nucleotide sequence of each of these cloned DNA fragments was subsequently determined and compared to the H. influenzae Rd genome sequence (14) by using TBLAST. This analysis revealed that two of the cloned PCR products were fragments of the H. influenzae 23S rRNA gene. The third PCR product, which was 152 bp long, encompassed the 3′ and 5′ ends of the rpoE and mclA genes, which encode the ςE subunit of RNA polymerase and a negative posttranslational regulator or anti-sigma factor, respectively (Fig. 2). Together with a third gene, rseB, which encodes a protein involved in negative regulation of ςE (possibly by modulating MclA activity), rpoE and mclA form an operon which is a prerequisite for the extracytoplasmic stress response in E. coli (8, 25).
FIG. 2.
Structure of the rpoE operon in H. influenzae Rd. rpoE encodes the alternative sigma factor ςE; mclA (homologous to rseA of E. coli) encodes the anti-sigma factor predicted to reside in the inner membrane; and rseB encodes a potential periplasmic protein thought to interact with mclA in negatively regulating ςE. The region cloned from the differential display gel and the coordinates in the genome sequence of Rd are shown.
Transcriptional activation of rpoE gene following adhesion and phagocytosis by macrophages.
To confirm that transcription of rpoE is upregulated by NTHi in the intracellualr environment of the macrophage, an lacZ transcriptional fusion to the promoter region of rpoE, was constructed. The level of rpoE expressed by intracellular NTHi could be quantified by using this construct. The location of the NTHi rpoE promoter was determined by comparison with the nucleotide sequence of the E. coli rpoE gene (30). Using this information, we designed a pair of primers which allowed PCR amplification of a 300-bp fragment encompassing the putative rpoE promoter region. To generate the rpoE-lacZ fusion, this fragment was then ligated to a 4-kb DNA fragment derived from PLK2 that carried a promoterless copy of the lacZ gene and a kanamycin resistance gene cassette. The fusion was then cloned into the HindIII site of ksgA, in the orientation opposite that of the open reading frame, to prevent transcriptional readthrough. ksgA is adjacent to lic2A and encodes a 16S RNA methyltransferase which confers sensitivity to the antibiotic kasugamycin. Mutations in this gene do not impair the ability of NTHi to survive in macrophages (data not shown) and provide a mechanism for introducing the rpoE-lacZ fusion into the genome of NTHi without disrupting the wild-type copy of rpoE. Accordingly, the rpoE-lacZ fusion was introduced into the chromosomal copy of ksgA by natural transformation, and transformants were selected by growth on BHI medium containing kanamycin. That the desired mutation had been introduced was confirmed by Southern hybridization analysis.
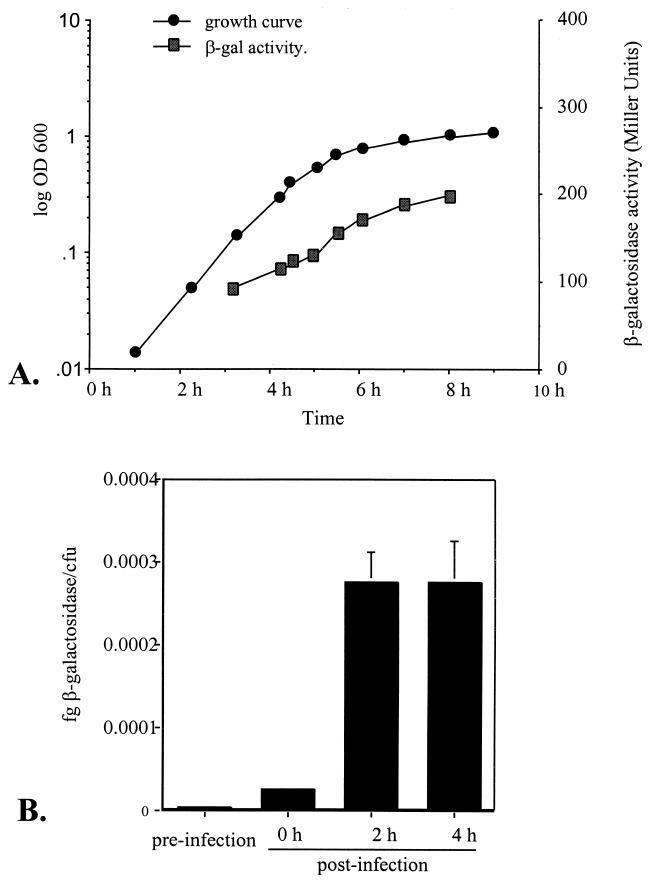
Before β-galactosidase activity in intracellular NTHi was monitored, expression of the rpoE-lacZ fusion was determined during growth of NT1008 in BHI broth in batch culture. This step was essential to ensure that any apparent change in the level of rpoE expression observed in intracellular bacteria was not solely attributable to growth-phase-dependent variation. As shown in Fig. 3A, a slight increase in rpoE transcription resulting in an approximately twofold increase in β-galactosidase activity was observed over a 5-h period between the mid-log phase and the stationary phase. Having established the range of β-galactosidase activity associated with broth-grown organisms, we then determined the level of rpoE expression induced following uptake of NTHi by macrophages. β-Galactosidase activity per CFU was measured at zero time and 2 and 4 h after phagocytosis by using Galacton Plus, a highly sensitive chemiluminescent substrate. All values recorded at these times were then compared to the β-galactosidase activity produced by bacteria resuspended in DMEM, the preinfection sample used to infect the macrophage monolayers. As shown in Fig. 3B, at zero time expression of the rpoE promoter had increased 10-fold compared to expression in the preinfection sample. At this time, immediately prior to addition of polymyxin B sulfate, the bacteria and macrophages had been incubated together for 1 h to allow phagocytosis to occur. The β-galactosidase activity at zero time was therefore the activity derived from all cell-associated bacteria, including both adherent and intracellular organisms. On the basis of previous studies (6), we estimated that at this stage 90% of the total population of cell-associated bacteria was bound to macrophages. By 2 h postinfection, when all extracellular, adherent organisms had been killed by the antibiotic, the β-galactosidase activity had increased dramatically to an average of 0.00027 fg/CFU, which was approximately 100-fold greater than the value obtained for the preinfection sample and 10-fold greater than the value obtained for cell-associated NTHi at zero time. This level of activity was retained at 4 h postinfection, suggesting that expression of rpoE had reached its maximal level in this environment (Fig. 3B). In contrast, uninfected macrophages and macrophages infected with wild-type NT1008 showed no measurable β-galactosidase activity. To confirm that the observed increases in β-galactosidase activity did not result from incubation of NTHi in DMEM, we compared expression of the rpoE promoter in bacteria incubated in BHI broth and expression of the rpoE promoter in bacteria incubated in DMEM. After 1 and 2 h of incubation in DMEM the levels of β-galactosidase activity were 2- and 1.6-fold greater, respectively, than the levels of β-galactosidase activity obtained for BHI broth-grown organisms. This verified that the observed increase in rpoE promoter activity was a direct result of the interaction of NTHi with macrophages.
FIG. 3.
(A) Regulation of ςE expression in BHI medium. The β-galactosidase activity of NT1008 rpoE′-′lacZ grown at 37°C in BHI medium was determined at different times. OD 600, optical density at 600 nm. (B) Regulation of ςE expression in the intracellular environment. The β-galactosidase activities produced by NT1008 rpoE′-′lacZ before infection, at zero time, and at 2 and 4 h after infection of J774A.1 cells are shown. At zero time the β-galactosidase activity was derived from total cell-associated bacteria, including both adherent and intracellular organisms. After this polymyxin B sulfate was added to kill all extracellular organisms. The error bars indicate the standard errors of the means based on the results of three independent experiments.
RpoE is required for intracellular survival of NTHi.
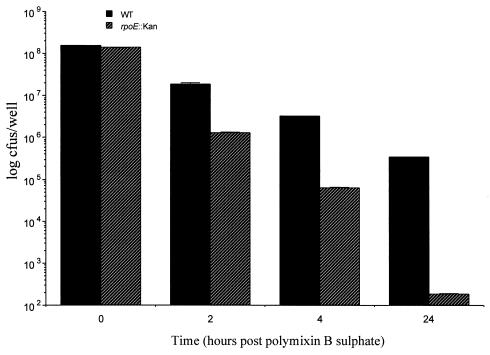
The observation that the rpoE gene is upregulated following phagocytosis of NTHi by macrophages suggests that ςE may play a role in the intracellular survival of this organism. To address this question, an NT1008 rpoE mutant was constructed by insertional inactivation of rpoE by using a kanamycin resistance gene cassette as described in Materials and Methods. Loss of a functional ςE had no apparent effect on the growth rate of NTHi, as judged by growth in BHI broth in batch culture. The doubling time of the rpoE mutant in BHI medium was indistinguishable from that of wild-type NTHi (data not shown). We therefore concluded that any differences in the intracellular survival rate of the mutant could not be attributed to reduced fitness caused by the rpoE mutation. To determine the effect of the ςE mutation on the ability of NTHi to persist in eukaryotic cells, the ability of the rpoE mutant to invade and survive in murine macrophage cell line J774A.1 or human epithelial cell line Hep2 was compared to that of wild-type NTHi. As shown in Fig. 4, comparable numbers of NT1008 rpoE::kan and wild-type NTHi CFU were recovered at zero time. At this time the number of organisms recovered reflected the total number of bacteria phagocytosed, and the results suggested that the rpoE mutation did not impair the ability of NTHi to invade macrophages. In contrast, by 2 h postinfection the number of intracellular NT1008 rpoE::kan CFU recovered from macrophages was more than 1 log lower than the number of NT1008 CFU recovered. A comparable decrease in the number of intracellular NT1008 rpoE::kan CFU recovered from macrophages was observed at 4 h postinfection. By 24 h only 200 CFU of NT1008 rpoE::kan per well were recovered from macrophages, compared to 5 × 105 CFU of NT1008 per well. This result indicated that the ςE mutant was deficient in the ability to survive in eukaryotic cells and provided compelling evidence that ςE plays a role in the survival of NTHi in macrophages.
FIG. 4.
Effect of an rpoE mutation on intracellular survival of NT1008. The ability of NT1008 rpoE::kan to survive in macrophages was examined by using mouse macrophage cell line J774A.1. The assay was performed as described in the text. The graph shows the numbers of viable bacteria inside the eukaryotic cells at zero time and at 2, 4, and 24 h after infection. Each bar represents the mean number of CFU based on the results of three independent infection experiments, and the error bars indicate the standard errors of the means. A Mann-Whitney nonparametric test was used to confirm that observed differences in the levels of intracellular survival of NT1008 and NTHi rpoE::kan were significantly different (P < 0.05). WT, wild type.
DISCUSSION
In this study we determined by using dd-RT-PCR that NTHi strain NT1008 exhibits differential gene expression following phagocytosis by macrophages. This suggests that this microorganism, like other intracellular bacteria, is able to exhibit an adaptive response to cope with the hostile environment of the macrophage. One of the genes upregulated was rpoE, which encodes the ςE subunit of RNA polymerase. By constructing an rpoE′-′lacZ fusion we confirmed that expression of rpoE is enhanced in response to the intracellular environment of the macrophage. This is in agreement with the recent findings of Graham and Clark-Curtiss, who showed that Mycobacterium tuberculosis had elevated transcript levels of a ςE homologue (ςH) during human macrophage infection (16). An increase in rpoE promoter activity was also observed at zero time, when both adherent and intracellular bacteria were still viable. It is therefore not possible to exclude the possibility that adhesion of NTHi to macrophages may also cause upregulation of the rpoE promoter. However, an alternative explanation is that the observed increase in promoter activity was solely attributable to the NTHi present in the population at zero time (6) which had been phagocytosed by macrophages. On the basis of previous studies we estimated that intracellular NTHi represented approximately 10% of the total cell-associated bacteria at zero time. If this was the case, then the level of β-galactosidase activity per CFU of intracellular bacteria would be predicted to be similar to that observed for NTHi 2 h following phagocytosis.
Alternative sigma factors provide a means of regulating gene expression to maintain homeostasis when the extracellular environment changes. In E. coli the ςE regulon is activated by extracytoplasmic stress resulting from the accumulation of immature outer membrane proteins or misfolded polypeptides in the periplasm (24, 25). Two-dimensional gel analysis of proteins suggests that there are at least 10 members of the ςE regulon. These include rpoE itself, htrA (or degP), which encodes a periplasmic serine protease, and fkpA, which codes for a periplasmic peptidyl-prolyl cis/trans isomerase. Under extreme stress conditions (50°C or 10% ethanol) ςE is also required in E. coli for expression of the classical heat shock sigma factor (ς32) (27, 31, 32). Upregulation of ςE in NT1008 after phagocytosis by macrophages would be expected to promote transcription of the genes of the ςE regulon in the intracellular environment. Indeed, it has been shown with other microorganisms that this does in fact occur. In Yersinia enterocolitica the GsrA stress protein (an HtrA homologue) and the HtrA protein of a Salmonella strain are both induced by macrophage phagocytosis (10, 37). These studies were the first studies to indicate that the ςE regulon may respond to stresses in the macrophage environment.
The activity of ςE in E. coli is tightly regulated; it is known that at the transcriptional level the rpoE gene is positively autoregulated and at the posttranslational level it is negatively regulated by the inner membrane spanning anti-sigma factor RseA and an accessory protein, RseB. RseB has been shown to interact with RseA and to downregulate the ςE pathway when it is overexpressed. rpoE, rseA, rseB, and a fourth gene, rseC (thought to encode a positive regulator of ςE in the absence of RseB), make up an operon in E. coli (7, 26). Since homologues of rseA (mclA) and rseB are present in H. influenzae and are organized in the same manner that they are organized in E. coli (there are two rseC homologues, but they are at different locations), it is highly likely that NTHi possesses a similar mechanism for regulation of ςE. In fact, homologous proteins have now been found in a number of bacterial species, indicating that there is a conserved mechanism of regulation for the ς factor and a cognate anti-sigma factor (reviewed by Missiakas and Raina [27]). However, in most cases the identity of the stimulus that leads to the release of the ς factor from the anti-sigma factor is unknown. ςE is positively autoregulated at the transcriptional level, and in E. coli release of ςE from the anti-sigma factor results in increased transcription of ςE. However, it remains to be seen whether the elevated levels of NT1008 ςE mRNA detected inside macrophages are due to release of ςE from MclA and, if so, what internal factors provoke this release.
The rpoE mutant strain of NT1008 exhibited a decreased ability to survive inside macrophages, indicating that genes of the ςE regulon are important in intracellular survival of this organism. It is unclear why this strain is less able to persist inside macrophages; presumably, genes of the ςE regulon are required to cope with some of the antimicrobial mechanisms inherent in these cells. S. enterica serovar Typhimurium strains with mutations in the ςE-regulated htrA and fkpA genes have been shown to exhibit decreased intracellular survival in macrophages (3, 20). The HtrA and FkpA proteins are probably required to cope with the accumulation of partially unfolded or denatured proteins in the extracytoplasmic compartments of the stressed intracellular bacteria. However, it has recently been shown that in S. enterica serovar Typhimurium single mutations in htrA and fkpA do not have as pronounced an effect on intracellular survival as a ςE mutation has. This suggests that other genes in the ςE regulon (or combinations of genes) may also be important in the survival process (20, 21). By identifying other genes of the ςE regulon we should get a better idea of how this stress response helps NTHi survive intracellularly.
Since until recently NTHi has been considered an extracellular organism, little work has been done to look at its interaction with host cells. In this study we found, for the first time, that the ςE locus of NTHi is clearly important for intracellular survival of this organism. Hopefully, by identifying other mRNAs that are upregulated in the macrophage environment we can start to build a picture of the mechanism(s) which this pathogen uses to persist in mammalian cells.
Acknowledgments
This work was funded by grants to N.J.H. from the Royal Society of Great Britain and The Leverhulme Trust.
Editor: D. L. Burns
REFERENCES
- 1.Alexander, H. E. 1965. The Haemophilus group, p.724–741. In R. J. Dubos and J. G. Hirsch (ed.), Bacterial and mycotic infections of man. Pitman Medical Publishing Co., Ltd., London, United Kingdom.
- 2.Barenkamp, S. J., P. A. Shurin, C. D. Marchant, R. B. Karasic, S. I. Pelton, V. M. Howie, and D. M. Granoff. 1984. Do children with recurrent Haemophilus influenzae otitis media become infected with a new organism or reacquire the original strain? J. Pediatr. 105:533–537. [DOI] [PubMed] [Google Scholar]
- 3.Baumler, A. J., J. G. Kusters, I. Stojiljkovic, and F. Heffron. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 62:1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730–732. [DOI] [PubMed] [Google Scholar]
- 5.Catlin, B. W., J. W. Bendler III, and S. H. Goodgal. 1973. The type b capsulation locus of Haemophilus influenzae: map location and size. J. Gen. Microbiol. 70:411–422. [DOI] [PubMed] [Google Scholar]
- 6.Craig, J. E., A. Cliffe, K. Garnett, and N. J. High. 2001. Survival of nontypeable Haemophilus influenzae in macrophages. FEMS Microbiol. Lett. 203:55–61. [DOI] [PubMed] [Google Scholar]
- 7.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. SigmaE is an essential sigma factor in Escherichia coli. J. Bacteriol. 179:6862–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. The ςE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of ςE. Mol. Microbiol. 24:373–385. [DOI] [PubMed] [Google Scholar]
- 9.Erickson, J. W., and C. A. Gross. 1989. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternate ς factor involved in high temperature gene expression. Genes Dev. 3:1462–1471. [DOI] [PubMed] [Google Scholar]
- 10.Everest, P. H., G. Frankel, J. Li, P. Lund, S. N. Chatfield, and G. Dougan. 1994. Expression of LacZ from the htrA, nirB and groE promoters in a Salmonella vaccine strain: influence of growth in mammalian cells. FEMS Microbiol. Lett. 126:97–102. [DOI] [PubMed] [Google Scholar]
- 11.Farley, M. M., D. S. Stephens, M. H. Miks, M. D. Cooper, J. V. Bricker, S. S. Mirra, and A. Wright. 1986. Pathogenesis of IgA1 protease-producing and -nonproducing Haemophilus influenzae in human nasopharyngeal organ cultures. J. Infect. Dis. 154:752–759. [DOI] [PubMed] [Google Scholar]
- 12.Figueroa, J., N. Wills, and L. Bossi. 1991. Common sequence determinants of the response of a prokaryotic promoter to DNA bending and supercoiling. EMBO J. 10:941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Butt, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. The genome of Haemophilus influenzae Rd. Science 269:496–512. [DOI] [PubMed] [Google Scholar]
- 15.Forsgren, J., A. Samuelson, A. Ahlin, J. Jonasson, B. Rynnel-Dagoo, and A. Lindberg. 1994. Haemophilus influenzae resides and multiplies intracellularly in human adenoid tissue as demonstrated by in situ hybridization and bacterial viability assay. Infect. Immun. 62:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557. [DOI] [PubMed] [Google Scholar]
- 18.Herriot, R. M., E. M. Meyer, and M. J. Vogt. 1970. Defined non-growth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.High, N. J., M. P. Jennings, and R. E. Moxon. 1996. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol. Microbiol. 20:165–174. [DOI] [PubMed] [Google Scholar]
- 20.Horne, S. M., T. J. Kottom, L. K. Nolan, and K. D. Young. 1997. Decreased intracellular survival of an fkpA mutant of Salmonella typhimurium Copenhagen. Infect. Immun. 65:806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, ςE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ketterer, M. R., J. Q. Shao, D. B. Hornick, B. Buscher, V. K. Bandi, and M. A. Apicella. 1999. Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Infect. Immun 67:4161–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach, A. J., J. B. Boswell, V. Asche, T. G. Nienhuys, and J. D. Mathews. 1994. Bacterial colonisation of the nasopharynx predicts very early onset and persistence of otitis media in Australian aboriginal infants. Pediatr. Infect. Dis. J. 13:983–989. [DOI] [PubMed] [Google Scholar]
- 24.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of ςE, an Escherichia coli heat-inducible ς-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618–2628. [DOI] [PubMed] [Google Scholar]
- 25.Missiakas, D., J.-M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli: SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871–884. [DOI] [PubMed] [Google Scholar]
- 26.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli ςE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355–371. [DOI] [PubMed] [Google Scholar]
- 27.Missiakas, D., and S. Raina. 1998. The extracytoplasic function sigma factors: role and regulation. Mol. Microbiol. 28:1059–1066. [DOI] [PubMed] [Google Scholar]
- 28.Moxon, E. R. 1989. Principles and practice of infectious diseases, p.506–510. Wiley Medical Publications, New York, N.Y.
- 29.Murphy, T. F., and M. A. Apicella. 1987. Nontypeable Haemophilus influenzae: a review of clinical aspects, surface antigens, and the human immune response to infection. Rev. Infect. Dis. 9:1–15. [DOI] [PubMed] [Google Scholar]
- 30.Oka, A., M. Sugisaki, and M. Takanmai. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217–226. [DOI] [PubMed] [Google Scholar]
- 31.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the ςE (ς24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat shock sigma factor, ςE, in Escherichia coli. EMBO J. 14:1032–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuelson, A., A. Freud, J. Jonasson, and A. A. Lindberg. 1995. Turnover of nonencapsulated Haemophilus influenzae in the nasopharynges of otitis-prone children. J. Clin. Microbiol. 33:2027–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, H. O., J. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538–540. [DOI] [PubMed] [Google Scholar]
- 35.St. Geme, J. W., III, and S. Falkow. 1990. Haemophilus influenzae adheres to and enters cultivated human epithelial cells. Infect. Immun. 58:4036–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swords, W. E., B. A. Buscher, I. K. Ver Steeg, A. Preston, W. A. Nichols, J. N. Weiser, B. W. Gibson, and M. A. Apicella. 2000. Non-typeable Haemophilus influenzae adhere to and invade human bronchial epithelial cells via an interaction of lipooligosaccharide with the PAF receptor. Mol. Microbiol. 37:13–27. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1997. The Yersinia enterocolitica GsrA stress protein, involved in intracellular survival, is induced by macrophage phagocytosis. Infect. Immun. 65:2190–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]