Abstract
Unlike animals which accumulate glutathione (γ-glutamyl-L-cysteinyl-glycine) alone as their major thiol antioxidant, several crops synthesize alternative forms of glutathione by varying the carboxy residue. The molecular basis of this variation is not well understood, but the substrate specificity of the respective GSs (glutathione synthetases) has been implicated. To investigate their substrate tolerance, five GS-like cDNAs have been cloned from plants that can accumulate alternative forms of glutathione, notably soya bean [hGSH (homoglutathione or γ-glutamyl-L-cysteinyl-β-alanine)], wheat (hydroxymethylglutathione or γ-glutamyl-L-cysteinyl-serine) and maize (γ-Glu-Cys-Glu). The respective recombinant GSs were then assayed for the incorporation of differing C-termini into γ-Glu-Cys. The soya bean enzyme primarily incorporated β-alanine to form hGSH, whereas the GS enzymes from cereals preferentially catalysed the formation of glutathione. However, when assayed with other substrates, several GSs and one wheat enzyme in particular were able to synthesize a diverse range of glutathione variants by incorporating unusual C-terminal moieties including D-serine, non-natural amino acids and α-amino alcohols. Our results suggest that plant GSs are capable of producing a diverse range of glutathione homologues depending on the availability of the acyl acceptor.
Keywords: glutathione synthetase, Glycine max (soya bean), homoglutathione synthetase, hydroxymethylglutathione, Triticum aestivum (wheat), Zea mays (maize)
Abbreviations: AABA, BABA and GABA, α-, β- and γ-aminobutyric acid respectively; ACV synthetase, L-δ-(α-aminoadipoyl)-L-cysteinyl-D-valine synthetase; BAIBA, β-aminoisobutyric acid; γ-EC, γ-glutamyl-L-cysteine; γ-ECE, γ-glutamyl-L-cysteine-glutamic acid; γ-ECS, γ-glutamyl-L-cysteine synthetase; ESI, electrospray ionization; GS, glutathione synthetase; hGS, homoglutathione synthetase; hGSH, homoglutathione; hmGSH, hydroxymethylglutathione; TOF, time-of-flight
INTRODUCTION
The majority of higher plants contain the tripeptide glutathione (γ-glutamyl-L-cysteinyl-glycine), which serves important functions as an antioxidant, a scavenger of reactive chemical species and a signalling agent [1,2]. In several economically important crops, glutathione is replaced either completely or in part by more unusual thiols. For example, in legume species including soya bean (Glycine max L.), hGSH (homoglutathione or γ-glutamyl-L-cysteinyl-β-alanine) is the dominant thiol [3]. In other legumes such as Medicago truncatula, both glutathione and hGSH are synthesized, accumulating in an organ-specific manner [4]. In ce- reals of the Poaceae family, such as wheat (Triticum aestivum L.), hmGSH (hydroxymethylglutathione or γ-glutamyl-L-cysteinylserine) co-accumulates with glutathione [5]. Maize (Zea mays L.) also has the capacity to accumulate γ-ECE (γ-glutamyl-L-cysteine-glutamic acid) on exposure to heavy metals [6].
Glutathione, hGSH and related variants are synthesized from γ-EC (γ-glutamyl-L-cysteine), which is formed from L-glutamate and L-cysteine in an ATP-dependent reaction catalysed by γ-ECS (γ-glutamyl-L-cysteine synthetase; EC 6.3.2.2). Glutathione is then synthesized from γ-EC and glycine by an ATP-dependent GS (glutathione synthetase; EC 6.3.2.3). Most GS enzymes selectively produce glutathione due to their tight acyl acceptor specificity for glycine as a substrate, but in legumes the enzyme has evolved to preferentially incorporate β-alanine, effectively becoming an hGS (homoglutathione synthetase). Thus the hGSs from M. truncatula and pea (Pisum sativum L.) selectively catalyse the synthesis of hGSH from γ-EC and β-alanine [7,8]. The relative abundance of glutathione and hGSH in legumes is then determined by the expression of either GS or hGS enzymes [9].
In wheat, the source of hmGSH is less clear-cut. While γ-EC remains the direct precursor, the serine could either be incorporated directly by an ATP-dependent GS-like reaction or could be derived through a post-synthetic modification of the glutathione molecule [5]. Similarly, the source of the glutamate analogue, γ-ECE, in maize is not known, with the possibilities being that it is derived from an alternative GS activity or from the degradation of phytochelatins, which are polymers of (γ-Glu-Cys)n-Glu-Cys-Gly [6]. To address whether or not the GSs of maize and wheat could determine the synthesis of alternative forms of glutathione, we have cloned enzymes from the respective cereals and determined their substrate preference, comparing the results obtained with an hGS from soya bean.
MATERIALS AND METHODS
Cloning of thiol synthetase cDNAs
DNA probes were prepared by PCR using a specific primer (MS-3, gCgAAgCCHCARMgAgARggHggAgg) based on aligned GS sequences [10]. PCR amplification was performed on cDNA prepared from total RNA prepared from either 10-day-old soya bean cell-suspension cultures (cv. Mandarin) [11], 10-day-old wheat shoots [12] or on plasmid DNA prepared from a mass excised cDNA library prepared from etiolated 10-day-old maize roots [13]. MS-3 was used in combination with the non-specific primer OG9 for soya bean [11], whereas, for maize and wheat, two specific primers were designed from GS sequences in the respective EST (expressed sequence tag) databases, namely ZmGS3′ (gCgCTgAgCTTgCCAgTCAAgTATAggC) and TaGS3′ (gCggTCgACCTCATCTgTgAggAgAATgC) respectively. Amplification products were cloned into pGEM-T Easy (Promega, Chilworth, Southampton, U.K.) and sequenced using an Applied Biosystems 373 DNA sequencer. For soya bean, the full-length GmGS cDNA was cloned as described previously [10]. The partial wheat (TaMS3-1) and maize (ZmMS3-1) clones were PCR-labelled with digoxigenin (Roche, Lewes, East Sussex, U.K.) and used to screen cDNA libraries prepared from shoots [14] and roots of etiolated seedlings [13] respectively. In each case, 200000 plaque-forming units were screened and positive clones were purified and sequenced on both strands.
Expression and purification of recombinant enzymes
PCR was used to engineer restriction sites for direct cloning of the coding sequences into pET-24a or pET-24d (Novagen, Madison, WI, U.S.A.) essentially as described for GmGS [10]. For the maize and wheat clones, the following primer pairs were used in the presence of 1 M betaine: ZmGS, ZmGS5′ Nde (CCAATACCTCATATgAgTgCCgCCATgCCg) and ZmGS3′ (gCgCTCgAgCTTgCCAgTCAAgTATAggC); TaGS1 either TaGSBNdensp (CCCTgAggCATATgggAgCCgAggCgC) and TaGS3′ (gCggTCgACCTCATCTgTgAggAgAATgC) or TaGSSigNde (gCgCATATgTCCTCTTgCgTCTCCTCCTCCC) and TaGS3′; TaGS2, TaGSCNde (CCACTgCCgCATATgAgCACCACC) and TaGSC3′ (gCgCCTCgAgCTTgTCggTCAggTATAAgC). The pET-based plasmids were resequenced and used to transform Rosetta™ DE3 (Novagen) bacteria. The production and purification of the recombinant GSs was then performed as described for GmGS [10]. In each case, the purity of the His-tagged proteins was confirmed by SDS/PAGE (10% polyacrylamide) before use.
Enzyme assays
Protein concentration was determined using the Bio-Rad dye-binding reagent, with γ-globulin standards, as recommended by the manufacturer. Enzyme assays were prepared in 250 mM Tris/HCl (pH 8.0) containing 20 mM MgCl2, 5 mM dithiothreitol, 10 mM ATP, 1 mM γ-EC and 100 mM (unless otherwise stated) of the co-substrate to be assayed, in a total volume of 100 μl. The reaction mixtures were incubated at 30 °C for up to 120 min, with enzyme or substrate omitted for controls. At time intervals, 20 μl samples were derivatized for 15 min at room temperature (21 °C) using 200 μl of 0.2 M Tris/HCl (pH 8.0) containing 0.24 μmol of monobromobimane. After adding 780 μl of 5% (v/v) acetic acid, 50 μl samples were analysed for fluorescent S-bimane derivatives by HPLC, with authentic glutathione and hGSH used to quantify tripeptide products [15]. After ensuring time and protein dependence for the assay, enzyme activity was expressed in pkat (=1 pmol of product formed s−1).
Reaction products were also analysed directly before bimane derivatization using a TOF (time-of-flight) mass spectrometer (Micromass LCT; Micromass, Manchester, U.K.) using ESI (electrospray ionization), in positive ion mode. The reaction mixture (20 μl) was injected on to a Jupiter C18 150 mm×2 mm 5 μm column (Phenomenex, Macclesfield, U.K.) in water/acetonitrile/formic acid (180:20:1, by vol.) at a flow rate of 0.2 ml·min−1. The eluate was analysed for the expected mass of ions in the range 100–800 Da using the supplied MassLynx software, after calibration with sodium iodide.
Determination of amino acid configuration in hmGSH
Samples of S-bimane-derivatized hmGSH were purified by HPLC from crude wheat leaf extracts or enzyme preparations after derivatization with monobromobimane, freeze-dried, resuspended in 10 mM Hepes buffer (pH 7.9) containing 0.2 mM EDTA, and digested with 1 unit of bovine γ-glutamyltranspeptidase for 2 h at 25 °C. The reaction products were analysed by reverse-phase HPLC [15] or on a Chirobiotic V 250 mm×4.6 mm column (Astec, Whippany, NJ, U.S.A.) using the same gradient conditions. The retention times of reaction products were compared with those for derivatized authentic L-Cys-L-Ser and L-Cys-D-Ser standards, prepared on an automated peptide synthesizer, and the identity of the peptides was confirmed by ESI-MS.
RESULTS
Cloning of GS-like sequences from soya bean, wheat and maize
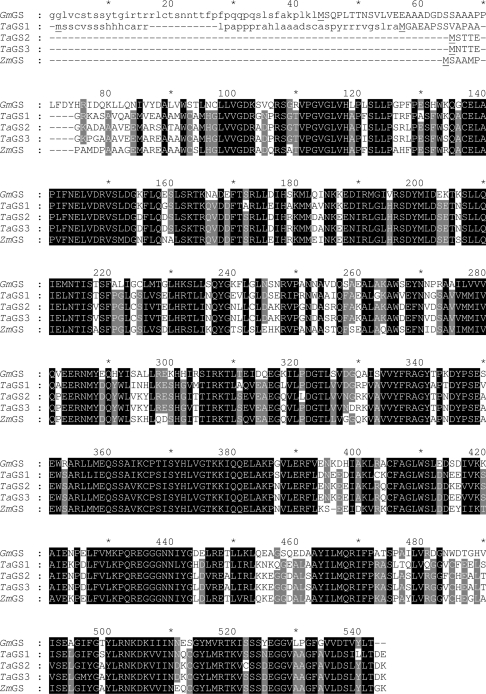
As described previously for the cloning of GmGS [10], PCR amplification generated a 354 bp sequence from wheat (TaMS3-1) and a 353 bp sequence from maize (ZmMS3-1), which were found to be closely related to other plant GS sequences when subjected to BLAST searches [16]. Following digoxigenin labelling of the PCR products, four positive clones (TaGS1–TaGS4) were obtained from a wheat library probed with TaMS3-1. When probed with ZmMS3-1, 17 positive clones, ZmGS1–ZmGS17, were obtained from the maize library. Following restriction enzyme analysis, the wheat clones were fully sequenced on both strands. A combination of restriction enzyme analysis and sequencing demonstrated all 17 maize clones to be essentially identical, except that some of the sequences were attenuated at the 5′-end. Three different wheat clone sequences were obtained, since TaGS1 and TaGS4 (=TaGS1) were identical. TaGS2 and TaGS3 differed in 13 out of 475 amino acids, but had homologous nucleotide sequences in both 5′- and 3′-untranslated regions. TaGS1 was only 77 and 79% identical at the amino acid level with the TaGS2 and TaGS3 clones respectively. The amino acid sequences of the four GS clones isolated from wheat and maize are shown aligned with GmGS for reference in Figure 1.
Figure 1. CLUSTAL W alignment of the predicted peptide sequences obtained for the GS cDNAs cloned from soya bean, wheat and maize.
Residues shaded black are identical or conserved in all five sequences and those shaded in grey, identical in only four of the five sequences. The methionine residues used as translation start codons in pET constructs have been underlined. The conserved alanine residues found within GS, but not within hGS protein sequences [7], can be seen at position 530–531 in the cereal crop sequences but not in the soya bean sequence. Putative N-terminal polypeptide extensions encoding cellular targeting signals are shown in lower-case letters.
The predicted amino acid sequences of the four cDNAs from the cereal species were more similar to GS rather than hGS sequences. ZmGS was most identical with a recently deposited maize cDNA putatively assigned to be a GS (EMBL accession no. AJ302784), which was predicted to encode a 45.9 kDa protein. However, ZmGS was 84 amino acids longer at its N-terminus than AJ302784 and contained four in frame methionine residues upstream, all of which scored highly as predicted translation start sites using NetStart 1.0 [17]. TaGS1, TaGS2 and TaGS3 were most identical with an unassigned rice cDNA clone (EMBL accession no. AK068792).
In common with GmGS, the TaGS1 protein contained an extended N-terminus in comparison with the other GS sequences cloned in the present study (Figure 1), and was predicted to be targeted to the chloroplast by both PSORT [18] and ChloroP [19]. Similarly, the rice cDNA AK068792, which most closely resembled TaGS1 in the database, also contained a homologous N-terminal extension predicted to target the putative peptide to the chloroplast. The other GS sequences from the cereals were not predicted to be targeted to subcellular compartments and therefore would be deposited in the cytosol.
Expression of recombinant GS polypeptides
The plant GS-coding sequences were subcloned into pET24 for the expression of the C-terminal His-tagged fusion proteins, using the first methionine residue in each case to initiate translation. TaGS3 could not be subcloned due to the difficulty in amplifying this cDNA with the required restriction enzyme sites. Similarly, difficulty was initially experienced in PCR amplification of the other wheat and maize clones, although the correct products were eventually obtained after adding betaine to the PCR. Since TaGS1 was predicted to contain an N-terminal signal peptide, two pET constructs were employed that either encoded the protein with the signal peptide or utilized an endogenous methionine residue at position 45, two amino acids downstream of the predicted cleavage site to initiate translation. When analysed by SDS/PAGE, the lysate from those induced bacteria harbouring the pET constructs were found to contain recombinant polypeptides with molecular masses between 50 and 60 kDa that were absent from control bacteria. As reported previously [8], considerable difficulty was experienced in expressing the recombinant plant GS proteins. This expression problem was overcome using Rosetta™ (DE3) cells grown in a medium containing 1% glucose with an overnight induction at 15 °C that improved both the solubility and yield of the recombinant GS proteins to between 5 and 20% of the total soluble protein extract.
Substrate selectivity of plant GSs
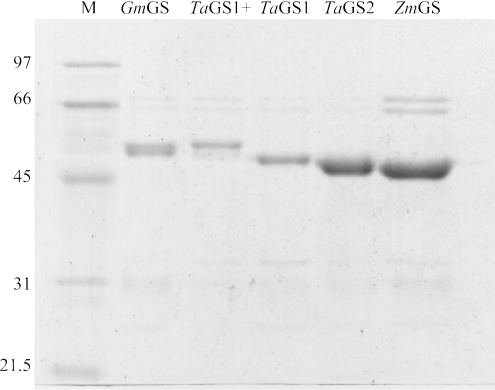
The His-tagged enzymes were purified using Ni-chelate affinity chromatography and purity was assessed by SDS/PAGE (Figure 2). When these purification runs were repeated with Escherichia coli extracts that did not contain recombinant GS proteins, no thiol synthetase activity was determined, confirming that the affinity-purified preparations were free of any contaminating bacterial GS enzyme. In the case of GmGS, depending on extraction conditions, the respective affinity-purified preparation could be resolved into a doublet of polypeptides. We presume that this was due to proteolytic processing of the recombinant GmGS, but on the basis of specific activity determinations between batches, this truncation did not affect activity. Purified protein (1 μg) was incubated with γ-EC in the presence of a representative range of acyl donor substrates including L-α-amino acids, D-amino acids and α-amino alcohols in the presence of ATP. Enzyme activity was determined by quantifying the amount of product formed after derivatization to the fluorescent S-bimane adducts and analysis by HPLC, with the respective molecular masses of the underivatized tripeptides confirmed by ESI-TOF MS (Table 1). Under all assay conditions, GS-catalysed product formation was directly proportional to enzyme concentration and incubation time over 120 min for all purified enzymes. Under standard assay conditions, incubations were carried out for 60 min. In the case of the TaGS1 clone, no significant differences were observed in the specific activities displayed when the recombinant protein was expressed with, or without, the signal peptide sequence. All activity data presented with respect to TaGS1 was obtained with recombinant enzyme expressed without the signal peptide sequence.
Figure 2. Analysis of purified recombinant GS polypeptides.
His-tagged proteins were purified on Ni-chelate affinity columns and analysed by Coomassie Blue staining after SDS/PAGE (10% polyacrylamide). From left to right: M, reference molecular-mass standards; GmGS (minus leader sequence); TaGS1+ (plus leader sequence); TaGS1 (minus leader sequence); TaGS2 (clone has no leader sequence); and ZmGS (clone has no leader sequence). Contaminating polypeptides around 60 and 30 kDa were present in all preparations and were presumably from the E. coli extract.
Table 1. Specific activities of purified recombinant thiol synthetases towards a range of substrates.
All assays were performed in triplicate with 1 mM γ-EC under standard assay conditions with 100 mM co-substrate, except for L-cysteine, which was used at 10 mM so as not to saturate the monobromobimane derivatization. Only those substrates with which an activity could be measured after correcting for enzyme-free controls are shown in the Table and the mass of the predicted reaction products, (M+H)+, was confirmed by MS in each case using NaI as calibrant. Other substrates assayed for which no enzyme activity could be detected include L-serine, D- and L-glutamic acid, L-valine, L-threonine, L-lysine, L-cysteine, L-arginine, L-asparagine, DL-BABA, 6-aminohexanoic acid and n-butylamine. Each value represents the mean±S.D. for three replicate experiments. NA, no detectable activity.
| Recombinant enzyme* | Tripeptide product | |||||
|---|---|---|---|---|---|---|
| Substrate | GmGS | TaGS1 | TaGS2 | ZmGS | HPLC retention time (min) | (M+H)+ |
| Glycine | 501±75 | 1620±13 | 2895±345 | 1781±416 | 8.5 | 308.09 |
| β-Alanine | 3306±469 | 56±5 | 403±77 | 591±29 | 10.9 | 322.11 |
| L-Alanine | NA | 23±5 | 564±169 | 177±12 | 10.3 | 322.11 |
| D-Alanine | NA | NA | 118±5 | 168±17 | 16.7 | 322.11 |
| AABA | NA | NA | 29±6 | NA | 16.9 | 336.12 |
| GABA | 2287±107 | 107±34 | 584±130 | 138±20 | 13.9 | 336.12 |
| BAIBA | 1641±106 | 10±2 | 16±3 | NA | 20.1 | 336.12 |
| L-2,3-Diaminopropionic acid | 1100±182 | 33±3 | 58±10 | 65±22 | 8.5 | 337.12 |
| Ethanolamine | 150±11 | NA | 17±1 | NA | 18.2 | 294.11 |
| 3-Amino-1-propanol | 88±8 | NA | 29±7 | NA | 23.0 | 308.13 |
| D-Serine | 35±2 | NA | 64±13 | NA | 8.6 | 338.10 |
| L-Ornithine | 21±1 | NA | 7±3 | NA | 10.1 | 365.15 |
*Activities are given in pkat (pmol of product formed·s−1).
Despite the presence of hGSH and hmGSH in place of glutathione in several plant species, the substrate selectivity of the respective GSs has received only limited attention. Initial experiments focused on the use of glycine and β-alanine as C-terminal substrates using a fixed concentration of 1 mM γ-EC. With the soya bean GS, the recombinant enzymes displayed a 6-fold higher specific activity with β-alanine as substrate compared with glycine (Table 1). With TaGS1, TaGS2 and ZmGS, glycine was the preferred substrate, although significant activity was also observed with β-alanine in each case. The purified recombinant enzymes were used for more detailed kinetic analysis to define the observed substrate preference between glycine and β-alanine (Table 2). The purified GmGS showed a significant specificity for β-alanine compared with glycine, as shown by an approx. 350-fold higher kcat/Km that is largely a consequence of an approx. 60-fold lower Km, indicating a strong ground state binding affinity discrimination. The kinetic constants determined were very similar to those reported for the respective enzyme partially purified from soya bean and runner bean [20]. In contrast, glycine was the preferred substrate with the TaGS and ZmGS recombinant enzymes. The >1000-fold specificity discrimination, as indicated by kcat/Km, for the maize and wheat GS enzymes was again largely a consequence of striking differences in Km. However, on the basis of kcat, a different pattern in their utilization of these two amino acids emerges (Table 2). While TaGS1 showed negligible turnover with β-alanine, TaGS2 had, in fact, a much greater Vmax with this substrate than with glycine.
Table 2. Kinetic characterization of purified recombinant GS enzymes.
Km data for both β-alanine and glycine were assayed with a constant γ-EC concentration (1 mM). Km data for γ-glutamylcysteine were assayed with a β-alanine concentration of 10 mM for GmGS and a glycine concentration of 10 mM for all other enzymes. For Vmax, activities are expressed as pkat·(mg of pure protein)−1. Kcat/Km values presented in the Table are with respect to the acyl acceptor. ND, kinetic constants could not be determined due to the very low activities determined. Vmax values are given in pkat·(mg of purified GS)−1.
| Co-substrate | |||
|---|---|---|---|
| Kinetic constant | Glycine | β-Alanine | |
| GmGS | Vmax (pkat·mg−1) | 364±36 | 2132±152 |
| Km (mM) | 19±6 | 0.32±0.08 | |
| Kcat/Km (M−1·s−1) | 107 | 37083 | |
| Km for γ-EC (mM) | ND | 0.06±0.04 | |
| TaGS1 | Vmax (pkat·mg−1) | 663±35 | ND |
| Km (mM) | 0.16±0.04 | ND | |
| Kcat/Km (M−1·s−1) | 24003 | ND | |
| Km for γ-EC (mM) | 0.18±0.07 | ND | |
| TaGS2 | Vmax (pkat·mg−1) | 2817±172 | 5660±3153 |
| Km (mM) | 0.07±0.02 | 170±136 | |
| Kcat/Km (M−1·s−1) | 212404 | 175 | |
| Km for γ-EC (mM) | 0.10±0.02 | ND | |
| ZmGS | Vmax (pkat·mg−1) | 1304±179 | 372.4±597 |
| Km (mM) | 0.18±0.08 | 592±1077 | |
| Kcat/Km (M−1·s−1) | 38060 | 3.3 | |
| Km for γ-EC (mM) | 0.03±0.02 | ND | |
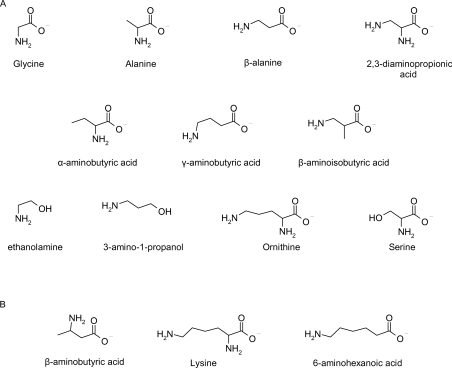
In soya bean, the presence of a β-alanine-selective GS correlated with the preferential accumulation of hGSH in this legume. It was therefore of interest to determine whether or not the accumulation of hmGSH in wheat and γ-ECE in maize could be similarly explained by an unusual amino acid selectivity of the respective GS enzymes. Therefore all enzymes were probed with the diverse array of C-terminal substrates shown in Figure 3. None among TaGS1, TaGS2 or ZmGS would accept L-serine or L-glutamate as co-substrate with γ-EC. When assayed with the other amino acid co-substrates, enzyme activity was only seen with L-alanine, notably with TaGS2. The utilization of L-alanine by the cereal GS enzymes then prompted the question as to what other amino derivatives (Figure 3) could serve as GS substrates for the wheat, maize and soya bean enzymes (Table 1). Putative reaction products were analysed by a combination of HPLC and MS (Figure 4).
Figure 3. Chemical structures of some potential substrates.
(A) Structures of substrates utilized in vitro by recombinant GS proteins from either G. max L., T. aestivum L. or Z. mays L. (B) Examples of substrates not utilized in vitro by any recombinant GS protein.
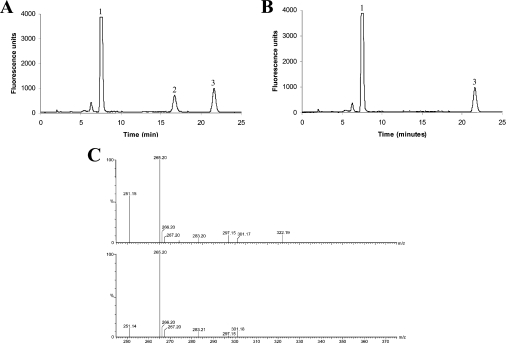
Figure 4. HPLC and MS analyses of a GS assay with a non-natural co-substrate.
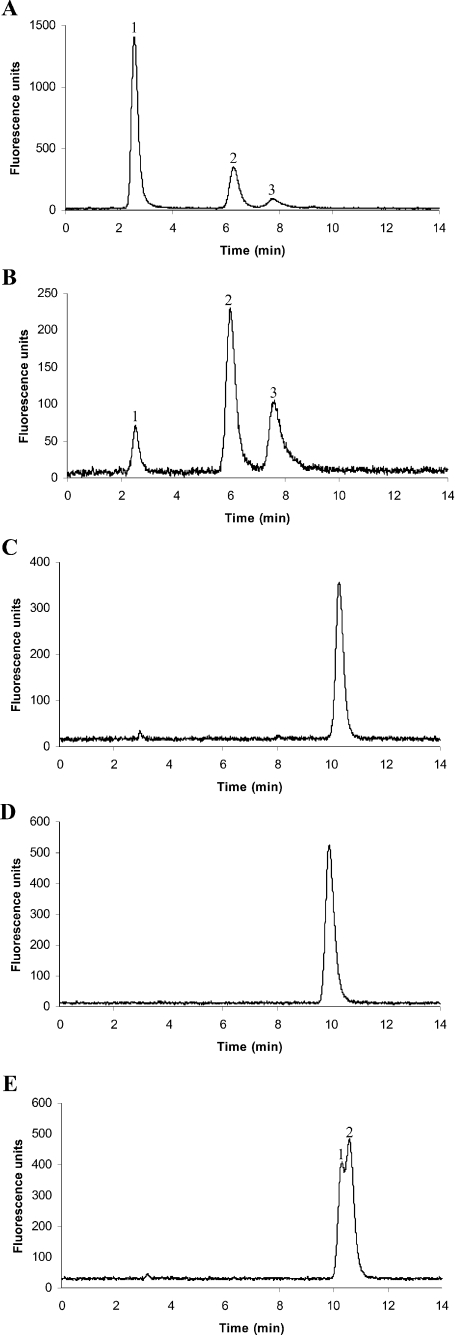
(A) HPLC fluorescence trace of S-bimane-derivatized reaction products of purified recombinant TaGS2 assay using γ-GC and D-alanine as substrates. Peak 1, excess γ-EC in the assay. Peak 2, novel reaction product (γ-Glu-Cys-D-Ala), which was not observed when TaGS2 was incubated with γ-EC minus D-alanine. Peak 3, bromobimane degradation product. (B) Control incubation of (A), with TaGS2 incubated with γ-GC only. (C) Upper and lower traces: mass spectral analysis of total reaction mixture shown in (A) and (B) respectively before derivatization with monobromobimane. The (M+H)+ at 251.1 corresponds to unchanged γ-GC, whereas the novel peak at 322.1 present in the upper trace but not in the lower trace corresponds to γ-Glu-L-Cys-D-Ala in trace (A).
TaGS1 showed a broadly similar selectivity for non-physiological amine substrates to GmGS, albeit with lower activities determined. TaGS2 proved to be the most versatile enzyme, acting on most of the substrates tested including D-alanine and D-serine, and being the only GS to utilize AABA (α-aminobutyric acid). ZmGS also catalysed the incorporation of L- and D-alanine but otherwise resembled TaGS1 in its selectivity and did not utilize D-glutamic acid. For reference, none of the recombinant GSs could utilize DL-BABA (DL-β-aminobutyric acid), 6-aminohexanoic acid or n-butylamine as substrates (Figure 3). A clear difference in the selectivity of GmGS and the wheat and cereal enzymes was the relative usage of GABA (γ-aminobutyric acid) and BAIBA (β-aminoisobutyric acid). All enzymes used GABA efficiently, but the introduction of a methyl group on the equivalent α-carbon in BAIBA provoked at least a 10-fold decrease in activity with the cereal GS, but only a 30% reduction in GmGS. These results are in contradiction to a previous report utilizing partially purified hGS from Phaseolus coccineus, which was 5-fold more active in utilizing BAIBA than GABA [20]. Although GABA has been shown to be a substrate for several GS enzymes, it may not be a universally acceptable substrate as previous reports showed that this acyl acceptor was tolerated by a partially purified GS from mung bean (Vigna radiata) but was not utilized by the respective enzyme from peas [21] or tobacco (Nicotiana tabacum) [22]. With the diamino acceptor 2,3-diaminopropionic acid, the site of peptide bond formation was not unequivocally demonstrated. However, based on the activity seen with BAIBA, it would seem most likely that where incorporation occurred the β-amino group was used. The utilization of β- and γ-amino groups was most markedly observed with GmGS, but in all cases further extension of the amino-group-bearing side chain beyond C-3 (ornithine) resulted in a total loss of activity [i.e. extension to C-4 (lysine) or C-5 (6-aminohexanoic acid)]. Replacement of the α-carboxy residue with an alcohol function (e.g. substitution of β-alanine with 3-amino-1-propanol and glycine with ethanolamine) also reduced incorporation, with only GmGS and TaGS2 capable of utilizing these substrates.
Active-site model based on chemical mapping
The use of a broad range of acyl acceptors allowed an assessment of the substrate specificity of the S1′ binding subsite in these peptide synthetases. For GmGS, three broad levels of selectivity were seen. The five substrates showing the highest activity (Table 1), namely glycine, β-alanine, GABA, BAIBA and L-2,3-diaminopropanoic acid, all have an amine substitution on the primary carbon. Unlike all the other GS enzymes examined, while GmGS was intolerant to α-substitution, β-substitution was acceptable as seen with BAIBA and L-2,3-diaminopropanoic acid. A secondary level of activity (21–150 pkat·mg−1), roughly an order of magnitude lower, was also observed for the simple substituted ethylamines [X-(CH2)2-NH2, where X=OH, CH2OH or CH2CH(NH2)CO2H], indicating that a certain amount of structural diversity and/or branching can be tolerated at the γ-position relative to the NH2 group. However, acceptors beyond a C-4 chain length (e.g. 6-aminohexanoic acid, Lys) are too large to occupy the GmGS subsite. For TaGS1, only one high-level acyl acceptor was identified in the screen, namely glycine (1620 pkat·mg−1). However, several moderate level acceptor substrates were turned over at a rate that was one order of magnitude lower (10–107 pkat·mg−1). From this pattern of substrate usage, it was concluded that TaGS1 must have a relatively short and narrow binding site since it could tolerate only minor α-branching tolerance (L-Ala), was stereoselective for the L-configuration (L-Ala not D-Ala) and had weak β-branching tolerance (BAIBA and L-2,3-diaminopropanoic acid), but had acceptance of unbranched ω-aminocarboxylic acids up to C-3 in length (Gly, β-Ala and GABA). For TaGS2, a broad substrate tolerance was seen. The optimal substrates (Table 1) Gly, β-Ala, L-Ala and GABA demonstrated tolerance of α-branching, even for the bulkier ethyl substituent, as seen in the activity of this enzyme towards AABA. Although an accompanying α-stereoselectivity was observed, this was relatively weak, with the ratio of L-Ala turnover to D-Ala turnover being 5:1. Limited activity (7–118 pkat·mg−1) was seen towards all other amines tested except those beyond a C-4 chain length, which were not tolerated, as was the case with all GS enzymes tested. Moderate β-branching tolerance was observed although this was substantially less than that shown by GmGS. ZmGS displayed high-level activity for just Gly and β-Ala (Table 1) with lesser activity (65–177 pkat·mg−1) observed for α-branched L- and D-Ala. Moderate activity towards GABA but not towards longer carbon chain nucleophiles indicated a shorter C-3-chain-binding subsite. Some weak tolerance of β-branching was also observed with L-2,3-diaminopropanoic acid, but not with BAIBA.
D-Serine as a substrate for TaGS
The lack of activity of TaGS1 and TaGS2 with L-serine suggested that neither enzyme was responsible for catalysing the direct formation of the all L-configuration hmGSH (i.e. γ-L-Glu-L-Cys-L-Ser). However, intriguingly, the activity seen with D-serine in TaGS2 demonstrated that it would be possible to synthesize the D-serine variant of hmGSH. Such a possibility could not be discounted, since the configuration of serine in hmGSH was not reported during its original identification in wheat plants [5]. Alternatively, γ-L-Glu-L-Cys-L-Ser may be synthesized in wheat by the TaGS after inversion of the D-serine to L-serine during or after ligation. An analogous co-ligation/epimerization is seen during tripeptide synthesis catalysed by ACV synthetase [L-δ-(α-aminoadipoyl)-L-cysteinyl-D-valine synthetase], a multifunctional enzyme, involved in penicillin biosynthesis [23]. By analogy to the synthesis of glutathione, ACV synthetase combines both γ-ECS and GS activities by sequentially synthesizing L-δ-(α-aminoadipoyl)-L-cysteine and then incorporating valine via a peptide bond. During this latter incorporation, L-valine is inverted to the D-configuration. The possibility that the incorporation of D-serine was associated with its epimerization was examined by preparing 1 μmol of the putative tripeptide reaction product. After S-derivatization with bromobimane, the conjugate was digested with γ-glutamyltranspeptidase and the product co-chromatographed with S-bimane derivatives of authentic L-Cys-L-Ser and L-Cys-D-Ser. The digested Cys-Ser-bimane product formed by TaGS2 co-chromatographed with the corresponding L-Cys-D-Ser-bimane standard rather than the L-Cys-L-Ser derivative, confirming that the serine had not undergone epimerization during ligation (Figure 5). hmGSH present in wheat plants was similarly S-bimane-derivatized and treated with γ-glutamyltranspeptidase after its purification by HPLC and was found to contain L-serine rather than D-serine.
Figure 5. HPLC analysis of wheat hmGSH and TaGS2 D-serine reaction product.
(A) HPLC trace of γ-glutamyltranspeptidase digest, run on a Chirobiotic V column, of the purified product of TaGS2 D-serine assay. MS identified peak 1 as undigested parent compound (S-bimane-derivatized γ-Glu-Cys-Ser: m/z+ 528.18), peak 2 as the S-bimane-derivatized Cys-Ser dipeptide product of the digest (m/z+ 399.13) and peak 3 as S-bimane-derivatized cys-teine (m/z+ 312.10). (B) HPLC trace of γ-glutamyltranspeptidase digest, run on a Chirobiotic V column, of hmGSH purified from wheat. MS confirmed the identity of each peak to be of identical mass to that shown in (A). (C) HPLC trace of purified peak 2 in (A), co-injected with S-bimane-derivatized L-Cys-D-Ser standard, on a Kingsorb C18 column. (D) HPLC trace of purified peak 2 in (B), co-injected with S-bimane-derivatized L-Cys-L-Ser standard, on a Kingsorb C18 column. (E) HPLC trace of S-bimane-derivatized L-Cys-L-Ser standard and S-bimane-derivatized L-Cys-D-Ser standard co-injected on to a Kingsorb C18 column. Peak 1 is S-bimane-derivatized L-Cys-L-Ser and peak 2 is S-bimane-derivatized L-Cys-D-Ser.
DISCUSSION
GSs are members of the ATP-grasp superfamily of ligases forming amide bonds in post-translationally synthesized peptides [24,25]. Family members include D-alanine:D-alanine ligase, ribosomal protein S6:glutamate ligase and an α-L-glutamate ligase involved in the biosynthesis of the coenzyme tetrahydrosarcinapterin [25]. The usage of differing amino acids in peptide formation has arisen through gene evolution, but our studies suggest that there is already latent diversity in the ability of the GS enzymes to synthesize diverse tripeptides. Thus, by expressing and assaying four recombinant GSs from three crop species, we have demonstrated differing substrate specificities and determined that these enzymes can catalyse the synthesis of a broad range of novel tripeptide products. This suggests that the nature of the thiol tripeptides formed in planta must be determined primarily by acyl acceptor availability in specific compartments, namely a readily available supply of glycine in maize and wheat and β-alanine in soya bean respectively. This is particularly relevant, since several of the amine acceptors tested in the present study are known to exist in plants and could therefore be incorporated into novel tripeptides. Thus GABA is a significant component of the free amino acid pool in plant species [26] and has been reported at concentrations as high as 1–2 μmol·(g of FW)−1, in soya bean leaves after biotic stress [27]. Similarly, AABA occurs in legumes, notably Lens sp. [28], and ethanolamine is known to accumulate in plants as an intermediate in choline synthesis after serine decarboxylation [29]. It is therefore possible that, if present in sufficiently high concentrations in a compartment along with a GS, these alternative amine acceptors could be incorporated into glutathione homologues in selected plant species. In the case of hGS, this diversification in the use of the C-terminus has resulted in the preferential incorporation of β-alanine and the resultant accumulation of hGSH in many legume species. The reasons for the selective use of β-alanine over glycine in hGS have received some attention. All cloned hGS enzymes to date, including the GS cloned from soya bean in the present study (Figure 1), have a substitution of leucine and proline for two alanine residues found in the putative glycyl binding domain of conventional GSs [30]. It has recently been demonstrated that mutation of the leucine alone, or both the leucine and proline residues, to the respective alanine residues (Figure 1) enhances the ability of an M. truncatula hGS to use glycine and synthesize glutathione [7]. However, as determined by comparison of the kinetic constants, GmGS exhibits a much greater preference for utilizing β-alanine over glycine as compared with the hGS of M. truncatula [7], suggesting that these two residues in the putative active site cannot be the only factors in determining hGS rather than GS specificity. Interestingly, when assayed with four structural isomers of aminobutyric acid, GmGS showed a marked preference for those substrates in which the amino group is on a terminal carbon rather than the α-carbon next to the carboxy group, which helps explain the preference for β-alanine over L-alanine. The kinetic mechanism of the Arabidopsis thaliana GS has recently been determined and shown to have a random Ter-reactant mechanism [31]. Studies with the Arabidopsis GS demonstrated that the order of binding of the first pair of substrates determined the affinity of binding to the third substrate. It would now be of interest to extend these studies to other acyl acceptors using different plant GSs.
Although it is possible that additional TaGS and ZmGS clones, encoding enzymes that could catalyse the incorporation of serine and glutamic acid respectively could have been overlooked in our screen, the available evidence increasingly suggests that hmGSH and γ-ECE arise from post-synthetic modifications of glutathione rather than from unique GSs that catalyse the ligation of serine rather than glycine at the C-termini. In the case of wheat, it has been suggested that hmGSH could arise by transpeptidation on the basis of the demonstration of hmGSH formation when a yeast carboxypeptidase was incubated with glutathione and L-serine [32]. Similarly, γ-ECE may well arise from the proteolytic transpeptidation of phytochelatins, which are polymers of (γ-Glu-Cys)n formed by plants on exposure to heavy metals [6,33].
Our studies demonstrate that, among the ATP-grasp ligase superfamily [24,25], the plant GSs are flexible peptide synthases. While these enzymes are limited to synthesizing γ-glutamyl linked glutathione analogues, this is of interest in regulating the activity of glutathione-dependent enzymes. For example, the overexpression of glutathione S-transferases in tumours is associated with the onset of multidrug resistance, and as such there has been considerable research effort to prepare inhibitory glutathione analogues for therapy [34]. It will now be of interest to screen the library of novel tripeptides produced by plant GSs for their ability to inhibit glutathione S-transferases and other glutathione-dependent proteins.
Acknowledgments
M. S. acknowledges financial support from the University of Durham through an Addison Wheeler Fellowship. We thank Dr J. Sanderson (Department of Chemistry, University of Durham) for the synthesis of L-Cys-L-Ser and L-Cys-D-Ser standards.
References
- 1.Noctor G., Arisi A. C. M., Jouanin L., Valadier M. H., Roux Y., Foyer C. H. Light-dependent modulation of foliar glutathione synthesis and associated amino acid metabolism in poplar overexpressing gamma-glutamylcysteine synthetase. Planta. 1997;202:357–369. [Google Scholar]
- 2.May M. J., Vernoux T., Leaver C., Van Montagu M., Inzé D. Glutathione homeostasis in plants: implications for environmental sensing and plant development. J. Exp. Bot. 1998;49:649–667. [Google Scholar]
- 3.Klapheck S. Homoglutathione: isolation, quantification and occurrence in legumes. Physiol. Plant. 1988;74:727–732. [Google Scholar]
- 4.Frendo P., Gallesi D., Turnbull R., Van de Sype G., Herouart D., Puppo A. Localisation of glutathione and homoglutathione in Medicago truncatula is correlated to a differential expression of genes involved in their synthesis. Plant J. 1999;17:215–219. [Google Scholar]
- 5.Klapheck S., Chrost B., Starke J., Zimmermann H. Gamma-glutamylcysteinylserine – a new homologue of glutathione in plants of the family Poaceae. Bot. Acta. 1992;105:174–179. [Google Scholar]
- 6.Meuwly P., Thibault P., Rauser W. E. Gamma-glutamylcysteinylglutamic acid – a new homolog of glutathione in maize seedlings exposed to cadmium. FEBS Lett. 1993;336:472–476. doi: 10.1016/0014-5793(93)80858-r. [DOI] [PubMed] [Google Scholar]
- 7.Frendo P., Jimenez M. J. H., Mathieu C., Duret L., Gallesi D., Van de Sype G., Herouart D., Puppo A. A Medicago truncatula homoglutathione synthetase is derived from glutathione synthetase by gene duplication. Plant Physiol. 2001;126:1706–1715. doi: 10.1104/pp.126.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iturbe-Ormaetxe I., Heras B., Matamoros M. A., Ramos J., Moran J. F., Becana M. Cloning and functional characterization of a homoglutathione synthetase from pea nodules. Physiol. Plant. 2002;115:69–73. doi: 10.1034/j.1399-3054.2002.1150107.x. [DOI] [PubMed] [Google Scholar]
- 9.Matamoros M. A., Moran J. F., Iturbe-Ormaetxe I., Rubio M. C., Becana M. Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol. 1999;121:879–888. doi: 10.1104/pp.121.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skipsey M., Cummins I., Andrews C. J., Jepson I., Edwards R. Manipulation of plant tolerance to herbicides through co-ordinated metabolic engineering of a detoxifying glutathione transferase and thiol co-substrate. Plant Biotech. J. 2005;3:409–420. doi: 10.1111/j.1467-7652.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- 11.Skipsey M., Andrews C. J., Townson J. K., Jepson I., Edwards R. Substrate and thiol specificity of a stress-inducible glutathione transferase from soybean. FEBS Lett. 1997;409:370–374. doi: 10.1016/s0014-5793(97)00554-1. [DOI] [PubMed] [Google Scholar]
- 12.Cummins I., Cole D. J., Edwards R. Purification of multiple glutathione transferases involved in herbicide detoxification from wheat (Triticum aestivum L.) treated with the safener fenchlorazole-ethyl. Pestic. Biochem. Physiol. 1997;59:35–49. [Google Scholar]
- 13.Dixon D. P., Cole D. J., Edwards R. Purification, regulation and cloning of a glutathione transferase (GST) from maize resembling the auxin-inducible type-III GSTs. Plant Mol. Biol. 1998;36:75–87. doi: 10.1023/a:1005958711207. [DOI] [PubMed] [Google Scholar]
- 14.Thom R., Cummins I., Dixon D. P., Edwards R., Cole D. J., Lapthorn A. J. Structure of a tau class glutathione S-transferase from wheat active in herbicide detoxification. Biochemistry. 2002;41:7008–7020. doi: 10.1021/bi015964x. [DOI] [PubMed] [Google Scholar]
- 15.Edwards R., Blount J. W., Dixon R. A. Glutathione and elicitation of the phytoalexin response in legume cell-cultures. Planta. 1991;184:403–409. doi: 10.1007/BF00195343. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen A. G., Nielsen H. Neural network prediction of translation initiation sites in eukaryotes: perspectives for EST and genome analysis. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:226–233. [PubMed] [Google Scholar]
- 18.Nakai K., Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emanuelsson O., Nielsen H., von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klapheck S., Zopes H., Levels H.-G., Bergmann L. Properties and localization of the homoglutathione synthetase from Phaseolus coccineus leaves. Physiol. Plant. 1988;74:733–739. [Google Scholar]
- 21.Macnicol P. K. Homoglutathione and glutathione synthetases of legume seedlings: partial purification and substrate specificity. Plant Sci. 1987;53:229–235. [Google Scholar]
- 22.Hell R., Bergmann L. Glutathione synthetase in tobacco suspension cultures: catalytic properties and localization. Physiol. Plant. 1988;72:70–76. [Google Scholar]
- 23.Byford M. F., Baldwin J. E., Shiau C. Y., Schofield C. J. The mechanism of ACV synthetase. Chem. Rev. 1997;97:2631–2649. doi: 10.1021/cr960018l. [DOI] [PubMed] [Google Scholar]
- 24.Dinescu A., Cundari T. R., Bhansali V. S., Luo J. L., Anderson M. E. Function of conserved residues of human glutathione synthetase – implications for the ATP-grasp enzymes. J. Biol. Chem. 2004;279:22412–22421. doi: 10.1074/jbc.M401334200. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Xu H. M., Graham D. E., White R. H. Glutathione synthetase homologs encode alpha-L-glutamate ligases for methanogenic coenzyme F-420 and tetrahydrosarcinapterin biosyntheses. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9785–9790. doi: 10.1073/pnas.1733391100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelp B. J., Bown A. W., McLean M. D. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- 27.Wallace W., Secor J., Schrader L. E. Rapid accumulation of gamma-aminobutyric acid and alanine in soybean leaves in response to an abrupt transfer to lower temperature, darkness, or mechanical manipulation. Plant Physiol. 1984;75:170–175. doi: 10.1104/pp.75.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozan P., Kuo Y. H., Lambein F. Amino acids in seeds and seedlings of the genus Lens. Phytochemistry. 2001;58:281–289. doi: 10.1016/s0031-9422(01)00200-x. [DOI] [PubMed] [Google Scholar]
- 29.Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W. I., Voelker D. R., Basset G., Hanson A. D. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 2001;276:35523–35529. doi: 10.1074/jbc.M106038200. [DOI] [PubMed] [Google Scholar]
- 30.Polekhina G., Board P. G., Gali R. R., Rossjohn J., Parker M. W. Molecular basis of glutathione synthetase deficiency and a rare gene permutation event. EMBO J. 1999;18:3204–3213. doi: 10.1093/emboj/18.12.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jez J. M., Cahoon R. E. Kinetic mechanism of glutathione synthetase from Arabidopsis thaliana. J. Biol. Chem. 2004;279:42726–42731. doi: 10.1074/jbc.M407961200. [DOI] [PubMed] [Google Scholar]
- 32.Okumura R., Koizumi Y., Sekiya J. Synthesis of hydroxymethylglutathione from glutathione and L-serine catalysed by carboxypeptidase Y. Biosci. Biotech. Biochem. 2003;67:434–437. doi: 10.1271/bbb.67.434. [DOI] [PubMed] [Google Scholar]
- 33.Meuwly P., Thibault P., Schwan A. L., Rauser W. E. Three families of thiol peptides are induced by cadmium in maize. Plant J. 1995;7:391–400. doi: 10.1046/j.1365-313x.1995.7030391.x. [DOI] [PubMed] [Google Scholar]
- 34.Lucente G., Luisi G., Pinnen F. Design and synthesis of glutathione analogues. Il Farmaco. 1998;53:721–735. doi: 10.1016/s0014-827x(98)00098-6. [DOI] [PubMed] [Google Scholar]