Abstract
Forkhead members of the ‘O’ class (FoxO) are transcription factors crucial for the regulation of metabolism, cell cycle, cell death and cell survival. FoxO factors are regulated by insulin-mediated activation of PI3K (phosphoinositide 3-kinase)–PKB (protein kinase B) signalling. Activation of PI3K–PKB signalling results in the phosphorylation of FoxO factors on three conserved phosphorylation motifs, which are essential for the translocation of FoxO factors from the nucleus to the cytosol. FoxO6, however, remains mostly nuclear due to the fact that its shuttling ability is dramatically impaired. FoxO1, FoxO3 and FoxO4 all contain an N- and C-terminal PKB motif and a motif located in the forkhead domain. FoxO6 lacks the conserved C-terminal PKB motif, which is the cause of the shuttling impairment. Since FoxO6 can be considered constitutively nuclear, we investigated whether it is also a constitutively active transcription factor. Our results show that FoxO6 transcriptional activity is inhibited by growth factors, independent of shuttling, indicating that it is not constitutively active. The PKB site in the forkhead domain (Ser184) regulated the DNA binding characteristics and the N-terminal PKB site acted as a growth factor sensor. In summary, FoxO6 is not a constitutively active transcription factor and can be regulated by growth factors in a Thr26- and Ser184-dependent manner, independent of shuttling to the cytosol.
Keywords: cytosol, FoxO6, growth factor, nucleo-cytoplasmic shuttling, transcriptional activity, translocation
Abbreviations: DBE, Daf-16 binding element; DMEM, Dulbecco's modified Eagle's medium; FCS, fetal calf serum; FoxO, forkhead members of the ‘O’ class; G-6-Pase, glucose-6-phosphatase; GFP, green fluorescent protein; HEK-293 cells, human embryonic kidney 293 cells; hiFCS, heat-inactivated FCS; IRU, insulin response unit; NES, nuclear export sequence; NLS, nuclear localization sequence; PBS-T, PBS containing 0.05% Tween 20; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B
INTRODUCTION
Forkhead members of the ‘O’ class (FoxO) are transcription factors that have been implicated in a multitude of biological processes including cell cycle, protection against oxidative stress, cell death and cell survival [1]. Their transcriptional activity is under the negative control of insulin/insulin-like signalling via the PI3K (phosphoinositide 3-kinase)–PKB (protein kinase B) pathway [2–4]. Activated PKB phosphorylates multiple FoxO residues, which results in the translocation of FoxO proteins from the nucleus to the cytosol, thereby automatically terminating its ability to induce target genes [1].
To date, the FoxO group has four mammalian members: FoxO1, FoxO3, FoxO4 and FoxO6. The degree of homology between these four members is high, especially in the forkhead domain, which contains the DNA-binding interface. FoxO members contain an N-terminal PKB motif, a PKB motif in the forkhead domain and a C-terminal PKB motif (Figure 1). The C-terminal PKB recognition sequence is not conserved in FoxO6 [5]. Phosphorylation of the C-terminal PKB residue in FoxO1 primarily depends on two neighbouring CK1 sites [6,7]. A constitutively phosphorylated DYRK1A site is located adjacent to the second CK1 site [8]. Together, this stretch of four phosphorylated serine residues facilitates nuclear export [5–7]. Since FoxO6 lacks all four serine residues, it is mainly nuclear under all conditions tested [5]. Insertion of the C-terminal PKB site, the CK1 sites and the DYRK1A site results in a gain of function mutant, which can shuttle from the nucleus to cytosol upon growth factor addition [5]. Shuttling is considered as the main negative regulator of FoxO-mediated transcriptional activity, although there are substantial data pointing to a shuttling-independent regulation of transcription activity [5,9].
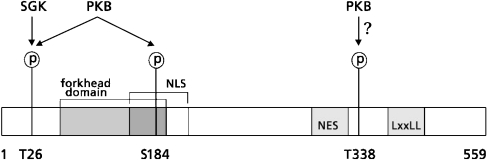
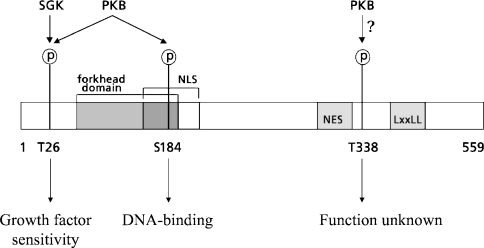
Figure 1. Schematic amino acid structure of FoxO6.
FoxO6 contains two conserved PKB motifs and one putative PKB motif. FoxO6 contains an N-terminal PKB site (Thr26), which is preferentially phosphorylated by SGK (serum and glucocorticoid regulated kinase), and a PKB site in the forkhead domain (Ser184), which is phosphorylated by PKB. Phosphorylation of Ser184 possibly obscures the NLS, which is located around the PKB motif. In the FoxO6 C-terminal region, a putative optimal PKB motif is present (Thr338) next to an NES. In the C-terminal region, FoxO6 contains an LXXLL motif implicated in nuclear receptor interactions.
Under conditions devoid of growth factors, the FoxO N- and C-terminal PKB sites are inaccessible and are in a non-phosphorylated state [10,11]. Insulin stimulation results in PKB-mediated phosphorylation of the PKB site in the forkhead domain, disruption of DNA binding and unmasking of the N- and C-terminal PKB sites [10–13]. Therefore the PKB site in the forkhead domain is considered as a gatekeeper of FoxO phosphorylation.
PKB-mediated phosphorylation of the N-terminal PKB site creates a docking motif for 14-3-3 proteins [2,11,14]. It has been suggested that a 14-3-3 dimer requires stable binding to the phosphorylated N-terminal PKB site before the other half of the dimer can bind to the phosphorylated PKB motif in the forkhead domain [1,15], which is not an optimal 14-3-3-binding motif by itself. The binding of a 14-3-3 dimer to a FoxO protein has several consequences as it blocks an intrinsic FoxO NLS (nuclear localization sequence) [16,17], prevents DNA binding [14,15] and mediates translocation to, and sequestration of FoxO factors in, the cytosol [17].
The C-terminal PKB site is not involved in 14-3-3 binding, but it is subjected to a sequence of hierarchical phosphorylation events [10,11]. Phosphorylation of the four residues containing the C-terminal PKB site increases the rate of export by mediating interactions with Ran-GTP and CRM1, possibly in co-operation with an NES (nuclear export sequence) located further downstream [6].
In the present study, we investigated the effect of growth factors on FoxO6-mediated transcriptional regulation. Since FoxO6 is constitutively nuclear, we examined whether its transcriptional activity was also constitutive. In addition, we focused on the role of the N-terminal PKB site and the PKB site in the forkhead domain in transcriptional regulation by growth factors. Since FoxO6 possesses a non-conserved optimal PKB motif in the C-terminal part (Thr338), we also studied the role of this non-conserved PKB motif.
Our results show that transcriptional activity of FoxO6 is efficiently regulated independent of shuttling to the cytosol and depends on intact FoxO DNA-binding sites. Transcriptional activity is regulated by Thr26 and Ser184, where Thr26 controls growth factor sensitivity and Ser184 determines the level of FoxO6 activity by regulating DNA-binding characteristics. Thr338 had no apparent role in the control of transcriptional activity or translocation. In summary, our results highlight the importance of Thr26 and Ser184 in the translocation-independent transcriptional regulation mediated by FoxO6 and suggests an additional level of FoxO regulation.
EXPERIMENTAL
FoxO–GFP (where GFP stands for green fluorescent protein) translational fusion
Mutations of Thr26, Ser184 and Thr338 to an alanine or aspartic residue were generated using site-directed mutagenesis by the same method as described in [5]. FoxO1, FoxO3, FoxO6 and the FoxO6-4Ser mutant protein were obtained as described previously [5].
Cell culture and transfection of HEK-293 cells (human embryonic kidney 293 cells)
HEK-293 cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) hiFCS (heat-inactivated fetal calf serum), 100 units/ml penicillin, 100 units/ml streptomycin and 2 mM L-glutamine in a humidified atmosphere with 5% CO2 at 37 °C. Cells were seeded in 12-well plates and grown for 24 h on glass coverslips. Cells were transfected with calcium phosphate precipitates containing 1.9 μg/well plasmid DNA (0.12 μg of target construct/1.78 μg of pBlueScript carrier DNA).
Forkhead translocation procedure
Twenty hours after transfection, cells were serum-starved for 24 h. Translocation was induced by replacing the serum-free medium with a medium supplemented with 10% hiFCS or 100 nM insulin. After 2 h of incubation, cells were fixed using 4% (w/v) paraformaldehyde in PBS for 10 min at room temperature (20 °C). Slips were embedded in Dabco-Mowiol and analysed by fluorescent microscopy. To ensure that the localization is analysed under similar conditions as during luciferase assays, control experiments after serum stimulation for 24 h were performed on all constructs (results not shown).
Luciferase assays
Cells were grown in six-well plates and transfected with 5 μg of plasmid DNA/well, including 1 μg of 6×DBE-Luc (where DBE stands for Daf-16 binding element; kindly provided by B. M. Burgering, University Medical Centre, Utrecht, The Netherlands) or 1 μg of the glucose-6-phosphatase-Luc or mutated glucose-6-phosphatase-Luc (both kindly provided by A. Barthel, University Hospital, Düsseldorf, Germany) with or without 0.3 μg of FoxO–GFP or empty vector and the appropriate amount of carrier plasmid. After transfection, cells were lysed and total GFP fluorescence was measured in 96-well plates using a FujiFilm FLA-5000 image reader to normalize the samples for transfection efficiency as described previously [5]. Each experiment was performed at least in triplicate.
FoxO6–DNA binding assay
Single-stranded oligosaccharides containing an optimal DBE (aattcaggattcctaggTTGTTTACattttaag) were end-labelled with [γ-32P]ATP and hybridized to the complementary oligosaccharides. HEK-293T cells were transfected as described above, and nuclear extracts were prepared as described previously [18]. Then, 40 fmol of probe was incubated with 1–5 μg of total protein for 30 min at 4 °C in 20 μl of binding buffer (40 mM Tris/HCl, pH 7.5, 5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 50 mM KCl, 10%, v/v, glycerol and 0.1% BSA) in the presence of 500 ng of sonicated salmon sperm and 500 ng of poly(dI-dC)·poly(dI-dC). Samples were loaded on to a 3.5% polyacrylamide gel containing 5% glycerol. Electrophoresis was performed in 0.25×TBE (90 mmol Tris/borate and 1 mmol EDTA) containing 5% glycerol at 4 °C for 120 min. GFP load was quantified using the FujiFilm FLA-5000 image reader. Gels were dried before autoradiography to visualize FoxO6–GFP/DNA binding.
Western blotting
Cells were grown in six-well plates and transfected with a total of 5 μg of plasmid DNA/well as described above. Then, cells were grown for 24 h in the presence of serum before they were starved for 24 h. Serum-starved cells were treated for 1 h with or without DMEM containing 10% FCS (fetal calf serum) before harvesting with lysis buffer containing 50 mM Tris, 1 mM EDTA, 1 mM EGTA, 0.5% Triton X-100, 1 mM PMSF, 100 mM sodium fluoride and 1 mM sodium vanadate on ice. Insoluble material was removed from the sample by centrifugation at 12000 g for 10 s. Concentrated SDS sample buffer containing 66 mM Tris/HCl (pH 6.8), 3% (w/v) SDS, 5% glycerol, 0.001% (w/v) Bromophenol Blue and 2% (v/v) 2-mercaptoethanol was added to the samples before the samples were heated for 15 min at 100 °C. Protein samples were separated by SDS/PAGE (9% polyacrylamide gel). After electrophoresis, protein was transferred on to nitrocellulose membranes (Amersham Biosciences) using a Bio-Rad Wet Blotting apparatus according to the manufacturer's instructions. Protein transfer and blotting efficiency were checked with ponseau-S. Blots were blocked for 1 h at room temperature in PBS-T (PBS containing 0.05% Tween 20) and 5% milk powder. Anti-phospho-Thr24/Thr32 FKHR/FKHRL1 (FOXO1/FOXO3) antibody (Cell Signaling Technology, Beverley, MA, U.S.A.) was diluted 1/1000 in PBS-T and incubated overnight. Secondary anti-rabbit antibody–horseradish peroxidase conjugate was diluted 1/50000 in PBS-T and incubated for 45 min before visualization with ECL® detection substrate (Amersham Biosciences) and HyperFilm (Amersham Biosciences).
After detection of phospho-Thr26, blots were stripped with PBS containing 2% SDS and 100 mM 2-mercaptoethanol for 10 min. Blots were blocked as described above before incubation with anti-GFP diluted 1/10000 in PBS-T for 1 h. The remainder of the procedure is as described above.
RESULTS
FoxO6 contains three putative RXRXXS/T phosphorylation motifs (where X denotes any amino acid and Ser/Thr denotes the residue phosphorylated by PKB), and two of the motifs are conserved among all other FoxOs. The two conserved PKB motifs are located at the N-terminus (Thr26) and in the forkhead domain (Ser184); the third FoxO6-specific PKB motif is located in the C-terminus (Thr338) (Figure 1). To explore the role of each individual PKB motif in FoxO6-mediated transcriptional activity, we created alanine and aspartic acid mutants, to mimic a non-phosphorylated or phosphorylated state respectively, and studied their response to growth factors.
Thr26 regulates translocation
Before studying the individual role of each FoxO6 PKB site in the regulation of transcriptional activity, we analysed their role in the regulation of growth factor-induced translocation. FoxO6–GFP-transfected cells were serum-starved for 24 h to inactivate the PI3K–PKB pathway before treatment with either insulin or FCS (serum). Subsequently, we analysed the intracellular FoxO6–GFP localization. Serum starvation resulted in a fully nuclear localization of wild-type FoxO6–GFP whereas insulin or serum treatment resulted in an increase in cytosolic localization as described in [5] (Figure 2). Mutation T26A (Thr26→Ala) or S184A (FoxO6 T26A and FoxO6 S184A) completely disrupted insulin- or serum-induced translocation to the cytosol, which has been reported earlier [5]. Mutation S184D (FoxO6 S184D) did not disrupt translocation (Figure 2). Surprisingly, the mutation T26D (FoxO6 T26D) abolished the effect of insulin or serum on FoxO6 shuttling and FoxO6 remained exclusively nuclear. A double mutation of T26D and S184D (FoxO6 T26D+S184D) rendered FoxO6 nuclear, which suggests that the mutation T26D is dominant over the mutation S184D (Figure 2).
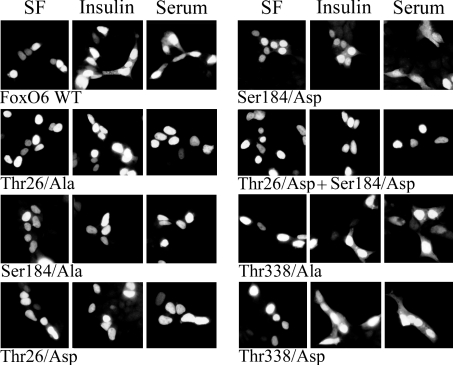
Figure 2. FoxO6 is mainly nuclear.
Cells were transfected with FoxO6–GFP or FoxO6 mutants fused to GFP. After transfection, cells were serum-starved and subsequently treated with insulin or serum. Wild-type FoxO6 translocates, to a low level, to the cytosol after insulin or serum treatment, which can be seen by the increase in cytosolic fluorescence. The mutation T26A, S184A or T26D completely prevents translocation. The mutation S184D does not prevent growth factor-induced translocation, whereas a double mutation of T26D and S184D does prevent translocation. Mutants T338A and T338D were indistinguishable from wild-type FoxO6.
A FoxO6-specific putative PKB motif is located next to the putative NES. Possibly, phosphorylation of Thr338 influences the functionality of this NES. To assess the role of Thr338 in growth factor-induced translocation, we analysed the alanine and aspartic acid mutants of this site. Mutation T338D or T338A did not affect growth factor-induced translocation from the nucleus to cytosol, nor did the serum-starved condition differ from wild-type FoxO6–GFP (Figure 2).
In summary, FoxO6 and the PKB site mutants of FoxO6 are localized mainly in the nucleus. Ser184 and Thr26 are both required for the small extent of shuttling to the cytosol, whereas Thr338 has no apparent function in shuttling.
Growth factor inhibition of transcriptional activity is mainly independent of shuttling
To analyse the role of shuttling from the nucleus to cytosol in the regulation of transcriptional activity, we compared wild-type FoxO6 with a FoxO6–FoxO3 chimaeric protein that has superior shuttling capabilities [5]. Both wild-type FoxO6 and the FoxO6-4Ser mutant have a comparable reduction in luciferase activity after the application of growth factors; however, the FoxO6-4Ser mutant translocates to the cytosol, whereas wild-type FoxO6 remains nuclear (Figure 3). These results indicate that profound shuttling is not required for the regulation of FoxO6 transcriptional activity.
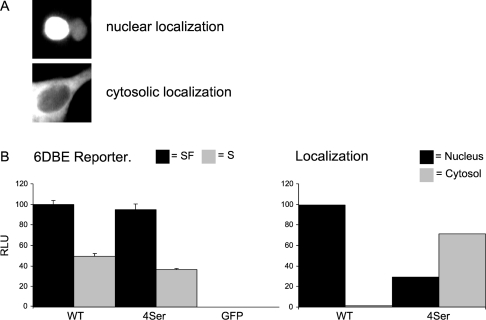
Figure 3. Growth factor-induced inhibition of transcriptional activation is independent of FoxO6 translocation.
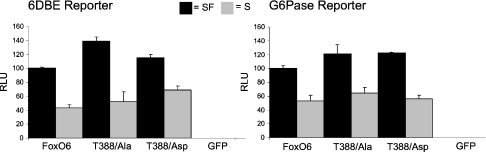
(A) Intracellular FoxO–GFP localization. Typical nuclear localization (upper panel) and a typical cytosolic FoxO–GFP localization (lower panel). (B) Left panel: the activity of FoxO6 was compared with a FoxO6–FoxO3 chimaeric protein that has the ability to shuttle extensively. Both wild-type FoxO6 (WT) and the chimaeric protein (4Ser) had comparable levels of transcriptional activity on the 6DBE reporter under serum (S) and serum-free (SF) conditions. Right panel: intracellular localization of WT and 4Ser was quantified under serum-treated conditions. WT was almost exclusively localized in the nucleus whereas the 4Ser mutant was mainly localized in the cytosol, using the same criteria as shown in (A). Nucleus, mainly nuclear localization; cytosol, mainly cytosolic localization.
Ser184 has a role in FoxO–DNA binding
Before investigating the role of Thr26 and Ser184 in the regulation of FoxO6-mediated transcriptional activity, we examined DNA binding capabilities of wild-type FoxO6 and mutants of FoxO6. Wild-type FoxO6 bound to the specific FoxO-binding element to a level comparable with that of FoxO6 T26A, FoxO6 T26D and FoxO6 S184A (Figure 4). The FoxO6 S184D, however, displayed strongly reduced binding, which suggests a role for Ser184 in the regulation of FoxO6–DNA interactions. The apparent increased binding observed with the T26A mutant was not reproducible, as small variations in binding efficiency occur. Taken together, the aspartic residue at position 184, mimicking phosphorylation of Ser184, can cause dissociation from the target DNA.
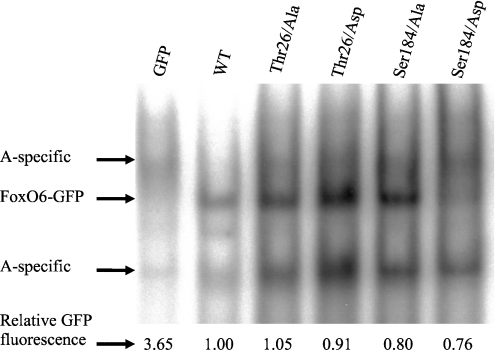
Figure 4. FoxO6 S184D has decreased DNA-binding characteristics.
Cells were transfected with GFP alone or alanine and aspartic acid FoxO6 mutants of Thr26. Subsequently, nuclear extracts were analysed for their ability to bind to a 32P-labelled optimal FoxO-binding element. Only the FoxO6 S184D mutant displays reduced DNA-binding abilities when compared with wild-type FoxO6. The other mutants display similar binding compared with wild-type FoxO6. The control lane (GFP) contains nuclear extract from cells transfected with the GFP plasmid alone. Samples were analysed for total GFP content as described in the Experimental section, which represents the total FoxO6–GFP present in the sample.
Transcriptional activity of FoxO6 is dependent on FoxO-binding sites
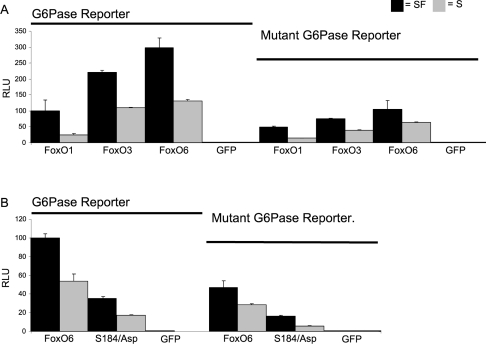
FoxO1 activity on the G-6-Pase (glucose-6-phosphatase) reporter depends on a stretch of multiple IRUs (insulin response units) [19,20]. Mutation of the IRUs within this promoter diminishes FoxO1 activity on this promoter [19]. To address whether FoxO6 activated the G-6-Pase reporter identically with FoxO1 and requires intact FoxO-binding sites, we monitored FoxO6 activity on wild-type and mutated G-6-Pase reporter constructs. FoxO1, FoxO3 and FoxO6 indeed have lower activity on the mutated promoter, indicating that binding to the IRUs is necessary for maximal activity (Figure 5A). Striking is the fact FoxO1 displays the lowest transcriptional activity on this reporter compared with FoxO3 and FoxO6. Although the mutant promoter contains mutated binding sites, FoxO6 still retains the ability to activate this reporter, comparable with FoxO1 on the wild-type promoter, suggesting that FoxO6 still retains the ability to bind to the mutant G-6-Pase reporter. Since the mutant G-6-Pase reporter still showed activity, we analysed the DNA-binding-deficient mutant FoxO6 S184D to validate that the activity is caused by FoxO6 binding to the target DNA. Indeed, FoxO6 S184D had lowest activity when compared with wild-type, suggesting that wild-type FoxO6 still binds to the mutant G-6-Pase reporter (Figure 5B). This further demonstrates that FoxO6 DNA binding is necessary to activate this mutant G-6-Pase promoter.
Figure 5. FoxO6 requires DNA binding for transcriptional activity.
(A) FoxO1, FoxO3 and FoxO6 were transfected together with wild-type (left) and mutant (right) G-6-Pase promoter-luciferase constructs. Serum application decreased FoxO activity on both wild-type and mutant promoters. (B) FoxO6 Ser184 influences transcriptional activity. Wild-type FoxO6 and FoxO6 S184D were transfected together with the wild-type or mutant G-6-Pase reporter. Activity of FoxO6 was greatly reduced on the mutant reporter and was further reduced by mutating S184D.
The FoxO6 mutant S184A does not inhibit growth factor sensitivity
Next, we investigated the role of Ser184 in FoxO6-mediated transcription, with and without serum stimulation. Mutation S184D greatly reduced the activity of FoxO6 under serum-free and serum conditions. This is consistent with the fact that this mutant is impaired in its ability to bind DNA. Interestingly, the FoxO6 S184A mutant displayed a higher activity when compared with wild-type FoxO6 (Figure 6). Moreover, this activity could be inhibited by growth factor addition. This suggests that the S184A mutant may still be regulated through phosphorylation.
Figure 6. Ser184 regulates the level of FoxO6 activity.
FoxO6 wild-type or alanine or aspartic acid mutants of Ser184 were transfected, together with a six times optimal FoxO-binding site reporter fused to luciferase (6DBE) or the G-6-Pase promoter fused to luciferase (G-6-Pase). Serum application (S) resulted in a decrease in transcriptional activity of the wild-type FoxO6 and Ser184 mutants. Both 6DBE and G-6-Pase reporters used showed comparable patterns of activity in response to FoxO6 and FoxO6 mutants. The S184A mutant displayed higher activity under serum and serum-free (SF) conditions when compared with wild-type FoxO6, whereas the S184D mutant showed lower activity. The GFP control does not display any transcriptional activity.
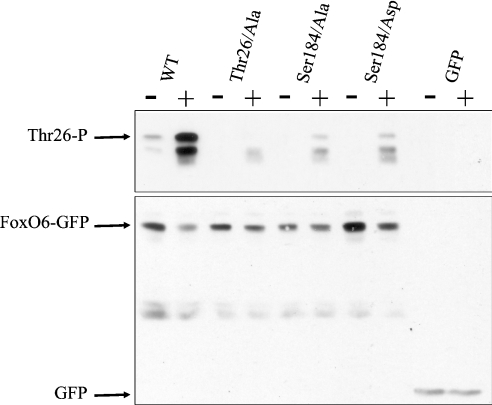
Phosphorylation on the PKB site in the forkhead domain of FoxO1 is required for phosphorylation of the N- and C-terminal PKB sites to occur [9,10]. Therefore the PKB site in the forkhead domain has been attributed as a ‘gatekeeper’ of FoxO phosphorylation. Since FoxO6 S184A could still be regulated by growth factors, we investigated whether Ser184 in FoxO6 also functions as ‘gatekeeper’. Using a phospho-specific antibody directed against phosphorylated Thr26, we studied the possible ‘gatekeeper’ function of Ser184 (Figure 7). Whereas no phosphorylation is observed under serum-free conditions, growth factor addition results in a large increase in phosphorylation of Thr26 in wild-type FoxO6. No phosphorylation of FoxO6 on Thr26 could be measured when cells were transfected with FoxO6 T26A, which confirms antibody specificity. Interestingly, the mutation S184A indeed greatly diminishes growth factor-induced phosphorylation of Thr26, but does not prevent it completely (Figure 7; S184A). Moreover, the S184D mutant displays similar behaviour. In conclusion, the mutation S184A or S184D diminishes the degree of phosphorylation of Thr26. However, in both cases, a small level of phosphorylation was observed, indicating that this may underlie the inhibition of transcriptional activity after growth factor addition.
Figure 7. The PKB site in the forkhead domain partly functions as a gatekeeper of phosphorylation.
Cells were transfected with wild-type (WT) FoxO6 or alanine or aspartic acid mutants of Thr26 and Ser184. Cells were serum-starved for 24 h before treatment with serum for 1 h. FoxO6 proteins were analysed for their phosphorylation on Thr26 using a phospho-specific antibody. Serum application induced a large increase in phosphorylation of Thr26 when compared with serum-free conditions. The antibody is specific for Thr26 since a mutation T26A is no longer recognized by the antibody. Mutation S184A greatly diminished phosphorylation of Thr26. Mutation S184D, however, also greatly diminished phosphorylation of Thr26. An increase in phosphorylation can still be observed in both Ser184 mutants after the application of serum. The specific band indicating Thr26 phosphorylation is indicated by an arrow. As a control, the same blot was stripped and subsequently analysed for its content of FoxO–GFP using a specific GFP antibody (arrow).
Thr26 regulates growth factor sensitivity
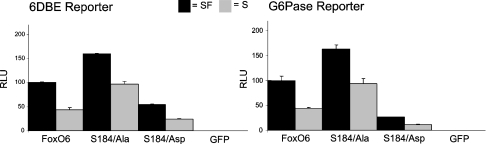
Ser184 did not regulate FoxO6 growth factor sensitivity, but did regulate phosphorylation of Thr26 to some extent. Therefore we further examined the role of Thr26 in the regulation of FoxO6. A FoxO6 T26A mutation resulted in a significant increase in FoxO6-mediated transcriptional activity under serum-free conditions, as compared with wild-type FoxO6 (Figure 8). Notably, under serum conditions, this mutant did not display a lower activity compared with serum-free conditions. Interestingly, FoxO6 T26D mutation led to the same result. In conclusion, phosphorylation of FoxO6 Thr26 appears to mediate growth factor sensitivity and mediates the inhibition of transcriptional activity independent of translocation.
Figure 8. Mutation of Thr26 impairs growth factor-induced inhibition of FoxO6 transcriptional activity.
When compared with wild-type FoxO6, the mutation T26A or T26D disrupted the growth factor-induced decrease in transcriptional activity. Activities on 6DBE and G-6-Pase reporters were comparable.
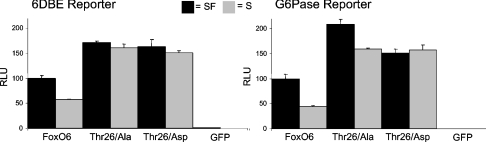
Thr338 has no apparent function in transcriptional activity
Finally, we analysed the role of Thr338 in FoxO6-mediated transcriptional activity. Thr338 displayed no apparent function in FoxO6 translocation. Thr338 is located in the putative transactivation domain of FoxO6, which suggests that it may mediate effects directly on FoxO6-mediated transcriptional activity. Although FoxO6 T338A and FoxO6 T338D mutants had slightly higher activity on both the 6DBE and G-6-Pase reporters, no clear function in the regulation of FoxO6-mediated transcriptional activity could be observed (Figure 9).
Figure 9. Thr338 is not involved in the regulation of FoxO6 transactivation activity.
Alanine or aspartic acid mutants of Thr338 were analysed for their transcriptional activity on a 6DBE or G-6-Pase luciferase construct. The T338A mutant displayed higher activity on the 6DBE and G-6-Pase reporter as compared with wild-type FoxO6 and activity of T338A could be decreased by serum application. A T338D mutant displayed higher activity under serum conditions on the 6DBE but not the G-6-Pase reporter. Overall, the Thr388 mutants behaved similar to wild-type FoxO6 in response to serum application and serum starvation.
DISCUSSION
Phosphorylation of specific FoxO residues by PKB triggers translocation to the cytosol [1]. FoxO6, however, is mainly nuclear. We have previously shown that the FoxO6 shuttling impairment is due to the absence of a stretch of conserved phosphorylation sites located in the C-terminal part. Since FoxO6 is mainly nuclear, we tried to determine whether FoxO6-mediated transcriptional activity is constitutive. In addition, we examined the role of the FoxO6 PKB residues Thr26 and Ser184 in the regulation of transcriptional activity. Our results show that FoxO6-mediated transcriptional activity is not constitutive and is regulated by growth factors, independent of translocation to the cytosol. Transcriptional activity is regulated by two phosphorylation sites: Ser184, which regulates DNA binding, and Thr26, which is required for growth factor-mediated inhibition of FoxO6.
Regulation of FoxO6 transcriptional activity does not require nucleo-cytoplasmic shuttling
Shuttling has been suggested as the main regulator of FoxO transcriptional activity. By physically removing the FoxO transcription factor from the nucleus, it can no longer be transcriptionally active. However, by comparing the activity profiles of a mutant FoxO6–FoxO3 chimaeric protein with wild-type FoxO6 on a FoxO reporter construct, we have clearly shown that shuttling is not the main regulator of FoxO6 function (Figure 3). Apparently, phosphorylation of PKB residues in FoxO6 is sufficient to suppress transcriptional activity without removal of the protein from the nucleus. This suggests that all the components needed for the negative regulation of FoxO6 are present in the nucleus or can be recruited there after growth factor stimulation.
Ser184 partly functions as a gatekeeper of FoxO6 phosphorylation
Phosphorylation of the PKB site in the forkhead domain inhibits FOXO1–DNA binding [12] and transcriptional activity. Accordingly, we show that FoxO6 S184D has the lowest DNA-binding capabilities. The low transcriptional activity of FoxO6 S184D presumably results from its low affinity for target DNA (Figure 4). Besides regulating target DNA binding, the PKB site in the forkhead domain has been shown to regulate the phosphorylation of the N- and C-terminal PKB sites in FOXO1 and has therefore been referred to as the gatekeeper of FoxO phosphorylation [9–12]. By mutating the PKB site in the forkhead domain, FOXO1 does not respond to growth factors and its transcriptional activity cannot be inhibited. In analogy, the ‘gatekeeper’ hypothesis partly applies to FoxO6. A FoxO6 S184A mutant diminishes phosphorylation of Thr26, which confirms the gatekeeper hypothesis (Figure 7). However, growth factor addition causes a small increase in Thr26 phosphorylation in the Ser184 mutants. This may explain the decrease in transcriptional activity of FoxO6 S184A after growth factor addition (Figure 6).
Thr26 mediates growth factor sensitivity
Since the N-terminal PKB site forms a 14-3-3 docking motif and negatively regulates CBP (CREB-binding protein)/p300 binding (where CREB stands for cAMP-response-element-binding protein) [2,21,22], we investigated its role in FoxO6-mediated transcriptional activity. Our results suggest that Thr26 is involved in sensing growth factors and is the dominant residue in regulating FoxO6 translocation and transcriptional activity (Figures 2 and 8). The mutation T26A completely prevents growth factor-induced translocation, which may be the result of its inability to bind 14-3-3 proteins (Figure 2). FoxO6 T26A-mediated transcriptional activity could not be inhibited by growth factor application and, as a consequence, transcriptional activity remained as high as that under serum-free conditions (Figure 8). This suggests that Thr26 functions as a growth factor sensor. Surprisingly, the T26D mutant was indistinguishable from the T26A mutant. Possibly, the aspartic residue may not fully mimic a phosphorylated residue and thus provides an invalid binding motif for 14-3-3 proteins. In addition to this, an important cofactor in FoxO-mediated transcription, CBP/p300, has been reported to interact specifically with the FoxO N-terminal region encompassing the PKB phosphorylation site [21]. Since phosphorylation of the N-terminal PKB site is required to disrupt the binding of CBP/p300 to this region, a Thr26-Ala-FoxO6 mutant, should be constitutively active, as is indeed observed in our experiments.
In conclusion, we have shown that inhibition of FoxO6-mediated transcriptional activity does not depend on shuttling to the cytosol, but is efficiently regulated in the nucleus. In addition to this, we have elucidated the individual role of Thr26 and Ser184 in mediating growth factor signals to FoxO6. Ser184 regulates DNA binding and provides a set point for the level of transcriptional activity (Figure 10). Thr26 senses the presence of growth factors and is therefore required for growth factor-mediated regulation of transcriptional activity (Figure 10). Presumably, Thr26 and Ser184 control FoxO6 transcriptional activity by regulating 14-3-3 and CBP/p300 binding, importantly, without the requirement of shuttling to the cytosol.
Figure 10. Role of PKB sites in the transcriptional regulation mediated by FoxO6.
Thr26 functions as a growth factor sensor, whereas Ser184 regulates DNA binding, thereby functioning as a set point for the level of transcriptional activity. The function of Thr338 is unknown.
References
- 1.van der Heide L. P., Hoekman F. M., Smidt M. P. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell (Cambridge, Mass.) 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 3.Kops G. J., de Ruiter N. D., de Vries-Smits A. M., Powell D. R., Bos J. L., Burgering B. M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature (London) 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 4.Rena G., Guo S., Cichy S. C., Unterman T. G., Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 1999;274:17179–17183. doi: 10.1074/jbc.274.24.17179. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs F. M., van der Heide L. P., Wijchers P. J., Burbach J. P., Hoekman M. F., Smidt M. P. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 2003;278:35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 6.Rena G., Woods Y. L., Prescott A. R., Peggie M., Unterman T. G., Williams M. R., Cohen P. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002;21:2263–2271. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rena G., Bain J., Elliott M., Cohen P. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 2004;5:60–65. doi: 10.1038/sj.embor.7400048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods Y. L., Rena G., Morrice N., Barthel A., Becker W., Guo S., Unterman T. G., Cohen P. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai W. C., Bhattacharyya N., Han L. Y., Hanover J. A., Rechler M. M. Insulin inhibition of transcription stimulated by the forkhead protein Foxo1 is not solely due to nuclear exclusion. Endocrinology. 2003;144:5615–5622. doi: 10.1210/en.2003-0481. [DOI] [PubMed] [Google Scholar]
- 10.Nakae J., Barr V., Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rena G., Prescott A. R., Guo S., Cohen P., Unterman T. G. Roles of the forkhead in rhabdomyosarcoma (FKHR) phosphorylation sites in regulating 14-3-3 binding, transactivation and nuclear targeting. Biochem. J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo S., Rena G., Cichy S., He X., Cohen P., Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X., Gan L., Pan H., Guo S., He X., Olson S. T., Mesecar A., Adam S., Unterman T. G. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J. Biol. Chem. 2002;277:45276–45284. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
- 14.Cahill C. M., Tzivion G., Nasrin N., Ogg S., Dore J., Ruvkun G., Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J. Biol. Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- 15.Obsil T., Ghirlando R., Anderson D. E., Hickman A. B., Dyda F. Two 14-3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14-3-3 proteins and inhibition of DNA binding. Biochemistry. 2003;42:15264–15272. doi: 10.1021/bi0352724. [DOI] [PubMed] [Google Scholar]
- 16.Brownawell A. M., Kops G. J., Macara I. G., Burgering B. M. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell. Biol. 2001;21:3534–3546. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet A., Kanai F., Stehn J., Xu J., Sarbassova D., Frangioni J. V., Dalal S. N., DeCaprio J. A., Greenberg M. E., Yaffe M. B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber E., Matthias P., Muller M. M., Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayala J. E., Streeper R. S., Desgrosellier J. S., Durham S. K., Suwanichkul A., Svitek C. A., Goldman J. K., Barr F. G., Powell D. R., O'Brien R. M. Conservation of an insulin response unit between mouse and human glucose-6-phosphatase catalytic subunit gene promoters: transcription factor FKHR binds the insulin response sequence. Diabetes. 1999;48:1885–1889. doi: 10.2337/diabetes.48.9.1885. [DOI] [PubMed] [Google Scholar]
- 20.Schmoll D., Walker K. S., Alessi D. R., Grempler R., Burchell A., Guo S., Walther R., Unterman T. G. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J. Biol. Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 21.Mahmud D. L., G-Amlak M., Deb D. K., Platanias L. C., Uddin S., Wickrema A. Phosphorylation of forkhead transcription factors by erythropoietin and stem cell factor prevents acetylation and their interaction with coactivator p300 in erythroid progenitor cells. Oncogene. 2002;21:1556–1562. doi: 10.1038/sj.onc.1205230. [DOI] [PubMed] [Google Scholar]
- 22.van der Heide L. P., Smidt M. P. Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem. Sci. 2005;30:81–86. doi: 10.1016/j.tibs.2004.12.002. [DOI] [PubMed] [Google Scholar]