Abstract
The human cytochrome P450 2J2 (CYP2J2) generates cytoprotective epoxyeicosatrienoic acids from arachidonic acid. Expression of CYP2J2 is decreased in hypoxia, and the resultant decrease in CYP2J2-derived epoxyeicosanoids may contribute to the pathogenesis of cardiac ischaemia. Recent studies have indicated that AP-1 (activator protein-1) regulates CYP2J2 expression in normoxia and hypoxia. Down-regulation of CYP2J2 in hypoxic HepG2 cells was closely associated with the up-regulation of c-fos and transient transfection analysis demonstrated that c-Fos abolishes the activation of CYP2J2 by the AP-1 protein c-Jun. Deletion of the region between nt −122 and −50 upstream of the start codon in CYP2J2 prevented c-Jun transactivation. In this study we demonstrate that the sequence at −105/−95 is a major regulatory element that binds c-Jun and has a prominent role in CYP2J2 gene transactivation. Mutagenesis of both the −105/−95 region and the previously identified element at −56/−63 was required for complete loss of transactivation by c-Jun; separate mutagenesis of the −105/−95 element or, to a lesser extent, the −56/−63 element resulted in a partial loss of gene activation. In contrast to the behaviour of the −56/−63 element, c-Jun homodimers and c-Fos/c-Jun heterodimers bound to the −105/−95 element. These findings demonstrate that the c-Jun-responsive module between −122 and −50 in the CYP2J2 proximal promoter contains an atypical AP-1 element at −105/−95 that has a major role in c-Jun transactivation and acts in conjunction with the −56/−63 element to regulate expression.
Keywords: c-Jun, CYP2J2, cytochrome P450, activator protein-1 (AP-1)
Abbreviations: AP-1, activator protein-1; ATF, activating transcription factor; CYP, cytochrome P450; EET, epoxyeicosatrienoic acid; EMSA, electrophoretic mobility-shift assay; poly(dI-dC), poly(2′-deoxyinosinic-2′-deoxycytidylic acid); Sp1, specificity protein 1
INTRODUCTION
Hepatic cytochromes P450 (CYPs) oxidize hydrophobic chemicals from exogenous and endogenous sources to polar products that are more readily excreted [1]. CYP2J2 exhibits a broader tissue distribution than most CYPs, which suggests that the enzyme may have important physiological functions. CYP2J2 is highly expressed in many tissues including liver [2], cardiovasculature [3], lung [4], pancreas [5] and the small intestine [6]. CYP2J2 catalyses the epoxidation of arachidonic acid to EETs (epoxyeicosatrienoic acids) [2] that influence a range of processes in tissues, including relaxation of vascular smooth muscle [7], modulation of inflammation [3], cytoprotection against hypoxic injury [8] and inhibition of apoptosis [9]. Thus, in aortic endothelial cells, transfection of CYP2J2 cDNA or treatment with EETs prior to culture at low oxygen tension promoted cell survival [8]. The anti-apoptotic effects of EETs have been linked to activation of the phosphoinositide 3-kinase/Akt cell survival cascade [9].
Jun, Fos, ATF (activating transcription factor) and maf (macrophage-activating factor) proteins are members of the AP-1 (activator protein-1) bZIP (family of leucine zipper) proteins [10–12]. AP-1 is activated by phorbol esters [13], cytokines [14], growth factors [10,15] and low oxygen tension [16–19] and has a critical role in cell survival and apoptosis in response to extracellular stimuli [15,20]. Previous studies have implicated AP-1 in the regulation of CYP2J2 in HepG2 cells cultured under normoxic and hypoxic conditions [21]. The down-regulation of CYP2J2 in hypoxic HepG2 cells was associated with up-regulation of c-fos. In transfection studies, c-Jun strongly enhanced transcriptional activity of the CYP2J2 promoter, which was abolished by c-Fos. In normoxia c-Jun homodimers activated CYP2J2 expression whereas, in hypoxia, the up-regulation of c-fos led to c-Jun–c-Fos heterodimerization that mediated CYP2J2 down-regulation. An AP-1-like element at −56/−63 bp within the CYP2J2 proximal promoter contributed to c-Jun-dependent activation; EMSA (electrophoretic mobility-shift assay) analysis confirmed the binding of c-Jun, but not c-Fos, to the element [21]. Deletion of the upstream region between −152 and −50 bp relative to the CYP2J2 start codon abolished c-Jun-responsiveness, but mutagenesis of the −56/−63 bp element only partially decreased activation by c-Jun [21]. Thus, additional elements in the 5′-flank were implicated in CYP2J2 regulation by AP-1 proteins. The present study identifies a second sequence at −105/−95 that has a prominent role in c-Jun-dependent transactivation of CYP2J2. These findings establish that the CYP2J2 proximal promoter contains a c-Jun-responsive module which comprises two distinct elements that regulate transcription by c-Jun.
EXPERIMENTAL
Materials and plasmids
The pGL3basic luciferase reporter vector, pCMV-β-galactosidase vector, Steady-Glo™ Luciferase assay system, β-galactosidase reporter lysis assay system and recombinant human c-Jun protein were purchased from Promega Corp. (Annandale, NSW, Australia). Active Motif recombinant human c-Fos protein was purchased from Bioscientific (Gymea, NSW, Australia). The c-Jun and c-Fos expression plasmids were a gift from Dr K. Imakawa (Laboratory of Animal Breeding, Faculty of Agriculture, University of Tokyo, Japan). The ABI Prism BigDye™ Terminator Cycle Sequencing Ready Reaction Kit and [32P]dCTP were purchased from PerkinElmer (Rowville, Vic., Australia). The Stratagene QuikChange™ Site-Directed Mutagenesis Kit was obtained from Integrated Sciences (Willoughby, NSW, Australia), and the FuGENE™ 6 transfection reagent was purchased from Roche Diagnostics (Castle Hill, NSW, Australia). All oligonucleotides were synthesized by Geneworks (Adelaide, SA, Australia). The ProbeQuant™ G-50 Micro Columns and Megaprime™ DNA Labelling System were obtained from Amersham Biosciences (Castle Hill, NSW, Australia). Protease inhibitors were from Sigma–Aldrich (Castle Hill, NSW, Australia). Enzymes were obtained from Roche Diagnostics unless otherwise specified. Antibodies directed against c-Jun, c-Fos and ubiquitin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A).
Cell culture
The HepG2 cell human hepatoma cell line was purchased from A.T.C.C. (Manassas, VA, U.S.A.). Media, antibiotics and fetal calf serum were from Thermo Trace (Noble Park, Victoria, Australia). HepG2 cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% heat-inactivated fetal calf serum, 1% L-glutamine, 1% penicillin/streptomycin antibiotic mix, 26 mM sodium hydrogen bicarbonate and 20 mM Hepes. Cells were cultured at 37 °C in an atmosphere of 95% air and 5% CO2 and were passaged twice a week. Cells at passage 4 were used in all experiments.
Construction of CYP2J2 reporter plasmids
A 2.4 kb fragment of the 5′-flanking region of the CYP2J2 gene was subcloned into the pGL3basic luciferase reporter vector [2J2(−2341/+98)] as described previously [21]. The construct 2J2(−152/+98), which contains the proximal CYP2J2 promoter sequence from −152 to +98 bp, was prepared from 2J2(−2341/+98) by digestion with SmaI. To create the 5′-truncated constructs 2J2(−122/+98), 2J2(−82/+98) and 2J2(−49/+98), SmaI recognition sites were introduced in 2J2(−152/+98) by site-directed mutagenesis (Stratagene QuikChange™ Kit) using the oligonucleotides shown in Table 1, followed by digestion and re-ligation. The mutagenized (mt) constructs 2J2(−152/+98; mt −56/−63), 2J2(−152/+98; mt −106/−96), 2J2(−152/+98; mt −56/−63; mt −106/−96), 2J2(−152/+98; mt −148/−142), 2J2(−152/+98; mt −141/−134) and 2J2(−152/+98; mt −133/−126) were prepared similarly using the oligonucleotides in Table 1. Sequences of all constructs were confirmed (ABI Prism Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit) and plasmid DNAs for use in transfection were prepared using a Qiagen Midiprep kit.
Table 1. Sequences of oligonucleotides used in site-directed mutagenesis to create CYP2J2 5′-truncation and mutation constructs.
Mutated nucleotides are underlined.
| Oligonucleotide | Sense/antisense | Sequence (5′—3′) |
|---|---|---|
| 2J2-mt-(−56/−63) | Sense | GAGGCGGGGCGGGAAAAAAAACCTGCTGGGACCG |
| 2J2-mt-(−56/−63) | Antisense | CGGTCCCAGCAGGTTTTTTTTCCCGCCCCGCCTC |
| 2J2-mt-(−148/−142) | Sense | GCGTGCTAGCCCGCTTAAAAAGCGCCTGGCATC |
| 2J2-mt-(−148/−142) | Antisense | GATGCCAGGCGCTTTTTAAGCGGGCTAGCACGC |
| 2J2-mt-(−141/−134) | Sense | AGCCCGGGAATCCTTTTAAAAGCATCTTCGCAGG |
| 2J2-mt-(−141/−134) | Antisense | CCTGCGAAGATGCTTTTAAAAGGATTCCCGGGCT |
| 2J2-mt-(−133/−126) | Sense | AATCCAGCGCCTGAAAAAAAAGCAGGGTGCTGCG |
| 2J2-mt-(−133/−126) | Antisense | CGCAGCACCCTGCTTTTTTTTCAGGCGCTGGATT |
| 2J2-mt-(−106/−96) | Sense | GGTGCTGCGAAGGGGGAAAAAAAAAAGCGGGGCACGGCTGG |
| 2J2-mt-(−106/−96) | Antisense | CCAGCCGTGCCCCGCTTTTTTTTTTCCCCCTTCGCAGCACC |
| 2J2-mt-(−122/SmaI) | Sense | CTGGCATCTTCGCACCCGGGTGCTGCGAAGG |
| 2J2-mt-(−122/SmaI) | Antisense | CCTTCGCAGCACCCGGGTGCGAAGATGCCAG |
| 2J2-mt-(−82/SmaI) | Sense | CGGGGCACGGCTCCCGGGAGCGAGGCG |
| 2J2-mt-(−82/SmaI) | Antisense | CGCCTCGCTCCCGGGAGCCGTGCCCCG |
| 2J2-mt-(−49/SmaI) | Sense | CCGTCGCCTGCTCCCGGGACCGCCGCC |
| 2J2-mt-(−49/SmaI) | Antisense | GGCGGCGGTCCCGGGAGCAGGCGACGG |
Transient transfections and luciferase and β-galactosidase assays
HepG2 cells (6×105 cells/well in 6-well plates) were co-transfected with CYP2J2 promoter constructs (1 μg/well) and a pCMV-β-galactosidase expression plasmid (0.5 μg/well; to control for transfection efficiency) using FuGENE™ 6. c-Jun and c-Fos expression plasmids were added at 0.5 μg/well. The plasmid DNA mixture was incubated with FuGENE™ 6 (FuGENE™/DNA, 3:2) for 20 min at room temperature and then added to wells. After 24 h, the medium was replaced and cells were harvested 48 h later. Luciferase activity was measured by scintillation spectrometry using the Steady-Glo™ Luciferase assay system, and β-galactosidase activity was determined colorimetrically using the Reporter Lysis assay system. All transfections were performed in duplicate wells in at least three separate experiments.
Preparation of nuclear extracts
Nuclear extracts were prepared from untransfected HepG2 cells or cells that had been transfected with c-Jun expression plasmid (0.5 μg/6×105 cells) [22]. Nuclear extracts were frozen in liquid N2 and stored at −80 °C until used in EMSAs. Protein was determined by the Lowry method using BSA as standard [23].
EMSAs
Oligonucleotides used as probes or competitors in EMSAs are shown in Table 2. Complementary oligonucleotides were annealed, end-labelled with [32P]dCTP (Megaprime™ DNA labelling system) and purified (ProbeQuant™ G-50 micro columns). Binding reactions containing 50–200 fmol of the end-labelled probes and 30 μg of nuclear protein were incubated for 20 min at room temperature and 10 min at 4 °C in a buffer containing 50 mM NaCl, 10 mM Tris/HCl (pH 7.5), 1 mM MgCl2, 0.5 mM EDTA (pH 8.0), 0.5 mM dithiothreitol, 4% glycerol and 1 μg of poly(dI-dC) [poly(2′-deoxyinosinic-2′-deoxycytidylic acid)]. For reactions with recombinant proteins, 0.6 μg of recombinant human c-Jun protein, or a mixture of 0.15 μg of recombinant human c-Fos protein and 0.15 μg of recombinant human c-Jun protein, was used with 0.2 μg of poly(dI-dC). Protein–DNA complexes were electrophoresed on 5% polyacrylamide gels in TBE buffer (90 mM Tris/HCl, 90 mM boric acid, 2.5 mM EDTA) at 100 V for ∼1.5 h at 4 °C. In competition experiments, 200-fold excess unlabelled probe was included in the binding reactions, with the Stat5 consensus sequence from the β-casein promoter [24] used as a negative control. In supershift experiments, either nuclear protein or recombinant protein was incubated with rabbit polyclonal anti-c-Jun or anti-c-Fos antibodies (2 μg) for 1 h at 4 °C prior to the binding reaction; an anti-ubiquitin antibody was used as a negative control. Following electrophoresis, gels were dried and autoradiographed.
Table 2. Sequences of double-stranded oligonucleotides used in EMSA analysis.
All oligonucleotides have a 3′-GGG overhang for labelling with [32P]dCTP with exceptions as follows: the AP-1 consensus and 2J2-A (−152/−103) oligonucleotides have a 5′-CTAG overhang on the sense strand and a 5′-GATC overhang on the antisense strand; the Stat5 antisense oligonucleotide has a 5′-G overhang. Mutated nucleotides are underlined. The AP-1 consensus sequence in the sense strand of the AP-1 consensus oligonucleotide, the Stat5 element in the Stat5 β-casein promoter oligonucleotides, and the c-Jun binding site at −105/−95 bp in the sense strand of probe 5 (−105/−86) are indicated in bold.
| Oligonucleotide | Sense/antisense | Sequence (5′–3′) |
|---|---|---|
| 2J2-A (−152/−103) | Sense | CTAGCCCGGGAATCCAGCGCCTGGCATCTTCGCAGGGTGCTGCGAAGGGGCGG |
| 2J2-A (−152/−103) | Antisense | GATCCCCGCCCCTTCGCAGCACCCTGCGAAGATGCCAGGCGCTGGATTCCCGGG |
| 2J2-B (−127/−79) | Sense | TCGCAGGGTGCTGCGAAGGGGCGGGCTGGGAGGCGGGGCACGGCTGGGAGGG |
| 2J2-B (−127/−79) | Antisense | TCCCAGCCGTGCCCCGCCTCCCAGCCCGCCCCTTCGCAGCACCCTGCGAGGG |
| 2J2-C (−102/−50) | Sense | CTGGGAGGCGGGGCACGGCTGGGAGCGAGGCGGGGCGGGGACCGTCGCCTGCTGGG |
| 2J2-C (−102/−50) | Antisense | AGCAGGCGACGGTCCCCGCCCCGCCTCGCTCCCAGCCGTGCCCCGCCTCCCAGGGG |
| 1 (−152/−128) | Sense | CCCGGGAATCCAGCGCCTGGCATCTGGG |
| 1 (−152/−128) | Antisense | AGATGCCAGGCGCTGGATTCCCGGGGGG |
| 2 (−137/−119) | Sense | CTGGCATCTTCGCAGGGTGGG |
| 2 (−137/−119) | Antisense | ACCCTGCGAAGATGCCAGGGG |
| 3 (−127/−106) | Sense | TCGCAGGGTGCTGCGAAGGGGCGGG |
| 3 (−127/−106) | Antisense | GCCCCTTCGCAGCACCCTGCGAGGG |
| 4 (−114/−97) | Sense | CGAAGGGGCGGGCTGGGAGGG |
| 4 (−114/−97) | Antisense | TCCCAGCCCGCCCCTTCGGGG |
| 5 (−105/−86) | Sense | GGGCTGGGAGGCGGGGCACGGGG |
| 5 (−105/−86) | Antisense | CGTGCCCCGCCTCCCAGCCCGGG |
| 6 (−94/−77) | Sense | CGGGGCACGGCTGGGAGCGGG |
| 6 (−94/−77) | Antisense | GCTCCCAGCCGTGCCCCGGGG |
| 7 (−85/−69) | Sense | GCTGGGAGCGAGGCGGGGGG |
| 7 (−85/−69) | Antisense | CCCGCCTCGCTCCCAGCGGG |
| 8 (−77/−60) | Sense | CGAGGCGGGGCGGGGACCGGG |
| 8 (−77/−60) | Antisense | GGTCCCCGCCCCGCCTCGGGG |
| 9 (−68/−48) | Sense | GCGGGGACCGTCGCCTGCTGGGGG |
| 9 (−68/−48) | Antisense | CCAGCAGGCGACGGTCCCCGCGGG |
| mt 5 (−105/−86; mt −105/−96) | Sense | AAAAAAAAAAGCGGGGCACGGGG |
| mt 5 (−105/−86; mt −105/−96) | Antisense | CGTGCCCCGCTTTTTTTTTTGGG |
| AP-1 consensus | Sense | CTAGTGATGAGTCAGCCGGATC |
| AP-1 consensus | Antisense | GATCGATCCGGCTGACTCATCA |
| Stat5 β-casein promoter | Sense | GGACTTCTTGGAATTAAGGGA |
| Stat5 β-casein promoter | Antisense | GTCCCTTAATTCCAAGAAGTCC |
Statistical analysis
Results are expressed as the means±S.E.M. Differences between experimental groups were detected using the Student's t test. P<0.05 was considered to be statistically significant. All data were derived at least in duplicate from at least three independent experiments.
RESULTS
c-Jun-responsive sequences between −152 and −50 bp in the CYP2J2 proximal promoter
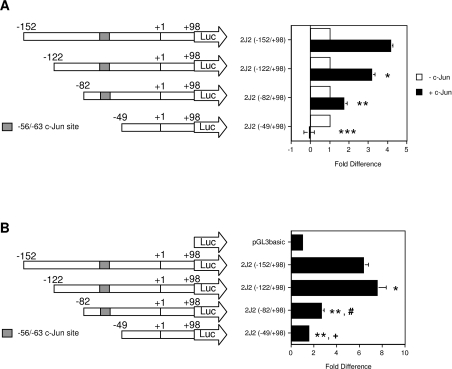
Deletion of the −152 to −50 bp region from the CYP2J2 promoter [2J2(−49/+98); Figure 1A] eliminated c-Jun-dependent transactivation in HepG2 cells, indicating that activation of CYP2J2 by c-Jun is mediated by elements located within this region. Consistent with the proposed role of c-Jun in maintaining expression of CYP2J2 in resting cells, deletion of the region between −152 and −50 bp also significantly reduced basal promoter activity in HepG2 cells. [2J2(−49/+98) displayed a basal activity only 1.6±0.1-fold higher than that of the promoterless pGL3basic backbone, compared with 6.3±0.5-fold for 2J2(−152/+98); Figure 1B.] A previous report identified a c-Jun binding element in the 5′-flank of the CYP2J2 gene at −56/−63 bp, relative to the start codon in exon 1, that contributed to transactivation by c-Jun [21]. To identify additional c-Jun-responsive element(s) within the −152/−50 region, the reporter constructs 2J2(−122/+98) and 2J2(−82/+98) were prepared from 2J2(−152/+98). As shown in Figure 1(A), deletion of the region between −152 and −122 bp decreased c-Jun-dependent transactivation to 3.2±0.2-fold of control compared with 4.2±0.1-fold of control for 2J2(−152/+98), but did not diminish basal promoter activity (Figure 1B). In fact, 2J2(−122/+98) exhibited higher basal activity than 2J2(−152/+98) (Figure 1B). In contrast, the deletion construct 2J2(−82/+98) displayed a pronounced loss of both c-Jun-dependent transactivation (to 1.8±0.2-fold of control; Figure 1A), and basal transcriptional activity [2.7±0.2-fold that of the pGL3basic vector backbone, compared with 6.3±0.5-fold for 2J2(−152/+98); Figure 1B]. These results indicate that, in addition to the −56/−63 site, the CYP2J2 proximal promoter contains an important c-Jun-responsive element between −122 and −82 bp. The possibility that the −152 to −122 bp region contained a c-Jun-responsive element was also evaluated further.
Figure 1. A c-Jun-responsive region in the CYP2J2 proximal promoter.
The CYP2J2 promoter deletion constructs 2J2(−122/+98), 2J2(−82/+98) and 2J2(−49/+98) were prepared by 5′-truncation of 2J2(−152/+98) as described in the Experimental section. (A) HepG2 cells were co-transfected with CYP2J2 constructs (1 μg per well) and a c-Jun expression plasmid (0.5 μg per well) as indicated. A pCMV-β-galactosidase expression plasmid (0.5 μg per well) was included in each well to control for transfection efficiency. Luciferase activity was normalized to β-galactosidase activity. Normalized activity of constructs in the absence of c-Jun was set to 1-fold, and the activities of deletion constructs in the presence of c-Jun were expressed relative to activity in the absence of c-Jun. The results are expressed as the means±S.E.M. for at least three independent experiments. Significantly different, in the presence of c-Jun, from: *2J2(−152/+98) (P=0.0001), **2J2(−122/+98) (P<0.0001), and ***2J2(−82/+98) (P<0.0001). (B) Basal activities of CYP2J2 promoter deletion constructs. Promoter constructs or the empty pGL3basic vector were co-transfected (1 μg per well) into HepG2 cells with a β-galactosidase expression plasmid (0.5 μg per well). Luciferase activity was normalized to β-galactosidase activity and presented relative to the normalized activity of the empty pGL3basic vector (set to 1-fold). The results are expressed as the means±S.E.M. for at least three independent experiments. Significantly different from: *2J2(−152/+98) (P<0.05), **2J2(−152/+98) (P<0.0001), #2J2(−122/+98) (P<0.0001), and +2J2(−82/+98) (P<0.02).
Identification of c-Jun binding sequences within the −152 to −50 bp region of CYP2J2
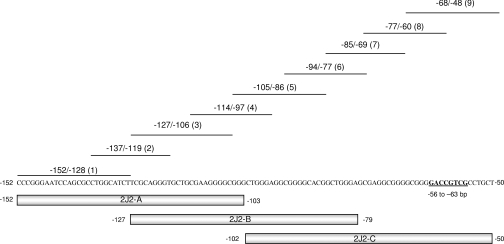
The Genomatix MatInspector Professional consensus sequence identification program [25] was used to identify potential AP-1 binding sites within the −152 to −50 region of CYP2J2. Although transfection studies indicated the presence of c-Jun-response elements within −152/−50 (Figure 1), only the −56/−63 AP-1-like element was identified by high- and low-stringency searching. Thus double-stranded oligonucleotides spanning three large overlapping regions of the −152 to −50 bp sequence (2J2-A, spanning −152 to −103 bp; 2J2-B, spanning −127 to −79 bp; and 2J2-C, spanning −102 to −50 bp; Figure 2; Table 2) were labelled with [32P]dCTP and used as probes in EMSA analysis with recombinant c-Jun protein. Competition experiments were performed with smaller oligonucleotides (Figure 2, probes 1–9; Table 2) to identify sequences important for c-Jun binding.
Figure 2. Probes and competitor sequences used in EMSA analysis to identify c-Jun binding regions in the −152 to −50 bp region of the CYP2J2 proximal promoter.
The probes 2J2-A (−152/−103), 2J2-B (−127/−79) and 2J2-C (−102/−50) which span the c-Jun responsive module (−152/−50) were end-labelled with [32P]dCTP. The sequences −152/−128 (probe 1), −137/−119 (probe 2), −127/−106 (probe 3), −114/−97 (probe 4), −105/−86 (probe 5), −94/−77 (probe 6), −85/−69 (probe 7), −77/−60 (probe 8) and −68/−48 (probe 9) were used as unlabelled competitors. The previously identified c-Jun binding site at −56 to −63 bp is highlighted [21]. Sequences of all probes are presented in Table 2.
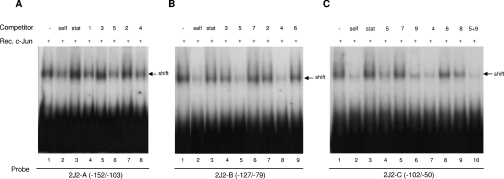
2J2-A (−152/−103 bp) elicited a significant binding reaction with recombinant c-Jun protein (Figure 3A, lane 1), which was diminished by excess unlabelled self probe but not by the unrelated Stat5 probe (lanes 2 and 3). Two hundred-fold excesses of probes 4 (−114/−97; lane 8), 5 (−105/−86; lane 6) and, to a lesser extent, 1 (−152/−128; lane 4), competed with c-Jun to bind 2J2-A, but probes 3 (−127/−106; lane 5) and 2 (−137/−119; lane 7) were ineffective.
Figure 3. Binding of the −152 to −50 bp CYP2J2 promoter region to c-Jun.
The double-stranded probes (A) 2J2-A (−152 to −103 bp of the CYP2J2 promoter), (B) 2J2-B (−127 to −79 bp of the CYP2J2 promoter) and (C), 2J2-C (−102 to −50 bp of the CYP2J2 promoter) were end-labelled with [32P]dCTP and incubated with recombinant c-Jun. Binding reactions that contained excess unlabelled self probe [2J2-A in (A), 2J2-B in (B) or 2J2-C in (C)], the non-specific Stat probe (Stat5 element from the β-casein promoter) and the smaller overlapping probes 1–9 (refer to Figure 2) are indicated. Retarded complexes are indicated by arrows.
The probe 2J2-B, which spans −127 to −79 bp of the CYP2J2 promoter, also bound strongly to recombinant c-Jun protein (Figure 3B, lane 1). Binding to 2J2-B (−127/−79) was competed by excess unlabelled self, but not the Stat5 probe (lanes 2 and 3). Binding of 2J2-B to recombinant c-Jun was effectively inhibited by 200-fold excess of the unlabelled smaller probes 5 (−105/−86; lane 5) and 4 (−114/−97; lane 8), but less so by probes 3 (−127/−106; lane 4), 7 (−85/−69; lane 6), 2 (−137/−119; lane 7) and 6 (−94/−77; lane 9).
A similar pattern of binding was seen with the probe 2J2-C (−102/−50 bp) and recombinant c-Jun protein (Figure 3C, lane 1). Binding of c-Jun was inhibited by 200-fold excess 2J2-C (−102/−50; self), but not by the Stat5 probe (lanes 2 and 3). The binding of recombinant c-Jun to probe 2J2-C (−102/−50) was decreased by 200-fold excess of the unlabelled probes 5 (−105/−86; lane 4), 9 (−68/−48; lane 6), 4 (−114/−97; lane 7) and to a lesser extent 8 (−77/−60; lane 9), but not by probes 7 (−85/−69; lane 5) or 6 (−94/−77; lane 8). The combination of the two most effective competitors (probes 5 and 9) inhibited complex formation efficiently (Figure 3C, lane 10). Taken together, these results implicate c-Jun binding regions at −114 to −86 bp, and possibly also between −152 and −128 bp. Binding in the −77 to −48 bp region was attributed to the previously characterized c-Jun/AP-1-like site at −56/−63 [21].
The −105/−95 sequence in the CYP2J2 5′-flank binds c-Jun and contributes to c-Jun-dependent activation
The nature of c-Jun binding to the −122/−82 bp region of CYP2J2 was explored further by EMSA analysis. Because probe 5 (−105/−86; Figure 4A) emerged as an extremely efficient competitor of c-Jun binding to 2J2-B (Figure 3), it was used directly in EMSA experiments. A pronounced shift was noted with both HepG2 nuclear protein and c-Jun recombinant protein (lanes 1 in Figures 4B and 4C respectively). The specificity of the shifted complex was confirmed in competition experiments with unlabelled probe 5 (self); the Stat5 probe did not compete (lanes 2 and 3, Figures 4B and 4C). Binding was inhibited by probe 4 but not by probe 6 (Figures 4B and 4C, lanes 4 and 5 respectively). Thus within the −105/−86 region, the sequence at −105/−95 is critical for c-Jun binding.
Figure 4. EMSA studies of c-Jun binding to the −114 to −86 bp region of the CYP2J2 promoter.
(A) Sequences of EMSA probes 4–6 and the c-Jun binding sequence at −114 to −86 bp in the CYP2J2 promoter. Nucleotides that are required for c-Jun binding are in bold and highlighted by asterisks. [32P]dCTP-labelled probe 5 was incubated with (B) nuclear protein fractions from untransfected HepG2 cells (UT) and (C) recombinant c-Jun protein (Jun) as described in the Experimental section. Some reactions included excess unlabelled probe 5 (self), Stat5 probe or the overlapping probes 4 (−114/−97 bp) or 6 (−94/−77) as indicated. Retarded complexes are indicated by arrows.
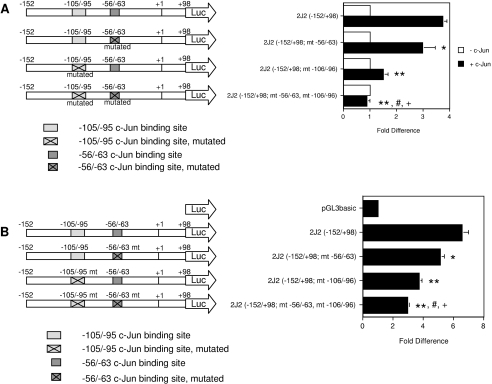
The functional role of the −105/−95 sequence in c-Jun-dependent transactivation was tested in transfection studies. The wild-type promoter construct 2J2(−152/+98) was strongly activated by c-Jun [to 3.7±0.2-fold of control (P<0.0001); Figure 5A]. Mutagenesis of the previously identified c-Jun binding site at −56/−63 bp and the −105/−95 sequence decreased c-Jun-responsiveness to 3.0±0.5-fold and 1.5±0.2-fold of respective control. Thus both elements mediate transactivation of CYP2J2 by c-Jun. Co-transfection studies with c-jun and c-fos were performed to evaluate the role of c-Fos at the elements. Consistent with results obtained previously with a construct containing 2341 bp of the CYP2J2 5′-upstream region [21], c-Jun-dependent activation of the proximal promoter construct 2J2(−152/+98) was abolished by co-transfection with c-fos [the activity of 2J2(−152/+98) was decreased from 3.7±0.2-fold of control with c-Jun to 0.5±0.05-fold of control with c-Fos/c-Jun; results not shown]. The residual c-Jun-dependent activity observed with the two mutated constructs 2J2(−152/+98; mt −56/−63) and 2J2(−152/+98; mt −106/−96) was also suppressed by co-transfection with c-fos. Thus the activity of 2J2(−152/+98; mt −56/−63) was decreased from 3.0±0.5-fold of control with c-Jun to −0.1±0.03-fold with c-Fos/c-Jun. Similarly, c-Fos decreased c-Jun-dependent activation of 2J2(−152/+98; mt −106/−96) from 1.5±0.2-fold of control to −1.4±0.2-fold of control (results not shown). These findings indicated that mutagenesis of either c-Jun-response element did not completely eliminate c-Jun-dependent activation, or c-Fos-mediated suppression, of the CYP2J2 promoter. On the other hand, mutagenesis of both elements rendered the promoter unresponsive to AP-1 [construct 2J2(−152/+98; mt −56/−63, mt −106/−96); Figure 5A]. Thus both the −56/−63 and −105/−95 elements contribute to c-Jun-dependent activation and c-Fos-dependent inhibition of the CYP2J2 promoter.
Figure 5. The −56/−63 bp and −105/−95 bp sequences mediate c-Jun-dependent activation of the CYP2J2 proximal promoter.
(A) Mutation of the −56/−63 bp and −105/−95 bp elements reduces c-Jun-dependent activation of the CYP2J2 promoter. The constructs 2J2(−152/+98; mt −56/−63) and 2J2(−152/+98; mt −106/−96) are mutated respectively at the previously identified c-Jun-binding site at −56 to −63 bp, and the newly identified c-Jun-binding sequence spanning −105 to −95 bp. In the construct 2J2(−152/+98; mt −56/−63, mt −106/−96), both the −56 to −63 bp and −105 to −95 bp sequences are mutated. CYP2J2 reporter constructs (1 μg per well) were co-transfected into HepG2 cells with a β-galactosidase expression plasmid (0.5 μg per well). A c-Jun expression plasmid was co-transfected at 0.5 μg per well as indicated. Luciferase activity was normalized as described in Figure 1(A). The results are expressed as the means±S.E.M. for at least three independent experiments. Significantly different, in the presence of c-Jun, from: *wild-type construct 2J2(−152/+98) (P<0.02), **wild-type construct 2J2(−152/+98) (P<0.0001), #mutated construct 2J2(−152/+98; mt −56/−63) (P<0.0001), and +mutated construct 2J2(−152/+98; mt −106/−96) (P<0.005). (B) Mutagenesis of the −56/−63 bp and −105/−95 bp elements reduces basal transcriptional activity of the CYP2J2 promoter. CYP2J2 reporter constructs or the empty pGL3basic vector were co-transfected (1 μg per well) into HepG2 cells with a β-galactosidase expression plasmid (0.5 μg per well). Luciferase activity was normalized to β-galactosidase activity and presented relative to the normalized activity of the empty pGL3basic vector (set to 1-fold). The results are expressed as the means±S.E.M. for at least three independent experiments. Significantly different from: * 2J2(−152/+98) (P<0.002), **2J2(−152/+98) (P<0.0005), #2J2(−152/+98; mt −56/−63) (P<0.005), and +2J2(−152/+98; mt −106/−96) (P<0.01).
Consistent with a role for c-Jun in maintenance of the basal expression of CYP2J2 in cells, separate mutagenesis of either the −56/−63 or −105/−95 elements significantly decreased basal activity of the CYP2J2 promoter in HepG2 cells. Mutagenesis of both elements produced the most pronounced effect, reducing basal promoter activity to only 2.9±0.1-fold of pGL3basic, compared with 6.6±0.4-fold for the wild type construct 2J2(−152/+98) (Figure 5B).
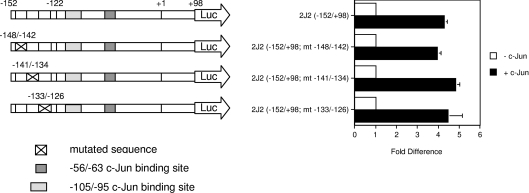
Finally, a series of additional reporter constructs, containing consecutive 8 bp mutations in the region −148 to −126, were prepared: 2J2(−152/+98; mt −148/−142), 2J2(−152/+98; mt −141/−134) and 2J2(−152/+98; mt −133/−126), to test whether the −152/−122 sequence also contained a c-Jun-responsive element. However, all of these constructs were similarly c-Jun-responsive in transfected cells (Figure 6), and did not exhibit impaired basal transcriptional activity (6.5–8.4-fold of pGL3basic; results not shown). These findings are consistent with the absence of a significant role for the −152/−122 bp region in basal and c-Jun-mediated regulation of the gene.
Figure 6. Mutation of the −152/−122 bp region does not influence c-Jun-dependent activation of the CYP2J2 promoter.
The constructs 2J2(−152/+98; mt −148/−142), 2J2(−152/+98; mt −141/−134) and 2J2(−152/+98; mt −133/−126) were mutagenized in the sequences spanning −148 to −142 bp, −141 to −134 bp, and −133 to −126 bp, respectively. 2J2 reporter constructs were co-transfected (1 μg per well) into HepG2 cells with a β-galactosidase expression plasmid (0.5 μg per well). A c-Jun expression plasmid was co-transfected at 0.5 μg per well as indicated. Luciferase activity was normalized as described in Figure 1(A). The results are expressed as the means±S.E.M. for at least three independent experiments.
Binding of c-Jun to the −105 to −95 bp element
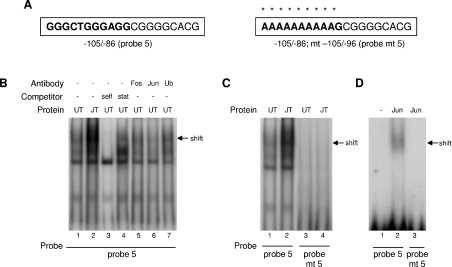
The binding of c-Jun to the −105/−95 sequence was investigated by EMSA analysis. Probe 5 (−105/−86; Figure 7A) generated a retarded complex with nuclear protein fractions from untransfected HepG2 cells (UT), which was more intense in fractions from c-Jun-transfected cells (JT; Figure 7B, compare lanes 1 and 2). The shift was competed by 200-fold excess of unlabelled probe 5 (self), but not by the Stat5 probe (Figure 7B, lanes 3 and 4). The complex was block-shifted by an anti-c-Jun antibody (Figure 7B lane 6), but not by an anti-c-Fos antibody (lane 5), or an antibody directed against ubiquitin (lane 7), thus confirming the specificity of c-Jun binding. Mutagenesis of the −105/−96 sequence (probe mt 5; Figure 7A) decreased the interactions with nuclear extracts from untransfected and c-Jun-transfected HepG2 cells, compared with those with the wild type probe (Figure 7C). These findings were supported with the use of recombinant human c-Jun protein, which interacted with probe 5 (Figure 7D, lane 2), but not with the mutant sequence (lane 3).
Figure 7. EMSA of the binding of c-Jun to the −105/−95 bp sequence in the CYP2J2 promoter.
(A) Sequences of the EMSA probes; probe 5 spans the −105/−95 bp element (bold) and probe mt 5 is mutated as indicated by asterisks. Probes were end-labelled and incubated with nuclear protein fractions from untransfected (UT) and c-Jun-transfected (JT) HepG2 cells (B, C) and with recombinant c-Jun protein (D). Some reactions included excess unlabelled probe 5 (self) or Stat5 probe, whereas others included antibodies directed against c-Jun, c-Fos or ubiquitin (Ub). Retarded complexes are indicated by arrows.
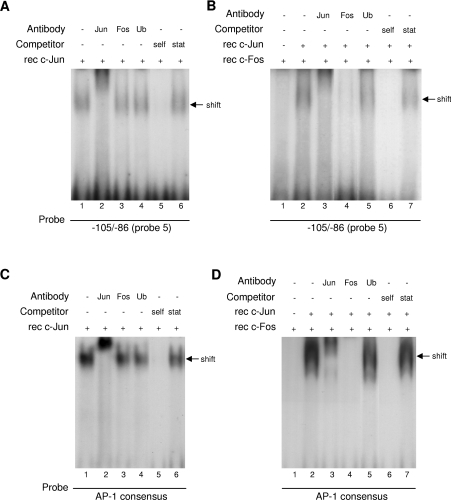
The capacity of the −105/−95 bp element to bind directly to c-Jun and c-Fos was tested by EMSA (Figure 8). A shifted complex was observed with recombinant c-Jun (Figure 8A, lane 1) that was competed by 200-fold excess of unlabelled self, but not the Stat5 probe (lane 5 and 6 respectively). As expected, the complex was supershifted by anti-c-Jun, but not anti-c-Fos or anti-ubiquitin, antibodies (lanes 2–4). A shift was also observed with the c-Fos/c-Jun protein combination (Figure 8B, lane 2) that was disrupted by antibodies directed against c-Jun and c-Fos, but not by anti-ubiquitin (Figure 8B, lanes 3–5); no shift was apparent with c-Fos protein alone (lane 1). Thus the −105/−95 sequence is able to bind c-Fos/c-Jun heterodimers and c-Jun homodimers. Specificity was confirmed by the competition with an excess of the self oligonucleotide, but not the Stat5 probe (Figure 8B, lanes 6 and 7). Also shown in Figure 8 (C and D) are control experiments conducted in parallel with an AP-1 consensus probe that interacted with both c-Jun homodimers (Figure 8C) and c-Fos/c-Jun heterodimers (Figure 8D). The specificity of these reactions with AP-1 was confirmed in supershift (Figure 8C, lanes 2–4 and Figure 8D, lanes 3–5) and unlabelled competition (Figure 8C, lanes 5 and 6 and Figure 8D, lanes 6 and 7) experiments; c-Fos alone did not bind to the consensus probe (Figure 8D, lane 1).
Figure 8. EMSAs of the binding of c-Jun and c-Fos/c-Jun to the −105/−95 bp element from the CYP2J2 promoter, and an AP-1 consensus probe.
Double-stranded probes containing the −105/−95 bp element (probe 5; A and B) and the AP-1 consensus sequence (C and D) were end-labelled with [32P]dCTP and incubated with recombinant (rec) c-Jun protein alone (A and C), or a combination of recombinant c-Jun and recombinant c-Fos protein (B and D). Some reactions included excess unlabelled probe 5 (self, A and B) or AP-1 consensus probe (self, C and D), or Stat5 probe; other reactions included antibodies directed against c-Jun, c-Fos or ubiquitin (Ub). Retarded complexes are indicated by arrows.
DISCUSSION
Because the number of reports describing the cytoprotective and anti-apoptotic properties of EETs is increasing, an understanding of the mechanisms that underlie CYP2J2 regulation is extremely important. Previous findings that CYP2J2 protein is decreased in endothelial cells cultured at low oxygen tension offers insight into mechanisms by which the gene is dysregulated [8]. The recent demonstration in hypoxic HepG2 cells that up-regulation of c-fos was associated with the down-regulation of CYP2J2 is also significant [21]. Thus, in transient transfection studies, the CYP2J2 gene was activated by c-Jun homodimers, but not by c-Fos/c-Jun heterodimers. Deletion of the −122/−50 bp region of the 5′-flank of the gene, relative to the start codon, abolished transactivation by c-Jun. These effects were partly dependent on an AP-1-like sequence at −56/−63 bp [21]. The present study has characterized an atypical c-Jun binding element at −105/−95 bp in the CYP2J2 5′-flank that has a major role in CYP2J2 activation by c-Jun.
Although sequence analysis of the −122/−50 bp region did not detect typical AP-1 binding elements, even under low-stringency searching, many AP-1-responsive genes do not contain consensus response elements in their promoter regions [26–28]. Despite the absence of a typical AP-1 element, the combination of functional analysis and EMSA analysis established the capacity of the −105/−95 bp region to interact directly with c-Jun. Mutation of this sequence impaired c-Jun-dependent transactivation and basal transcriptional activity of a CYP2J2 reporter construct and binding to c-Jun. The −105/−95 region of CYP2J2 contains the TGGGAGG sequence so that there is a match of only three nucleotides with the AP-1 consensus TGA(G/C)TCA motif [13]. Thus it is possible that c-Jun acts in concert with other transcription factors that bind in close proximity. Indeed, c-Jun interacts with several transcription factors including GATA-2 [29], GATA-4 [30], PU.1 [31], Ets family members [32], NF-κB (nuclear factor κB) [33], NFAT (nuclear factor of activated T-cells) [34], Smad family proteins [35] and Sp1 (specificity protein 1) [36] to activate gene transcription.
Of particular interest in the context of CYP2J2 gene regulation are the potential Sp1 binding motifs that overlap (−110/−100 bp and −98/−88 bp), or flank (−76/−66 bp and −71/−61 bp), the −105/−95 c-Jun-binding site. Indeed, a recent study implicated the −76/−66 sequence in Sp1-dependent transactivation of CYP2J2 [37]. This report, in conjunction with the present study, suggests that Sp1 and c-Jun may have the capacity to bind at adjacent elements. Indeed, activation of the monoamine oxidase B gene promoter by c-Jun and Sp1, and its inhibition by c-Fos, has been demonstrated [38].
EMSA analysis demonstrated that c-Jun, but not c-Fos, was present in the protein complex bound to the −105/−95 sequence in untreated HepG2 cells. This finding is consistent with the basal expression of c-jun, but not c-fos, in unstimulated HepG2 cells [21,39], and with the proposed role of c-Jun in maintenance of CYP2J2 expression [21]. Because c-Fos abolished the activation of the CYP2J2 promoter by c-Jun [21], the capacity of the −105/−95 element to interact with c-Fos and mediate suppression by c-Fos was examined in the present study. Transfection studies established that c-Fos exerts its suppressive effects at both the −56/−63 and −105/−95 c-Jun-response elements, as mutagenesis of either site alone did not eliminate c-Fos-mediated inhibition. However, mutagenesis of both elements rendered the CYP2J2 promoter unresponsive to AP-1. In EMSAs with the −105/−95 element, binding to both c-Jun homodimers and c-Fos/c-Jun heterodimers was observed, whereas the −56/−63 element was found previously not to interact with c-Fos/c-Jun heterodimers [21]. Taken together these findings suggest that the inhibitory effect of c-Fos on CYP2J2 expression involves divergent actions at the two elements within the c-Jun-responsive module.
Regulatory regions in target genes may consist of complex elements that encompass binding motifs for many transcription factors [12]. Complexes comprising multiple AP-1 proteins have been shown to induce different DNA bending patterns near AP-1 sites, which may alter interactions with transcription factors [12]. Binding of c-Jun to the −105/−95 bp sequence under basal conditions may facilitate interplay with other transcription factors. On the other hand, binding of c-Fos/c-Jun heterodimers at the −105/−95 element, under conditions in which c-Fos is highly expressed, may promote a conformation of the CYP2J2 promoter that is suboptimal for transactivation. An analogous situation has been reported in which transactivation of the osteocalcin promoter by the vitamin D receptor is impaired by the binding of c-Fos/c-Jun heterodimers at an AP-1 element adjacent to the vitamin D receptor response element [40]. Complex elements of the type identified in the present study enable fine control of gene transcription and responsiveness to multiple stimuli.
Findings to date suggest that c-Jun transactivates CYP2J2 and maintains expression in normoxic cells. Down-regulation of CYP2J2 can occur in response to c-Fos up-regulation and the formation of transcriptionally inactive c-Fos/c-Jun heterodimers [21]. This behaviour resembles the regulation of the human atrial natriuretic peptide gene promoter which is strongly induced by c-Jun, but diminished by over-expression of c-Fos and c-Jun. This occurred in atrial and ventricular cardiomyocytes, but not in rat pituitary tumour cells (GC cells) or cardiac mesenchymal cells [41]. Similarly, De Cesare et al. [39] reported that c-Fos inhibits c-Jun-dependent activation of the human urokinase gene promoter. Positive regulation was mediated by c-Jun/ATF-2 heterodimers at an AP-1-like element in the upstream enhancer region of the gene, whereas c-Jun/c-Fos heterodimers bound weakly to the element. It was proposed that inhibition by c-Fos was due to sequestration of c-Jun, which limited its availability for dimerization with ATF-2 [39]. Thus it is feasible that the response of AP-1-regulated promoters may relate to the concentration and subunit composition of the prevailing AP-1 complexes in cells. The present study adds to these reports and supports the notion that differential dimerization of AP-1 proteins gives rise to different effects on transcriptional activity of AP-1-responsive genes [11,12,42].
In summary, the present study describes an atypical AP-1 sequence at −105/−95 that has the major role in CYP2J2 transactivation by c-Jun. It now emerges that the −105/−95 sequence and the previously described −56/−63 element are critical features of a c-Jun-responsive module within the CYP2J2 proximal promoter that regulates expression of the gene.
Acknowledgments
This work was supported by a grant from the Australian National Health and Medical Research Council and equipment grants from The Wellcome Trust, U.K., and the Clive and Vera Ramaciotti Foundations. We thank Dr K. Imakawa for generously providing the c-Jun and c-Fos expression plasmids.
References
- 1.Roman R. J. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol. Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 2.Wu S., Moomaw C. R., Tomer K. B., Falck J. R., Zeldin D. C. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J. Biol. Chem. 1996;271:3460–3468. doi: 10.1074/jbc.271.7.3460. [DOI] [PubMed] [Google Scholar]
- 3.Node K., Huo Y., Ruan X., Yang B., Spiecker M., Ley K., Zeldin D. C., Liao J. K. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science. 1999;285:1276–1279. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeldin D. C., Foley J., Ma J., Boyle J. E., Pascual J. M. S., Moomaw C. R., Tomer K. B., Steenbergen C., Wu S. CYP2J subfamily P450s in the lung: expression, localization, and potential functional significance. Mol. Pharmacol. 1996;50:1111–1117. [PubMed] [Google Scholar]
- 5.Zeldin D. C., Foley J., Boyle J. E., Moomaw C. R., Tomer K. B., Parker C., Steenbergen C., Wu S. Predominant expression of an arachidonate epoxygenase in islets of Langerhans cells in human and rat pancreas. Endocrinology. 1997;138:1338–1346. doi: 10.1210/endo.138.3.4970. [DOI] [PubMed] [Google Scholar]
- 6.Zeldin D. C., Foley J., Goldsworthy S. M., Cook M. E., Boyle J. E., Ma J., Moomaw C. R., Tomer K. B., Steenbergen C., Wu S. CYP2J subfamily cytochrome P450s in the gastrointestinal tract: expression, localization, and potential functional significance. Mol. Pharmacol. 1997;51:931–943. doi: 10.1124/mol.51.6.931. [DOI] [PubMed] [Google Scholar]
- 7.Campbell W. B., Harder D. R. Endothelium-derived hyperpolarizing factors and vascular cytochrome P450 metabolites of arachidonic acid in the regulation of tone. Circ. Res. 1999;84:484–488. doi: 10.1161/01.res.84.4.484. [DOI] [PubMed] [Google Scholar]
- 8.Yang B., Graham L., Dikalov S., Mason R. P., Falck J. R., Liao J. K., Zeldin D. C. Overexpression of cytochrome P450 CYP2J2 protects against hypoxia-reoxygenation injury in cultured bovine aortic endothelial cells. Mol. Pharmacol. 2001;60:310–320. doi: 10.1124/mol.60.2.310. [DOI] [PubMed] [Google Scholar]
- 9.Chen J.-K., Capdevila J., Harris R. C. Cytochrome P450 expoxygenase metabolism of arachidonic acid inhibits apoptosis. Mol. Cell. Biol. 2001;21:6322–6331. doi: 10.1128/MCB.21.18.6322-6331.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angel P., Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 11.Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. U.S.A. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerppola T. K., Curran T. Selective DNA bending by a variety of bZIP proteins. Mol. Cell. Biol. 1993;13:5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell (Cambridge, Mass.) 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 14.Lafyatis R., Kim S.-J., Angel P., Roberts A. B., Sporn M. B., Karin M., Wilder R. L. Interleukin-1 stimulates and all-trans-retinoic acid inhibits collagenase gene expression through its 5′ Activator Protein-1-binding site. Mol. Endocrinol. 1990;4:973–980. doi: 10.1210/mend-4-7-973. [DOI] [PubMed] [Google Scholar]
- 15.Wisdom R., Johnson R. S., Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay R. S., Phelan M., Faller D. V. Hypoxia induces AP-1-regulated genes and AP-1 transcription factor binding in human endothelial and other cell types. Biochim. Biophys. Acta. 1995;1264:72–78. doi: 10.1016/0167-4781(95)00116-x. [DOI] [PubMed] [Google Scholar]
- 17.Rupec R. A., Baeuerle P. A. The genomic response of tumor cells to hypoxia and reoxygenation. Differential activation of transcription factors AP-1 and NF-κB. Eur. J. Biochem. 1995;234:632–640. doi: 10.1111/j.1432-1033.1995.632_b.x. [DOI] [PubMed] [Google Scholar]
- 18.Müller J. M., Krauss B., Kaltschmidt C., Baeuerle P. A., Rupec R. A. Hypoxia induces c-fos transcription via a mitogen-activated protein kinase-dependent pathway. J. Biol. Chem. 1997;272:23435–23439. doi: 10.1074/jbc.272.37.23435. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y. J., Corry P. M. Hypoxia-induced bFGF gene expression is mediated through the JNK signal transduction pathway. Mol. Cell. Biochem. 1999;202:1–8. doi: 10.1023/a:1007059806016. [DOI] [PubMed] [Google Scholar]
- 20.Mathas S., Hinz M., Anagnostopoulos I., Krappmann D., Lietz A., Jundt F., Bommert K., Mechta-Grigoriou F., Stein H., Dörken B., Scheidereit C. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-κB. EMBO J. 2002;21:4101–4113. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marden N. Y., Fiala-Beer E., Xiang S.-H., Murray M. Role of activator protein-1 in the down-regulation of the human CYP2J2 gene in hypoxia. Biochem. J. 2003;373:669–680. doi: 10.1042/BJ20021903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber E., Matthias P., Müller M. M., Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–270. [PubMed] [Google Scholar]
- 24.Gebert C. A., Park S. H., Waxman D. J. Regulation of STAT5b activation by the temporal pattern of growth hormone stimulation. Mol. Endocrinol. 1997;11:400–414. doi: 10.1210/mend.11.4.9904. [DOI] [PubMed] [Google Scholar]
- 25.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryseck R.-P., Bravo R. c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene. 1991;6:533–542. [PubMed] [Google Scholar]
- 27.Pestell R. G., Hollenberg A. N., Albanese C., Jameson J. L. c-Jun represses transcription of the human chorionic gonadotropin α and β genes through distinct types of CREs. J. Biol. Chem. 1994;269:31090–31096. [PubMed] [Google Scholar]
- 28.Chinenov Y., Kerppola T. K. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 29.Kawana M., Lee M.-E., Quertermous E. E., Quertermous T. Cooperative interaction of GATA-2 and AP-1 regulates transcription of the endothelin-1 gene. Mol. Cell. Biol. 1995;15:4225–4231. doi: 10.1128/mcb.15.8.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzig T. C., Jobe S. M., Aoki H., Molkentin J. D., Cowley A. W., Jr, Izumo S., Markham B. E. Angiotensin II type1a receptor gene expression in the heart: AP-1 and GATA-4 participate in the response to pressure overload. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7543–7548. doi: 10.1073/pnas.94.14.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behre G., Whitmarsh A. J., Coghlan M. P., Hoang T., Carpenter C. L., Zhang D.-E., Davis R. J., Tenen D. G. c-Jun is a JNK-independent coactivator of the PU.1 transcription factor. J. Biol. Chem. 1999;274:4939–4946. doi: 10.1074/jbc.274.8.4939. [DOI] [PubMed] [Google Scholar]
- 32.Bassuk A. G., Leiden J. M. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 33.Yang X., Chen Y., Gabuzda D. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-κB. J. Biol. Chem. 1999;274:27981–27988. doi: 10.1074/jbc.274.39.27981. [DOI] [PubMed] [Google Scholar]
- 34.McCaffrey P. G., Luo C., Kerppola T. K., Jain J., Badalian T. M., Ho A. M., Burgeon E., Lane W. S., Lambert J. N., Curran T., et al. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 35.Liberati N. T., Datto M. B., Frederick J. P., Shen X., Wong C., Rougier-Chapman E. M., Wang X.-F. Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4844–4849. [Google Scholar]
- 36.Kardassis D., Papakosta P., Pardali K., Moustakas A. c-Jun transactivates the promoter of the human p21WAF1/Cip1 gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- 37.Spiecker M., Darius H., Hankeln T., Soufi M., Sattler A. M., Schaefer J. R., Node K., Börgel J., Mügge A., Lindpaintner K., et al. Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation. 2004;110:2132–2136. doi: 10.1161/01.CIR.0000143832.91812.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong W. K., Ou X.-M., Chen K., Shih J. C. Activation of human monoamine oxidase B gene expression by a protein kinase C MAPK signal transduction pathway involves c-Jun and Egr-1. J. Biol. Chem. 2002;277:22222–22230. doi: 10.1074/jbc.M202844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Cesare D., Vallone D., Caracciolo A., Sassone-Corsi P., Nerlov C., Verde P. Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 40.Schüle R., Umesono K., Mangelsdorf D. J., Bolado J., Pike J. W., Evans R. M. Jun-Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell. 1990;61:497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- 41.Kovacic-Milivojevic B., Gardner D. G. Divergent regulation of the human atrial natriuretic peptide gene by c-jun and c-fos. Mol. Cell. Biol. 1992;12:292–301. doi: 10.1128/mcb.12.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki T., Okuno H., Yoshida T., Endo T., Nishina H., Iba H. Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucleic Acids Res. 1991;19:5537–5542. doi: 10.1093/nar/19.20.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]