Abstract
For animals, dietary protein is critical for the nutrition of the organism and, at the cellular level, protein nutrition translates into amino acid availability. Amino acid deprivation triggers the AAR (amino acid response) pathway, which causes enhanced transcription from specific target genes. The present results show that C/EBPβ (CCAAT/enhancer-binding protein β) mRNA and protein content were increased following the deprivation of HepG2 human hepatoma cells of a single amino acid. Although there was a modest increase in mRNA half-life following histidine limitation, the primary mechanism for the elevated steady-state mRNA was increased transcription. Transient transfection documented that C/EBPβ genomic fragments containing the 8451 bp 5′ upstream of the transcription start site did not contain amino-acid-responsive elements. However, deletion analysis of the genomic region located 3′ downstream of the protein coding sequence revealed that a 93 bp fragment contained an amino-acid-responsive activity that functioned as an enhancer. Exogenous expression of ATF4 (activating transcription factor 4), known to activate other genes through amino acid response elements, caused increased transcription from reporter constructs containing the C/EBPβ enhancer in cells maintained in complete amino acid medium. Chromatin immunoprecipitation demonstrated that RNA polymerase II is bound at the C/EBPβ promoter and at the 93 bp regulatory region in vivo, whereas ATF4 binds to the enhancer region only. Immediately following amino acid removal, the kinetics of binding for ATF4, ATF3, and C/EBPβ itself to the 93 bp regulatory region were similar to those observed for the amino-acid-responsive asparagine synthetase gene. Collectively the findings show that expression of C/EBPβ, which contributes to the regulation of amino-acid-responsive genes, is itself controlled by amino acid availability through transcription.
Keywords: activating transcription factor 3 (ATF3), activating transcription factor 4 (ATF4), amino acid response element (AARE), basic leucine-zipper (bZIP), CCAAT/enhancer-binding protein (C/EBP), nutrient starvation
Abbreviations: AAR, amino acid response; AARE, amino acid response element; ActD, actinomycin D; ATF, activating transcription factor; ASNS, asparagine synthetase gene; BAC, bacterial artificial chromosome; bZIP, basic leucine-zipper; C/EBP, CCAAT/enhancer-binding protein; ChIP, chromatin immunoprecipitation; CHOP, C/EBP homology protein; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LAP, liver-activating protein; LIP, liver-inactivating protein; MEM, minimal essential medium; NSRE, nutrient sensing response element; qPCR, quantitative real-time PCR; Pol II, RNA polymerase II; SV40, simian virus 40; UPRE, unfolded protein response element
INTRODUCTION
Dietary protein intake and a balanced amino acid supply is an important factor in the general nutrition of animals, especially during development [1,2] and may have an impact on lifespan as well [3]. At the level of individual tissues and cells, protein nutrition is represented by amino acid availability. Although the amino acid content in the bloodstream and protein degradation both provide some buffering capacity for variations in dietary protein/amino acid intake, fluctuations in the intracellular levels of individual amino acids may occur in response to diet, disease and metabolic need [4]. Indeed, animals possess an innate mechanism to detect and reject a diet with an imbalanced amino acid composition [5]. Obviously, the metabolic stance of the animal must be altered to adapt to these changes, both at the level of inter-organ metabolite fluxes and at the cellular level to change the flux through individual metabolic pathways. In this context, amino acids serve as signal-transduction messengers to transmit the nutritional status of the organism to individual cells. The signalling pathway that is triggered in response to amino-acid-deprivation is referred to as the ‘amino acid response’ (AAR).
One of the mechanisms by which cells respond to amino acid stress is by increasing transcription from specific genes that are targets of the AAR pathway. These genes contain AAREs (amino acid response elements) that mediate the enhancement of transcription [6–9]. The AARE sequences function as enhancer elements [10,11] and have a 9–10 bp core that can differ in sequence by one or two nucleotides between genes. Several of the enhancer binding proteins that assemble on AAREs have been identified, including ATF2 (activating transcription factor 2) [12], ATF4 [12,13], ATF3 [14,15], and C/EBPβ (CCAAT/enhancer-binding protein β) [16]. For the amino-acid-dependent control of the ASNS (asparagine synthetase gene), C/EBPβ has been documented to be a required component [16]. Increased C/EBPβ expression itself may represent a critical AAR pathway step just prior to AARE-containing target genes [17]. Consequently, the goal of the present studies was to investigate the mechanism by which the AAR pathway leads to increased C/EBPβ expression.
C/EBPβ is a member of a family of transcription factors that also includes C/EBPα, C/EBPγ, C/EBPδ, C/EBPϵ and CHOP (C/EBP homology protein) (reviewed in [18,19]). The C/EBP members dimerize with other bZIP (basic leucine-zipper) family members, as well as other transcription factors [20]. C/EBPβ plays a role in a wide range of important cellular processes, such as adipocyte differentiation, carbohydrate metabolism, inflammation and cellular proliferation [18,19,21]. The C/EBPβ mRNA is subject to differential translational start site selection from each of three methionine codons within the sequence, such that three protein isoforms are produced [22]. Although C/EBPβ post-translational modification and function has been studied extensively, investigation of the transcriptional control of the C/EBPβ gene itself is limited. A recent report from this laboratory, documenting the presence of an unfolded protein response element [23], is the only published information on genomic element identification for the human C/EBPβ gene. The rat C/ebpβ promoter has been studied, and Niehof et al. [24] have demonstrated that there are two CRE (cAMP-response element)-like sequences within the rat promoter that are necessary for maintaining basal transcription. Marten et al. reported that the rat liver mRNA content for C/ebpβ was increased in response to reduced dietary protein [25] and in rat hepatoma cells following incubation in amino-acid-limiting medium [26]. Subsequently, Siu et al. [16] showed that not only is C/EBPβ mRNA increased by histidine deprivation of HepG2 human hepatoma cells, but there is also increased C/EBPβ DNA binding activity in the nuclear extract of amino-acid-deprived HepG2 cells. More recently, Chen et al. [17] have documented by ChIP (chromatin immunoprecipitation) analysis that C/EBPβ, binds to the asparagine synthetase AARE in vivo and thus functions as an important component of the AAR pathway.
The present study was designed to survey other C/EBP family members for amino acid responsiveness and to investigate the mechanism by which C/EBPβ expression is induced in response to activation of the AAR pathway. The results documented that amino-acid-deprivation slowed the turnover rate of the C/EBPβ mRNA slightly, but increased transcription from the C/EBPβ gene by about 8-fold. Transient transfection of genomic fragments linked to a luciferase reporter gene demonstrated that the C/EBPβ promoter plays no major regulatory role with regard to the amino acid responsiveness. However, deletion analysis of a genomic region located 3′ downstream to the protein coding sequence of the intronless C/EBPβ gene revealed that a 93 bp fragment contained an amino-acid-responsive activity that functioned as an enhancer and that was required for the induction following AAR pathway activation. Mutagenesis of possible AAREs within the 93 bp region implicated a C/EBP-ATF composite site as a primary contributor, but the data suggest that multiple sites are required. Exogenous expression of ATF4 caused increased transcription from a reporter plasmid containing the 93 bp fragment, and ChIP analysis documented in vivo binding of ATF4 to this enhancer region of the C/EBPβ gene, but not to the promoter. Collectively, the data indicate that the increased transcription seen, following amino-acid-limitation, from the C/EBPβ gene is the mechanism by which elevated expression of this key regulator of the AAR pathway occurs.
MATERIALS AND METHODS
Chemicals
ActD (actinomycin D) was obtained from Sigma–Aldrich (St. Louis, MO, U.S.A.) and a stock solution (300 μM) was prepared in 100% (v/v) ethanol. An aliquot of the stock solution (or the solvent for the control) was added directly to the cells at a final ActD concentration of 5 μM.
Cell culture
Human hepatoma HepG2 cells were cultured in MEM (minimal essential medium, Mediatech, Herndon, VA, U.S.A.), pH 7.4, supplemented to contain a 1×non-essential amino acid solution (Mediatech), 4 mM glutamine, 25 mM NaHCO3, 100 μg/ml streptomycin sulphate, 100 units/ml penicillin G, 0.25 μg/ml amphotericin B and 10% (v/v) FBS (fetal bovine serum) (Gibco/Invitrogen, Carlsbad, CA, U.S.A.). Cells were maintained at 37 °C in a CO2/air (1:19) incubator. For all experiments, cell cultures were replenished with fresh medium and serum for 12 h prior to initiating all treatments to ensure that the cells were in the basal (‘fed’) state. Amino-acid-deprivation was induced by transfer of cells to a medium deficient in either one of two essential amino acids: amino-acid-complete MEM versus MEM lacking histidine (Gibco/Invitrogen), or amino-acid-complete DMEM (Dulbecco's modified Eagle's medium, Mediatech) versus DMEM lacking methionine (Gibco/Invitrogen). The induction of C/EBPβ expression after limitation for either of these amino acids was qualitatively similar and mechanistically the same. For the period of amino-acid-deprivation, all media were supplemented with 10% (v/v) dialysed FBS (Sigma).
RNA isolation and Northern blot analysis
HepG2 cells were cultured to 70–80% confluence in 60-mm-diameter dishes and then incubated for the indicated time in complete MEM or MEM lacking histidine. Total cellular RNA was isolated using an RNeasy® Mini Kit (Qiagen Inc., Valencia, CA, U.S.A.). 32P-radiolabelled cDNA probe synthesis and Northern analysis was performed as described by Aslanian et al. [27]. The cDNA probe for C/EBPβ was nt +1425 to +1632, which corresponds to a segment of the 3′ untranslated region, obtained by PCR and confirmed by sequencing. The cDNA probe for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was the entire protein coding sequence, obtained from Dr Anupam Agarwal, Division of Nephrology, Department of Medicine, University of Alabama at Birmingham, Birmingham, AL, U.S.A.
Subcloning of the human C/EBPβ gene
A BAC (bacterial artificial chromosome) clone (RP11-112L6) containing sequence from human chromosome 20 was obtained from the Wellcome Trust Sanger Institute, Hinxton, Cambridge, U.K. To obtain a C/EBPβ-containing genomic fragment, the BAC clone was digested with EcoRI and PvuI, the fragments were separated by preparative field inversion gel-electrophoresis, and then they were ligated into the EcoRI site of the pBluescript® II SK vector (Stratagene, La Jolla, CA, U.S.A.). Using the C/EBPβ cDNA probe described above, colony hybridization was used to screen the resulting DH5α colonies and an 11.5 kb C/EBPβ clone was obtained that contained nt −8451 to +3074, relative to the transcription start site (+1).
Deletion analysis
C/EBPβ fragments containing nt −8451/+157 and −1595/+157 were obtained by restriction endonuclease digestion of the −8451/+3074 clone, whereas the C/EBPβ promoter fragment (nt −325/+157) was prepared by PCR and checked by sequencing. The C/EBPβ sequences +1554/+1646, +1423/+2213 and +1423/+3541, which are 3′ to the protein coding sequence, were amplified by PCR using either the −8451/+3074 fragment or the original BAC clone as template. The promoter fragments were checked by sequencing and then ligated into the SmaI site, upstream of the firefly luciferase protein coding sequence within the pGL3-basic vector (Promega, Madison, WI, U.S.A.). The C/EBPβ gene downstream sequences were tested for amino acid responsiveness by ligation into the BamHI site, located 3′ to the luciferase protein coding sequence. To test the C/EBPβ genomic sequences in conjunction with the SV40 (simian virus 40) promoter, oligonucleotides were synthesized with BamHI linkers (Invitrogen) and ligated into the BamHI site of the pGL3-promoter vector (Promega).
Site-directed mutagenesis
Site-directed mutagenesis was performed using the Quik-Change® Site-Directed Mutagenesis Kit (Stratagene). Block substitutions were made within the C/EBPβ 3′ genomic sequence from nt +1423 to +2213, which was cloned into BamHI site downstream of the firefly luciferase reporter gene (pGL3-basic vector, Promega) under the control of the C/EBPβ promoter fragment nt −1593/+157. The mutations were confirmed by DNA sequencing. A C/EBP-ATF site (underlined), 5′-TTGATGCAATC-3′ (nt +1567/+1576), was mutated to 5′-CGAGGCGTTAT-3′. An UPRE (unfolded protein response element; underlined) and its 3′ flanking sequence, 5′-ACTGACGCAACCCACGTG-3′ (nt +1618/+1627), were changed to 5′-ACTGACTTCGATATTCTG-3′. A portion of the NSRE-2 (nutrient sensing response element-2) (underlined) and its 3′ flanking nucleotides, 5′-TGTAACTGTCAG-3′ (nt +1628/+1639), were replaced by 5′-TGGGGACTCAGT-3′. The C/EBPβ wild-type or mutated sequences were transiently transfected into HepG2 cells, as described below, and the enhancer activity was assayed by measuring the firefly luciferase activity.
Transient transfection and luciferase reporter assay
HepG2 cells were transfected at ≈50% confluence in 24-well plates (2×105 cells/well) using the SuperFect® transfection reagent according to the manufacturer's instructions (Qiagen, Valencia, CA, U.S.A.). For each transfection, 1 μg of the pGL3 Firefly luciferase reporter construct, driven by the indicated promoter, was co-transfected along with 0.5 ng of a reference Renilla (sea pansy) luciferase expression plasmid, phRL-SV40 (Promega). After transfection and a subsequent 18 h recovery in complete MEM medium containing 10% FBS, cells were then incubated for 12 h in either fresh complete MEM/DMEM or the corresponding medium lacking histidine/methionine, each supplemented with 10% dialysed FBS. After the incubation times indicated in each figure, the firefly and Renilla luciferase activities were assayed by the Dual-Luciferase Reporter System according to the manufacturer's directions (Promega). The data are expressed as the means±S.D. for three or four assays and each experiment was repeated with multiple batches of cells.
Immunoblot analysis
After incubation in amino-acid-complete or amino-acid-deficient medium for 0–24 h, total cell extracts were prepared for immunoblot analysis. Protein content was quantified by a Lowry assay and samples containing 30 μg of protein were separated on a precast Criterion™ Tris/HCl polyacrylamide gel (Bio-Rad, Hercules, CA, U.S.A.) or a standard 20 cm long 10% gel (for C/EBPβ). After electrotransfer to a Bio-Rad nitrocellulose membrane, the membrane was stained with Fast Green to check for equal loading and then incubated with 10% blocking solution [10% (w/v) Carnation non-fat dry milk and Tris-buffered saline/Tween (30 mM Tris base (pH 7.6), 200 mM NaCl and 0.1% Tween-20)] for 1 h at room temperature with mixing. Immunoblotting was performed using rabbit polyclonal antibodies against the α, β, γ, δ, or ϵ isoforms of C/EBP (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.) at an antibody concentration of 0.2–2 μg/ml in 10% dry milk blocking solution for 2 h at room temperature (≈21 °C) with mixing. The membrane was washed five times for 5 min in 5% blocking solution on a shaker and then incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD, U.S.A.) in 5%-dry-milk blocking solution at a 1:20000 dilution for 1 h at room temperature with mixing. The membrane was then washed five times for 5 min in 5% dry milk blocking solution and two times for 5 min in TBS/T (30 mM Tris base pH 7.6, 200 mM NaCl and 0.1% Tween-20). The bound secondary antibody was detected using an Enhanced Chemiluminescence kit (Amersham Biosciences, Piscataway, NJ, U.S.A.) and exposing the membrane to Biomax® MR film (Kodak, Rochester, NY, U.S.A.). To provide a demonstration of equal loading, some membranes were re-probed with a 1:5000 dilution of an antibody specific for actin (Sigma).
ChIP analysis
For ChIP analysis, HepG2 cells were seeded at 1.5×107/150-mm-diameter dish with complete MEM and then grown for 24 h. Cells were transferred to fresh MEM for 12 h before transfer to either complete MEM or MEM lacking histidine for the time period indicated in each Figure. Protein and DNA were cross-linked by adding formaldehyde directly to the culture medium to a final concentration of 1%, the reaction being stopped 10 min later by adding 2 M glycine to a final concentration of 0.125 M. Cross-linked chromatin was solubilized by sonication using a Sonic Dismembrator (Model 100, Fisher Scientific Co.) for five bursts of 40 s at power 5 with 2 min cooling on ice between each burst. An extract from 1×107 HepG2 cells was incubated with 2 μg of antibody. The antibody-bound complex was precipitated by Protein A–Sepharose beads (Amersham Biosciences). The DNA fragments in the immunoprecipitated complex were released by reversing the cross-linking at 65 °C for 5 h and purified using a QIAquick® PCR purification kit (Qiagen Inc.). Purified, immunoprecipitated DNA was analysed by qPCR (quantitative real-time PCR). Primers for the qPCR were designed using Vector NTI® Version 7.1 software (InforMax Inc., Frederick, MD, U.S.A.) to amplify the C/EBPβ proximal promoter sequence nt −411 to −481 (forward primer, 5′-GGGAGTCGTCACAGGCGTCAA-3′, and reverse primer, 5′-TCCCGTAAACTCCCACCTCCTC-3′) and the 3′ genomic sequence at nt +1607 to +1670 (forward primer, 5′-CGCAACCCACGTGTAACTGTCAG-3′, and reverse primer, 5′-CAGCAACAAGCCCGTAGGAACA-3′). To measure the transcription rate from the intronless C/EBPβ gene, the method of Sandoval et al. [28] was used, which relies on ChIP analysis to monitor Pol II (RNA polymerase II) binding at a region distal to the promoter. For this purpose primers were chosen within the proteincoding region of the gene (forward primer, 5′-AGAACGAGCGGCTGCAGAAGA-3′, and reverse primer, 5′-CAAGTTCCGCAGGGTGCTGC-3′). The qPCR analysis was performed using the DNA Engine Opticon® 2 system (Genetic Technologies, Miami, Fl, U.S.A.) and the product was detected with SYBR® Green I. Serial dilutions of input chromatin were used to generate a standard curve for determining the relative amount of product. Duplicates for both the standards and the samples were simultaneously amplified using the same reaction master mixture. The reactions were incubated at 95 °C for 15 min to activate the polymerase, followed by amplification at 95 °C for 15 s and 60 °C for 60 s for 35 cycles. After PCR, melting curves were acquired by stepwise increases in the temperature from 55 to 95 °C to ensure that a single product was amplified in the reaction. The results are expressed as the ratio to a 1:20 dilution of input DNA. Samples from at least three independent immunoprecipitations were analysed and results are reported as the means±S.E.M.
RESULTS
Effect of amino-acid-limitation on the protein content for members of the C/EBP family
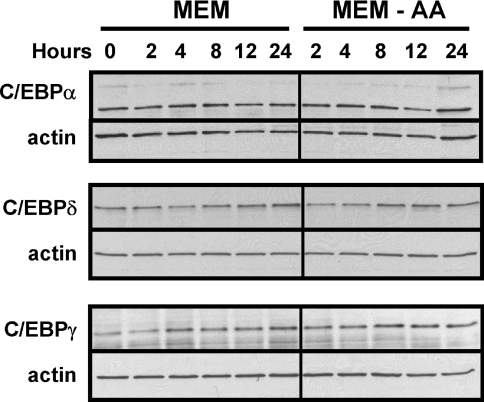
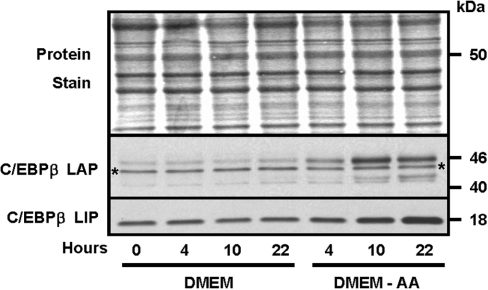
Previous work from our laboratory had shown that the abundance of C/EBPβ was increased in response to amino-acid-deprivation of HepG2 cells [17]. The CHOP gene is also known to be amino-acid-regulated [29]. Other members of the C/EBP family were surveyed for the response to amino-acid-limitation by using a total cell protein extract to immunoblot for each member of the family. Figure 1 shows that C/EBPα, C/EBPγ and C/EBPδ exhibited only minor changes in their abundance following amino-acid-deprivation. C/EBPϵ protein was not detectable, consistent with its primary role in myeloid cells. The protein content for the LAP (liver-activating protein) isoform of C/EBPβ was induced by methionine limitation, and the protein was detected as a collection of bands migrating from 40–45 kDa (Figure 2). The multiple bands may be the result of post-translational modification, as the C/EBPβ protein is known to be the target of phosphorylation and SUMOylation (SUMO is small ubiquitin-related modifier 1) [18,21,30]. The C/EBPβ mRNA is subject to translational control, which leads to the synthesis of a truncated isoform, termed ‘liver-inactivating protein’ (LIP), from a downstream methionine codon [19,22]. As shown by the results presented in Figure 2, the LIP isoform is also induced following amino-acid-deprivation of cells, although the kinetics appear to be slightly different from those for LAP in that, by 22 h, the LAP content peaked and may actually have declined slightly, whereas the amount of LIP continued to increase to 22 h.
Figure 1. Amino-acid-limitation of HepG2 cells does not affect the protein content of C/EBPα, C/EBPγ or C/EBPδ family members.
HepG2 cells were maintained in complete MEM to reach 70–80% confluence and then transferred to fresh complete MEM (MEM) or MEM lacking histidine (MEM-AA) for the period of time indicated. Total cell extracts were isolated and subjected to immunoblotting, as described in the Materials and methods section. After probing the blot with an antibody specific for C/EBPα, C/EBPγ or C/EBPδ the membrane was probed with an antibody against actin to ensure equal loading between lanes. A representative blot for each protein is shown, but analysis was performed on multiple cell preparations with qualitatively similar results.
Figure 2. Amino-acid-deprivation increases HepG2 cell C/EBPβ protein content.
HepG2 cells were maintained in complete MEM to reach 70–80% confluence (time=0) and then transferred to fresh complete DMEM (DMEM) or DMEM lacking methionine (DMEM-AA) for 4, 10 or 22 h. Total cell extracts were isolated and subjected to immunoblotting as described in the Materials and methods section. Prior to probing the blot with an antibody specific for C/EBPβ, the membrane was stained with Fast Green (‘Protein Stain’) to ensure equal loading between lanes. The C/EBPβ antibody used is against the C-terminus and therefore detects both the LAP and LIP isoforms of the protein (see the text for details). A representative blot is shown, but analysis was performed on several independent cell preparations with similar results. The asterisks denote a non-specific band that has been reported by other laboratories for this antibody (Santa Cruz Biotechnology, catalogue number SC-150).
Effect of histidine deprivation on C/EBPβ mRNA content
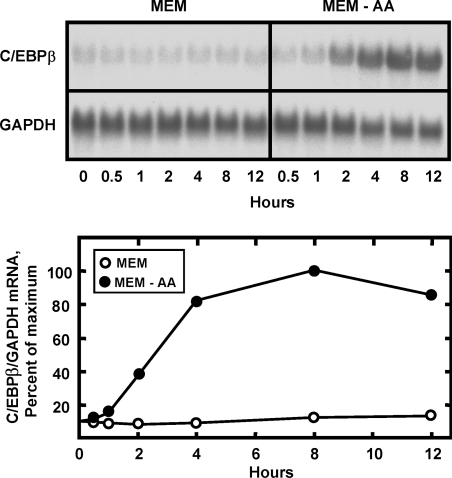
To investigate the mechanism responsible for the increased C/EBPβ protein expression, the C/EBPβ mRNA content was assayed in amino-acid-deprived HepG2 cells for 0–12 h (Figure 3). After a lag of about 30 min, the C/EBPβ mRNA level rose steadily over the next 8 h, resulting in an increase of about 9-fold, in the cells lacking the amino acid. Although C/EBPα mRNA content was reported to be increased by amino-acid-limitation of rat hepatoma cells [26], when the same blot shown in Figure 3 was stripped and re-probed for C/EBPα mRNA, the data revealed that the amount of C/EBPα mRNA actually declined slowly over the 12 h tested (results not shown).
Figure 3. Amino-acid-limitation increases the C/EBPβ mRNA content of HepG2 cells.
HepG2 cells were maintained in complete MEM to reach 70–80% confluence and then transferred to fresh complete MEM (MEM) or MEM lacking histidine (MEM-AA). Total RNA was isolated at the time indicated and Northern-blot analysis (15 μg/lane) was performed to measure the mRNA content for C/EBPβ or GAPDH. The GAPDH was used as a loading control and a negative control for amino-acid-dependent activation. The blots were digitized by using a phosphorimager, followed by quantification. The data were plotted as the normalized mRNA content (C/EBPβ/GAPDH) and the value for cells maintained for 8 h in the MEM-AA condition was set to be 100%.
Induction of C/EBPβ mRNA content by the AAR Is primarily due to increased transcription
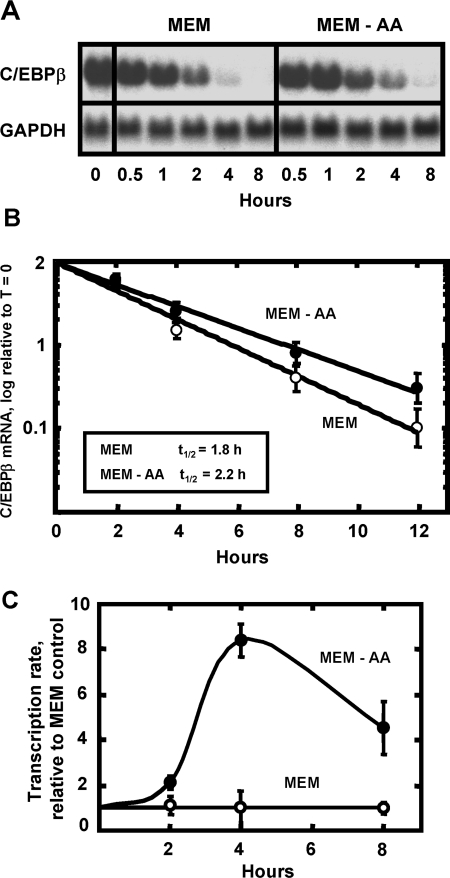
The mRNA for the cat-1 cationic amino acid transporter [31], the cell cycle regulatory proteins p21 and p27 [32], and ATF3 (Y-X. Pan and M. S. Kilberg, unpublished work) are stabilized by amino-acid-deprivation. To test for mRNA stability as a possible mechanism for the AAR enhancement of C/EBPβ mRNA, HepG2 cells were incubated in histidine-free MEM for 8 h to induce C/EBPβ mRNA content and then transferred to either fresh histidine-free MEM or complete MEM, both containing 5 μM ActD (Figures 4A and 4B). The results showed that the half-life of C/EBPβ mRNA in the cells incubated without histidine was increased slightly (2.2 versus 1.8 h), although it is not likely that this modest amount can account for the 9-fold increase in the steady-state mRNA (Figure 3). To assay the transcription rate from the C/EBPβ gene, cells were subjected to histidine deprivation for 2, 4 or 8 h and then ChIP analysis was performed to monitor Pol II binding to a region distal to the promoter. Sandoval et al. [28] demonstrated that measurement of Pol II binding within the coding region of a gene reflects the transcription rate. For the C/EBPβ gene, this analysis showed that the transcription rate was increased by about 8-fold in amino-acid-deprived cells relative to control cells (Figure 4C) and, therefore, increased transcription appears to account for most of the elevation in C/EBPβ mRNA following amino acid stress.
Figure 4. The elevation in C/EBPβ mRNA content following AAR activation is primarily due to increased transcription.
HepG2 cells were maintained in MEM lacking histidine for 8 h to reach maximal induction of C/EBPβ mRNA (time=0). The cells were washed, then incubated in fresh amino-acid-complete MEM plus 5 μM Act D (MEM) or fresh MEM lacking histidine in the presence of ActD (MEM-AA). Total RNA was isolated at the time indicated and, for (A) Northern-blot analysis (15 μg/lane) was performed to measure the mRNA content for C/EBPβ and GAPDH (as a loading control). (B) Rather than Northern blotting, qPCR was performed to quantify the mRNA turnover and the data were plotted as the logarithm of mRNA content versus time after transfer to the ActD-containing media. (C) The transcription rate for HepG2 cells incubated in either complete MEM or MEM lacking histidine (MEM-AA) was analysed using the ChIP approach described in the text. The data were plotted as the fold increase relative to the MEM control and each point represents the mean±S.E.M. for three independent assays, each performed in duplicate.
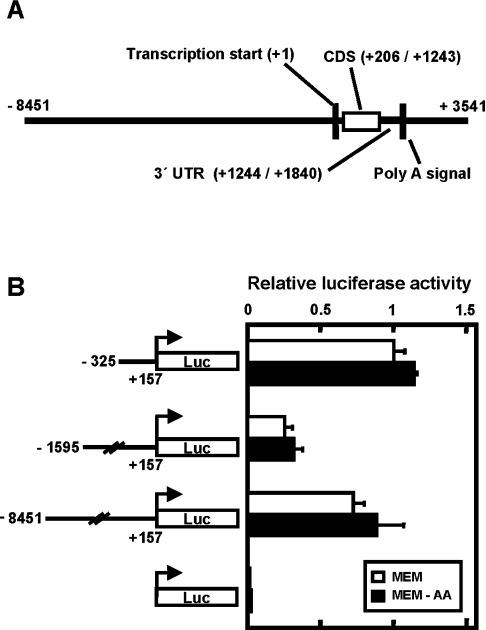
The C/EBPβ promoter region alone is not sufficient to mediate induction via the AAR
Human C/EBPβ is an intronless gene is located on chromosome 20 [33], and relative to the transcription start site (designated as +1), the first of three translation start sites is at nt +206 and the universal translation stop codon is at nt +1243 (Figure 5A). A polyadenylation signal (5′-AATAAA-3′) is located approximately 1.8 kbp downstream from the transcription start site. To investigate the potential role of the proximal promoter region in mediating C/EBPβ gene induction in response to amino-acid-limitation, a fragment (nt −325/+157) corresponding to the human C/EBPβ proximal promoter was tested (Figure 5B). This promoter fragment resulted in basal transcription, but no increase under the histidine-limited condition. To test the possibility that an AARE was located further 5′ upstream, longer genomic fragments (nt −1595/+157 and −8451/+157) were examined, but similar results were obtained, in that neither a 1.7 nor a 8.5 kbp fragment mediated a response to amino-acid-limitation (Figure 5B).
Figure 5. The C/EBPβ 5′ upstream region is not sufficient to mediate induction following amino-acid-deprivation.
(A) illustrates the human C/EBPβ genomic structure. The transcription start site for the C/EBPβ mRNA is indicated as nucleotide +1 and all other features are labelled accordingly. The protein coding sequence of this intronless gene is labelled as ‘CDS’ and the untranslated region is labelled as ‘UTR’. (B) Transcription driven by the three different C/EBPβ 5′ upstream regions was monitored for the response to histidine deprivation after transient transfection of HepG2 cells with C/EBPβ-luciferase reporter constructs. Approx. 18 h after transfection, the cells were incubated for 12 h in either complete MEM or MEM lacking histidine (MEM-AA) and then extracts were assayed for firefly and Renilla luciferase activity. ‘Relative luciferase activity’ represents the firefly activity normalized for transfection efficiency with Renilla luciferase expression driven by the SV40 promoter. The value of the relative luciferase activity of the C/EBPβ promoter fragment nt −325/+157 in the MEM condition was set to 1. All data are expressed as the mean±S.D. for triplicate determinations.
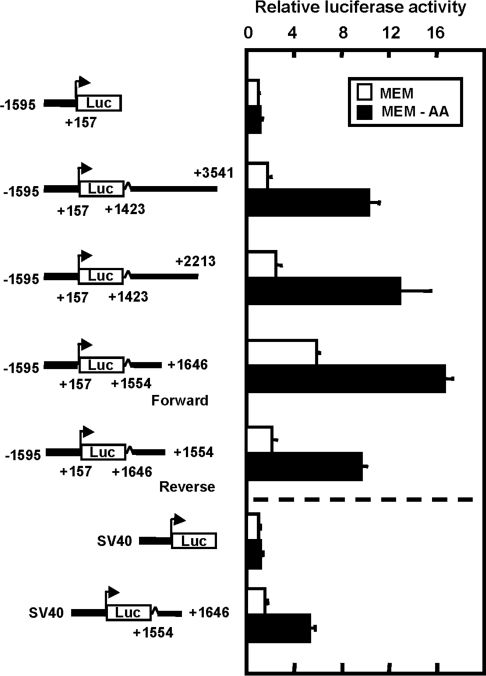
C/EBPβ genomic sequence 3′ to the protein coding region is essential for the AAR activation
Given that the C/EBPβ promoter did not support induction by amino-acid-deprivation and that C/EBPβ is an intronless gene, the 3′ genomic region was tested for amino acid responsiveness (Figure 6). Sequentially deleted 3′ genomic fragments were ligated downstream of the firefly luciferase reporter gene driven by a C/EBPβ promoter fragment containing nt −1595 to +157. The C/EBPβ 3′ sequence from nt +1423 to +3541 mediated inducible reporter gene expression when cells were deprived of histidine. When this 2.1 kbp genomic sequence was deleted from its 3′ end to a 790 bp fragment covering nt +1423 to +2213, the induction was maintained (Figure 6). Further deletion of this fragment to a 93 bp sequence covering nt +1554 to +1646 still resulted in activated transcription, even though the basal rate, measured in amino-acid-compete MEM medium, was increased as well. To test the hypothesis that the 93 bp 3′ genomic fragment could confer amino acid responsiveness to an unrelated promoter, the C/EBPβ promoter sequence was replaced with the SV40 promoter (Figure 6). The SV40 promoter alone was inert to histidine deprivation, but the presence of the 93 bp C/EBPβ genomic sequence induced transcription by nearly 4-fold that of MEM control. The 93 bp fragment was functional when placed either 5′ or 3′ relative to the C/EBPβ promoter and the distance from the promoter had no effect (results not shown). Furthermore, the 93 bp fragment could be reversed in orientation without loss of amino acid responsiveness (Figure 6). These results suggest that this fragment contains an AARE and that the functional properties fulfil the requirements of enhancer activity.
Figure 6. The genomic region downstream of the protein coding sequence is required for induction of the C/EBPβ gene via the AAR pathway and functions as an enhancer.
Sequentially deleted human C/EBPβ genomic fragments from the region 3′ to the protein coding sequence were ligated downstream of the firefly luciferase reporter gene under the control of the C/EBPβ promoter fragment nt −1595/+157 or the SV40 promoter. The cells were subjected to amino-acid-limitation 18 h after transfection. After incubation of the cells in MEM or MEM lacking histidine (MEM-AA) for 12 h, the transcription regulatory action of the C/EBPβ 3′ genomic sequences was measured by monitoring the firefly luciferase activity. To correct for the transfection efficiency between wells, the firefly activity was normalized with that of Renilla luciferase driven by the SV40 promoter (‘relative luciferase activity’). The transcription activity of the C/EBPβ promoter fragment nt −1595/+157 alone under the MEM condition was set to 1. All data are expressed as the mean±S.D. for assays in triplicate.
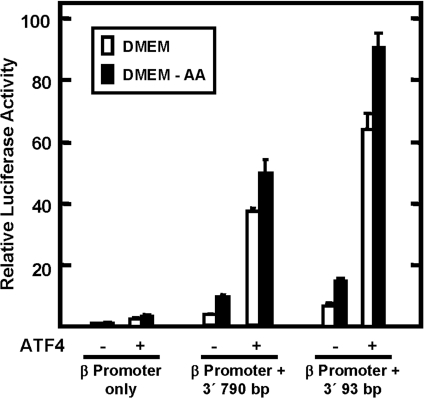
ATF4 regulates transcription from the C/EBPβ gene downstream enhancer
Increased expression of the bZIP transcription factor ATF4 is an important factor in the transcriptional activation of target genes of the AAR pathway [12,13,17]. To determine if the C/EBPβ gene was regulated by ATF4, HepG2 cells were transiently co-transfected with an ATF4 expression plasmid and a luciferase reporter plasmid containing the C/EBPβ promoter (nt −1595 to +157) and the 790 or 93 bp C/EBPβ downstream fragment containing the enhancer activity. Approx. 48 h after transfection, the cells were incubated for 12 h in either amino-acid-complete DMEM or DMEM lacking methionine (Figure 7). The data showed that the C/EBPβ-promoter-only construct was relatively inert to ATF4. However, when either downstream fragment was present, transcription was increased significantly by ATF4 expression in control cells (incubated in complete MEM). For those cells in which activation of C/EBPβ enhancer-driven transcription was induced by incubation in methionine-free medium, exogenous ATF4 caused a slightly larger increase, but the absolute amount of the increase was not as great as that for the control (MEM) cells (Figure 7). These results show that amino-acid-limitation and exogenous ATF4 activation are not additive, a result consistent with the interpretation that ATF4 mediates the AAR pathway. Collectively, the results document that the 93 bp C/EBPβ fragment contains an amino-acid-responsive activity.
Figure 7. The amino-acid-responsive genomic fragments of the C/EBPβ gene mediate the transcriptional activation by elevated ATF4.
The 790 bp and 93 bp 3′ C/EBPβ genomic fragments were placed downstream of the firefly luciferase reporter gene, which was under the control of the C/EBPβ promoter nt −1595/+157. As indicated, the HepG2 cells were co-transfected with the reporter constructs with or without an expression plasmid encoding ATF4. Empty pcDNA3.1 vector was used to keep the total amount of transfected plasmid constant. At 18 h after transfection, the cells were incubated for 12 h in complete DMEM or DMEM lacking methionine (DMEM-AA). The transcriptional regulatory action of the C/EBPβ 3′ genomic sequences is reported as the relative luciferase activity (firefly/Renilla). The transcription activity of the C/EBPβ promoter fragment nt −1595/+157 alone under the DMEM condition was set to 1. All data are expressed as the mean±S.D. for assays performed in triplicate.
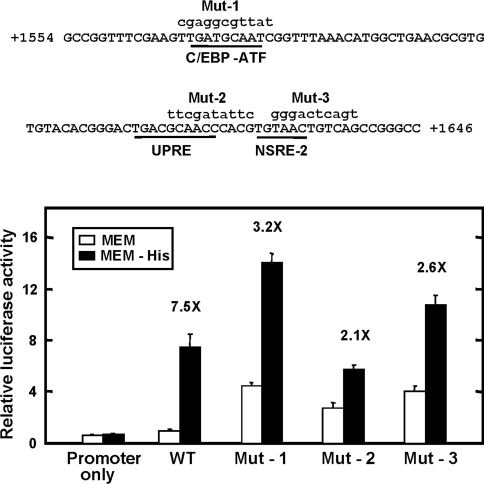
Mutagenesis of the 93 bp C/EBPβ downstream region
Computer analysis revealed that within the 93 bp C/EBPβ 3′ genomic region there is a 9 bp sequence, present at nt +1568 to +1576 (5′-TGATGCAAT-3′), that is identical with the AARE within the genes for CHOP (CHOP/GADD153) [34] and the System A transporter SNAT2 [11]. Sequences similar to this one are referred to as ‘C/EBP-ATF composite sites’ [35,36] and have been shown to mediate the amino-acid-response pathway through binding of ATF4 [13,17]. For the C/EBPβ gene, this C/EBP-ATF sequence is completely conserved across the human, rat and mouse species [23]. However, mutating the core and flanking nucleotides of the C/EBP-ATF sequence had only a partial effect (7.5-fold versus 3.2-fold) on the induced transcription following amino-acid-deprivation (Figure 8) or on the increased transcription after transient expression of exogenous ATF4 (results not shown). A similar result was obtained when the core sequence was completely deleted from the human C/EBPβ 3′ genomic region (results not shown). In fact, this sequence may have a repressive effect, in that mutation resulted in an increase in the absolute values for both the basal and the enhanced transcription, relative to the wild-type sequence (Figure 8). The 93 bp fragment also contains a second C/EBP-ATF like sequence at nt +1614 to +1623 (5′-TGACGCAAC-3′) that we have shown previously functions as a mammalian UPRE [23]. Mutation of this sequence also enhanced the basal rate of transcription relative to the wild-type and caused a suppression of amino-acid-regulated transcription to a level 2.1-fold over the MEM control value (Figure 8). The AARE of ASNS is made up of two distinct sites, NSRE-1 and NSRE-2 [6,10]. A sequence identical with NSRE-2 is present within the C/EBPβ 93 bp fragment at nt +1628 to +1633 (Figure 8). Mutation of this site resulted in a partial suppression of the amino-acid-dependent induction (7.5-fold versus 2.6-fold), and once again, in large part due to an increase in the transcription rate for those cells maintained in amino-acid-complete medium. Collectively, the results indicate that multiple sites contribute to the regulation of the C/EBPβ gene by amino-acid-deprivation and more extensive analysis of this region will be required to fully characterize what are likely to be complex interactions that occur between these sequences that contribute to the AAR activity.
Figure 8. The C/EBPβ 93 bp sequence contains cis-elements that contribute to amino acid control.
Top panel: the sequence of the 93 bp C/EBPβ genomic region containing nt +1554 to +1646. Potential AAREs, based on other nutrient responsive genes (see the text for details), are underlined. Block mutation of these nutrient response elements was performed in the context of the C/EBPβ 3′ genomic fragment nt +1423/+2213 which was inserted downstream of the firefly luciferase reporter gene driven by the C/EBPβ promoter fragment nt −1595/+157. Mutated sequences for the C/EBP-ATF site (Mut-1), the UPRE site (Mut-2), and the NSRE-2 site (Mut-3) are shown in lower-case letters. Analysis of these mutations is shown in the bottom panel. HepG2 cells were transfected with the reporter constructs and the effect of amino-acid-limitation tested by incubation in either complete MEM or MEM lacking histidine (MEM-His) for 12 h. The relative luciferase activity (the ratio of the firefly to Renilla luciferase activity) for the wild-type (WT) construct in the MEM condition was set to 1. All data are expressed as the mean±S.D. The fold increase in transcription following amino-acid-limitation is shown for each construct.
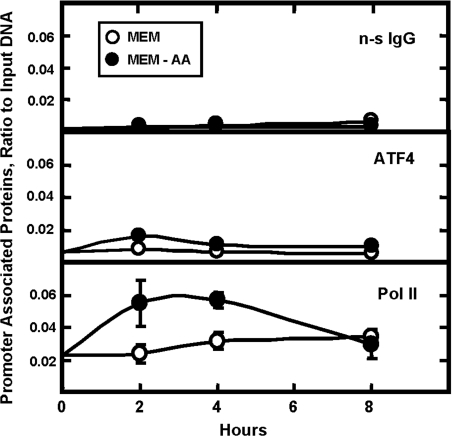
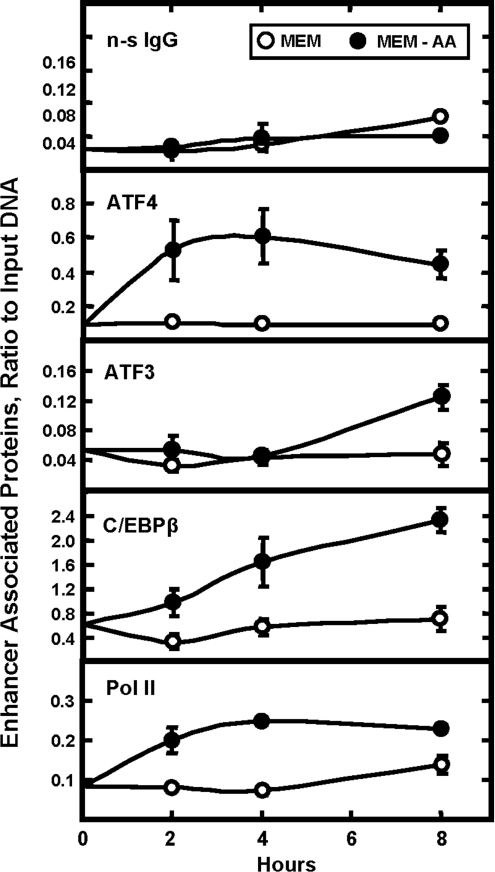
ChIP analysis of the C/EBPβ proximal promoter and the downstream genomic region containing enhancer activity
To assess transcription factor binding in vivo, HepG2 cells were incubated in complete MEM or MEM lacking histidine for 0–8 h and then ChIP analysis was performed on the C/EBPβ gene (Figure 9). A rabbit anti-chicken IgG was used as the negative control to illustrate the background noise of the assay. When Pol II was immunoprecipitated, a significant level of binding to the C/EBPβ proximal promoter was detected and the level of that binding was increased after amino-acid-deprivation of the cells (Figure 9). In contrast, when binding for ATF4 was tested (with or without amino-acid-limitation), the amount of C/EBPβ promoter immunoprecipitated was nearly the same as for the non-specific anti-chicken IgG. When the immunoprecipitated DNA was tested for the presence of sequence overlapping with the 93 bp regulatory region, amino-acid-dependent binding of Pol II was still detected (Figure 10). However, for ATF4 binding, the results differed considerably from those for the promoter. For this 3′ downstream regulatory area of the gene, a basal level of ATF4 binding, well above background, was observed in the amino-acid-replete cells and the amount of ATF4 bound increased by about 6-fold between 2–4 h after histidine deprivation (Figure 10).
Figure 9. Kinetic analysis of protein binding to the C/EBPβ promoter following amino-acid-deprivation.
HepG2 hepatoma cells were incubated in complete MEM or MEM lacking histidine (MEM-AA) for 0–8 h. ChIP analysis was performed using a rabbit anti-chicken IgG antibody (non-specific control; n-s IgG), Pol II or ATF4. The qPCR was performed using primers to specifically amplify the promoter nt −411 to −481. Data were plotted as the ratio to the value obtained with a 1:20 dilution of input DNA. Each point represents the mean value of three independent experiments each performed in duplicate, and the error bars represent the S.E.M. Where not shown, the error bars are contained within the symbol.
Figure 10. ChIP analysis of the C/EBPβ genomic region containing the amino-acid-responsive enhancer activity.
HepG2 hepatoma cells were incubated in complete MEM or MEM lacking histidine (MEM-AA) for 0–8 h. ChIP analysis was performed using a rabbit anti-chicken IgG antibody (non-specific control, n-s IgG), Pol II, ATF3, C/EBPβ or ATF4. The qPCR was performed using primers to specifically amplify the genomic sequence from nt +1607 to +1670. Data were plotted as the ratio to the value obtained with a 1:20 dilution of input DNA. Each point represents the mean value of three independent experiments each performed in duplicate, and the error bars represent the S.E.M. Where not shown, the error bars are contained within the symbol.
The bZIP transcription factors ATF3 as well as C/EBPβ itself have been implicated in the regulation of AARE-containing genes (reviewed in [37]). Consequently, binding of these two factors to the C/EBPβ enhancer region was tested and both were shown to be present. At the AARE within the human ASNS, relative to the rapid increase in ATF4 binding within 45 min after amino acid removal, recruitment of ATF3 and C/EBPβ is delayed several hours, and these two factors are thought to hold in check the level of transcriptional induction [17]. For the C/EBPβ 3′ regulatory region, amino-acid-dependent binding of both ATF3 and C/EBPβ was readily detected (Figure 10), and the kinetics were similar to those for ASNS. That is, a high level of constitutive C/EBPβ binding was observed in cells maintained in amino-acid-complete MEM and, after amino acid removal from the medium, the amount associated with the enhancer steadily increased during the 8 h investigated. The binding of ATF3 in the amino-acid-replete cells was low, but was enhanced by amino-acid-deprivation after a lag of about 4 h (Figure 10). This time course for ATF3 recruitment is consistent with the hypothesis that it is functioning as a transcriptional repressor at the AARE [17], because it coincides with a slow decline in ATF4 binding and with a suppression of the transcription rate from the C/EBPβ gene (Figure 4).
DISCUSSION
The experiments described in the present paper have led to these novel observations. (1) Stabilization of the C/EBPβ mRNA cannot account for the magnitude of the increase in steady-state mRNA following amino-acid-limitation; rather, transcriptional control of the gene appears to be the primary regulatory mechanism. (2) The C/EBPβ 5′ upstream genomic region, up to 8.45 kbp, is not sufficient to induce C/EBPβ expression following amino-acidlimitation. (3) The regulatory element(s) necessary for the induction of C/EBPβ gene are located within a 93 bp genomic sequence that is 3′ to the protein coding sequence. (4) The 93 bp fragment has enhancer-like activity in that it conveys amino acid responsiveness to a heterologous promoter, and it is functional regardless of location and orientation. (5) The amino-acid-responsive fragment of the C/EBPβ gene responds to activation by ATF4. (6) Multiple cis-elements contribute to the activation of the gene. 7) Despite the unique location of the amino-acid-responsive activity, ChIP analysis of the C/EBPβ gene documents a temporal relationship between ATF4, Pol II, C/EBPβ and ATF3 binding that has been seen for other amino-acid-responsive genes [17].
In 1994, Marten et al. [26] reported that, in H4-II-E rat hepatoma cells, the mRNA content for both C/EBPβ and C/EBPα was increased in response to depletion of phenylalanine, methionine, leucine or tryptophan from the culture medium. In the present study, it was established that the C/EBPβ response was similar in HepG2 cells, but in these cells C/EBPα mRNA content actually declined in response to amino-acid-limitation. The difference may be due to cell type, species differences or to experimental conditions, because Marten et al. incubated their cells in serum-free, amino-acid-depleted medium for 24 h, whereas the present experiments were for 12 h duration or less and the medium contained 10% dialysed FBS. Interestingly, Claeyssens et al. [38] have shown that C/EBPα and C/EBPδ bind to a glutamine responsive element within the promoter of the GAPDH gene. Their results illustrate that members of the C/EBP family also contribute to the transcriptional control of genes induced by the presence of amino acids, in their case, glutamine and certain essential amino acids.
The present results demonstrate that the increase in C/EBPβ mRNA expression following AAR pathway activation is primarily due to increased transcription. Interestingly, this transcriptional activation is not mediated through the C/EBPβ promoter, but rather by a genomic sequence 3′ to the translation stop codon. A few examples of previously identified AARE sequences have been located in the proximal promoter regions of CHOP [7] and ASNS [6], in the first exon of the cat-1 transporter gene [8], in the promoter of the xCT amino acid transporter gene [9], and in the first intron of the SNAT2 transporter gene [11]. Deletion analysis of the C/EBPβ 3′ genomic sequence demonstrated that the nt +1554 to +1646 sequence contains a regulatory element that activates transcription in response to amino-acid-deprivation and to elevated ATF4. This 93 bp sequence confers amino acid responsiveness to an otherwise inert SV40 promoter and can also function in an orientation- and location-independent manner, suggesting that it contains an enhancer-like activity. Mutagenesis within the 93 bp fragment showed the amino-acid-dependent regulation of the C/EBPβ gene may be complicated in that multiple sites appear to contribute. Further analysis will be necessary to definitively characterize the role of each of these elements.
C/EBPβ has been documented to contribute to the regulation of the human ASNS gene following amino-acid-deprivation, but its exact role is not well understood. By EMSA (electrophoretic mobility-shift assay) and ChIP analysis it was shown that C/EBPβ binds to the AARE in the ASNS gene and that the binding activity is increased in amino-acid-deprived HepG2 cells [16,17]. Furthermore, overexpression of the naturally occurring dominant-negative isoform of C/EBPβ, LIP, blocks the increase in transcription from the ASNS promoter [16]. Overexpression of the C/EBPβ LAP isoform is more complex. Overexpression of C/EBPβ LAP by itself causes induction of basal transcription from the ASNS promoter and further induction of the enhanced transcription after amino-acid-deprivation [16]. However, when co-expressed with ATF4 and ATF3, C/EBPβ appears to act in concert with ATF3 to hold in check the induction of the ASNS gene by ATF4 [17]. Accordingly, after amino-acid-deprivation, increased expression of endogenous C/EBPβ and its binding to the ASNS promoter in vivo peaks at a time when the transcription rate from the ASNS gene is beginning to subside [17]. This pattern of C/EBPβ and ATF3 binding was also observed in the present study at the C/EBPβ gene 3′ regulatory region. Thus future studies aimed at understanding the control of expression from the C/EBPβ gene itself and the function of C/EBPβ at AARE sites within a number of genes will be important in advancing our understanding of the AAR pathway.
Acknowledgments
This work was supported by a grant (DK-52064) to M.S.K. from the Institute of Diabetes, Digestive and Kidney Diseases, the National Institutes of Health. We thank other members of the laboratory for technical advice, reagents and helpful discussions.
References
- 1.Holemans K., Aerts L., Van Assche F. A. Fetal growth restriction and consequences for the offspring in animal models. J. Soc. Gynecol. Invest. 2003;10:392–399. doi: 10.1016/s1071-5576(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 2.Burrin D. G., Davis T. A. Proteins and amino acids in enteral nutrition. Curr. Opin. Clin. Nutr. Metab. Care. 2004;7:79–87. doi: 10.1097/00075197-200401000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman J. A., Malloy V., Krajcik R., Orentreich N. Nutritional control of aging. Exp. Gerontol. 2003;38:47–52. doi: 10.1016/s0531-5565(02)00149-3. [DOI] [PubMed] [Google Scholar]
- 4.Koehnle T. J., Russell M. C., Morin A. S., Erecius L. F., Gietzen D. W. Diets deficient in indispensable amino acids rapidly decrease the concentration of the limiting amino acid in the anterior piriform cortex of rats. J. Nutr. 2004;134:2365–2371. doi: 10.1093/jn/134.9.2365. [DOI] [PubMed] [Google Scholar]
- 5.Hao S., Sharp J. W., Ross-Inta C. M., McDaniel B. J., Anthony T. G., Wek R. C., Cavener D. R., McGrath B. C., Rudell J. B., Koehnle T. J., Gietzen D. W. Uncharged tRNA and sensing of amino acid deficiency in mammalian piriform cortex. Science (Washington D.C.) 2005;307:1776–1778. doi: 10.1126/science.1104882. [DOI] [PubMed] [Google Scholar]
- 6.Barbosa-Tessmann I. P., Chen C., Zhong C., Siu F., Schuster S. M., Nick H. S., Kilberg M. S. Activation of the human asparagine synthetase gene by the amino acid response and the endoplasmic reticulum stress response pathways occurs by common genomic elements. J. Biol. Chem. 2000;275:26976–26985. doi: 10.1074/jbc.M000004200. [DOI] [PubMed] [Google Scholar]
- 7.Bruhat A., Jousse C., Carraro V., Reimold A. M., Ferrara M., Fafournoux P. Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol. Cell. Biol. 2000;20:7192–7204. doi: 10.1128/mcb.20.19.7192-7204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez J., Lopez A. B., Wang C., Mishra R., Zhou L., Yaman I., Snider M. D., Hatzolgou M. Transcriptional control of the arginine/lysine transporter, cat-1, by physiological stress. J. Biol. Chem. 2003;278:50000–50009. doi: 10.1074/jbc.M305903200. [DOI] [PubMed] [Google Scholar]
- 9.Sato H., Nomura S., Maebara K., Sato K., Tamba M., Bannai S. Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem. Biophys. Res. Commun. 2004;325:109–116. doi: 10.1016/j.bbrc.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Zhong C., Chen C., Kilberg M. S. Characterization of the nutrient sensing response unit in the human asparagine synthetase promoter. Biochem. J. 2003;372:603–609. doi: 10.1042/BJ20030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palii S. S., Chen H., Kilberg M. S. Transcriptional control of the human sodium-coupled neutral amino acid transporter system A gene by amino acid availability is mediated by an intronic element. J. Biol. Chem. 2004;279:3463–3471. doi: 10.1074/jbc.M310483200. [DOI] [PubMed] [Google Scholar]
- 12.Averous J., Bruhat A., Jousse C., Carraro V., Thiel G., Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 13.Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 14.Pan Y.-X., Chen H., Siu F., Kilberg M. S. Amino acid deprivation and endoplasmic reticulum stress induce expression of multiple ATF3 mRNA species which, when overexpressed in HepG2 cells, modulate transcription by the human asparagine synthetase promoter. J. Biol. Chem. 2003;278:38402–38412. doi: 10.1074/jbc.M304574200. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., Wek R. C. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 2004;24:1365–1377. doi: 10.1128/MCB.24.3.1365-1377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siu F. Y., Chen C., Zhong C., Kilberg M. S. CCAAT/enhancer-binding protein beta (C/EBPβ) is a mediator of the nutrient sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2001;276:48100–48107. doi: 10.1074/jbc.M109533200. [DOI] [PubMed] [Google Scholar]
- 17.Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J. Biol. Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 18.Ramji D. P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calkhoven C. F., Muller C., Leutz A. Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 20.Newman J. R., Keating A. E. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science (Washington D.C.) 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 21.Buck M., Chojkier M. Signal transduction in the liver: C/EBPβ modulates cell proliferation and survival. Hepatology. 2003;37:731–738. doi: 10.1053/jhep.2003.50155. [DOI] [PubMed] [Google Scholar]
- 22.Descombes P., Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell (Cambridge, Mass.) 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 23.Chen C., Dudenhausen E. E., Pan Y., Zhong C., Kilberg M. S. Human CCAAT/enhancer-binding protein beta (C/EBPβ) gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J. Biol. Chem. 2004;279:27948–27956. doi: 10.1074/jbc.M313920200. [DOI] [PubMed] [Google Scholar]
- 24.Niehof M., Kubicka S., Zender L., Manns M. P., Trautwein C. Autoregulation enables different pathways to control CCAAT/enhancer binding protein beta (C/EBPβ) transcription. J. Mol. Biol. 2001;309:855–868. doi: 10.1006/jmbi.2001.4708. [DOI] [PubMed] [Google Scholar]
- 25.Marten N. W., Sladek F. M., Straus D. S. Effect of dietary protein restriction on liver transcription factors. Biochem. J. 1996;317:361–370. doi: 10.1042/bj3170361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marten N. W., Burke E. J., Hayden J. M., Straus D. S. Effect of amino acid limitation on the expression of 19 genes in rat hepatoma cells. FASEB J. 1994;8:538–544. doi: 10.1096/fasebj.8.8.8181673. [DOI] [PubMed] [Google Scholar]
- 27.Aslanian A. M., Fletcher B. S., Kilberg M. S. Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem. J. 2001;357:321–328. doi: 10.1042/0264-6021:3570321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval J., Rodriguez J. L., Tur G., Serviddio G., Pereda J., Boukaba A., Sastre J., Torres L., Franco L., Lopez-Rodas G. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 2004;32:e88. doi: 10.1093/nar/gnh091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruhat A., Jousse C., Wang X.-Z., Ron D., Ferrara M., Fafournoux P. Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J. Biol. Chem. 1997;272:17588–17593. doi: 10.1074/jbc.272.28.17588. [DOI] [PubMed] [Google Scholar]
- 30.Kim J., Cantwell C. A., Johnson P. F., Pfarr C. M., Williams S. C. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 2002;277:38037–38044. doi: 10.1074/jbc.M207235200. [DOI] [PubMed] [Google Scholar]
- 31.Yaman I., Fernandez J., Sarkar B., Schneider R. J., Snider M. D., Nagy L. E., Hatzoglou M. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung-Pineda V., Pan Y., Chen H., Kilberg M. S. Induction of p21 and p27 expression by amino acid deprivation of HepG2 human hepatoma cells involves mRNA stabilization. Biochem. J. 2004;379:79–88. doi: 10.1042/BJ20031383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takiguchi M. The C/EBP family of transcription factors in the liver and other organs. Int. J. Exp. Pathol. 1998;79:369–391. doi: 10.1046/j.1365-2613.1998.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruhat A., Averous J., Carraro V., Zhong C., Reimold A. M., Kilberg M. S., Fafournoux P. Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 2002;277:48107–48114. doi: 10.1074/jbc.M206149200. [DOI] [PubMed] [Google Scholar]
- 35.Wolfgang C. D., Chen B. P., Martindale J. L., Holbrook N. J., Hai T. gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell. Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT/enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 1999;339:135–141. [PMC free article] [PubMed] [Google Scholar]
- 37.Kilberg M. S., Pan Y. X., Chen H., Leung-Pineda V. Nutritional control of gene expression: How mammalian cells respond to amino acid limitation. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Claeyssens S., Gangneux C., Brasse-Lagnel C., Ruminy P., Aki T., Lavoinne A., Salier J. P. Amino acid control of the human glyceraldehyde 3-phosphate dehydrogenase gene transcription in hepatocyte. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G840–G849. doi: 10.1152/ajpgi.00060.2003. [DOI] [PubMed] [Google Scholar]