Abstract
Recent data indicate that PPARγ (peroxisome proliferator-activated receptor γ) could be involved in the modulation of the amyloid cascade causing Alzheimer's disease. In the present study we show that PPARγ overexpression in cultured cells dramatically reduced Aβ (amyloid-β) secretion, affecting the expression of the APP (Aβ precursor protein) at a post-transcriptional level. APP down-regulation did not involve the pathway of the secretases and correlated with a significant induction of APP ubiquitination. Additionally, we demonstrate that PPARγ was able to protect the cells from H2O2-induced necrosis by decreasing Aβ secretion. Taken together, our results indicate a novel mechanism at the basis of the neuroprotection shown by PPARγ agonists and an additional pathogenic role for Aβ accumulation.
Keywords: Alzheimer's disease, non-steroidal anti-inflammatory drugs (NSAIDs), peroxisome proliferator-activated receptor γ (PPARγ), amyloid-β, amyloid precursor protein (APP)
Abbreviations: AICD, cytoplasmic APP (amyloid precursor protein) intracellular domain; Aβ, amyloid-β; AD, Alzheimer's disease; BACE, beta-site APP-cleaving enzyme or β-secretase; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HEKAPP+, human embryonic kidney cells overexpressing APP; N2a, mouse Neuro-2a cells; NCBI, National Center for Biotechnology Information; NSAIDs, non-steroidal anti-inflammatory drugs; PI, propdium iodide; PPARγ, peroxisome proliferator-activated receptor γ; RT-PCR, reverse-transcription PCR; RXR, retinoic X receptor; sAPPα, APP secreted by the cell
INTRODUCTION
AD (Alzheimer's disease) is a progressive and fatal neurodegenerative disorder characterized by the deposition of extracellular Aβ (amyloid-β) plaques and the formation of intracellular tangles in the central nervous system. Aβ plaques are mainly composed of Aβ peptides, which are protein fragments derived by proteolytic cleavage of the APP (amyloid precursor protein), an integral membrane protein processed by several different secretases. BACE (β-secretase or beta-site APP-cleaving enzyme) is responsible for the N-terminal cleavage of Aβ, whereas γ-secretase generates the C-terminal end of the peptide and the release of Aβ into the extracellular compartment. The inflammatory response to Aβ accumulation has been proposed to contribute to the pathogenesis of AD and to increase neuronal damage. This view is corroborated by a number of epidemiological studies providing evidence of a protective effect of long-term medication with NSAIDs (non-steroidal anti-inflammatory drugs) against the neurodegenerative disorder [1–3]. These epidemiological studies are now well supported by data showing that in vivo treatment of APP transgenic mice with NSAIDs significantly diminished amyloid deposition [4] and improved behaviour [5]. Moreover, NSAIDs were shown to affect directly the generation of Aβ, suggesting a new mechanism of action behind the protective effect of NSAIDs [6]. A potential target of NSAIDs is PPARγ (peroxisome proliferator-activated receptor γ) [7], a ligand-activated transcription factor and a member of the nuclear receptor superfamily [8,9]. Recent reports indicate that PPARγ agonists down-regulate Aβ generation, although the mechanism of this phenomenon still remains controversial. Sastre and colleagues [10] reported the finding that PPARγ agonists modulate processing of APP through regulation of β-secretase, whereas Camacho and co-workers [11] showed that activation of PPARγ directly affects the stability of Aβ externally added to the cells, suggesting the activation of a rapid clearance mechanism. In addition, it has been argued that NSAIDs may interact directly with the γ-secretase to affect amyloid production [12].
The aim of the present study was to understand the effective role played by PPARγ in the amyloidogenic process causing AD. We report that, in cultured cells, overexpression of PPARγ dramatically decreased Aβ production, concomitantly increasing APP ubiquitination. Moreover, we demonstrate that the reduction of Aβ secretion protected the cells from H2O2-induced necrosis, suggesting a novel mechanism at the basis of the anti-inflammatory properties of PPARγ and an additional pathogenic significance for Aβ accumulation.
MATERIALS AND METHODS
Cloning of PPARγ and RXR (retinoic X receptor)
PPARγ full length was amplified according to mRNA sequence NM_015869 [NCBI (National Center for Biotechnology Information)] and then cloned into pcDNA3.1 expression vector (Invitrogen, Carlsbad, CA, U.S.A.) using 5′-GCGCGCGGTACCATGGGTGAAACTCTGGGAGATTC-3′ (forward) and 5′-GCGCGCCTCGAGCTAGTACAAGTCCTTGTAGATCTC-3′ (reverse) primers with KpnI and XhoI restriction sites in their 5′ end and 3′ end respectively. RXR full length was amplified according to mRNA sequence NM_002957 (NCBI) using 5′-GCGCGCGGTACCGAGTTAGTCGCAGACATGGACA-3′ (forward) and 5′-GCGCGCCTCGAGCTAAGTCATTTGGTGCGGCGC-3′ (reverse) primers, with KpnI and XhoI restriction sites in their 5′ end and 3′ end and then cloned into pcDNA3.1 expression plasmid.
Cell culture and transfections
HEK-293 cells were cultured in DMEM (Dulbecco's modified Eagle's medium; Invitrogen) supplemented with 10% (v/v) fetalbovine serum (Biofluids, Rockville, MD, U.S.A.) and penicillin/streptomycin. HEKAPP+ cells (HEK-293 cells stably transfected with APP695) were obtained from Dr Luciano D'Adamio (Department of Microbiology and Immunology, Albert Einstein College of Medicine, Bronx, New York, N.Y., U.S.A.) and grown in the above-described cell-culture medium supplemented with 5 μg/ml puromycin. N2a (mouse Neuro-2a cells) stably expressing human APP695 were obtained from Professor Peter Davies (Departments of Pathology and Neuroscience, Albert Einstein College of Medicine, Bronx, New York, NY, U.S.A.) and grown in DMEM/Opti-MEM® (1:1, v/v), with 0.1 mM non-essential amino acids, 200 μg/ml geneticin and 5% fetal bovine serum. Transient transfections were performed using SuperFect® (Qiagen, Hilden, Germany) at 2 μl/μg of DNA.
Luciferase assay
The plasmid (pCD-basic) containing the human CD36 promoter in front of the luciferase gene of pGL3-basic (Promega, Mannheim, Germany) has been previously described [13]. The DR1 plasmid contained a consensus PPARγ element derived from the 3-hydroxy-3-methylglutaryl-CoA reductase gene [14] in plasmid pTAL-Luc (BD Biosciences Clonetech, Palo Alto, CA, U.S.A.). CD36 and DR1 promoter plasmids, PPARγ expression vector and pRL-TK internal control vector were transfected into HEKAPP+ cells using SuperFect. After 3 h transfection, the cells were treated with the diabetes drug troglitazone (50 μM) (Alexis, Lausen, Switzerland) and harvested 24 h later. Promoter activity was measured by using the dual luciferase assay kit (Promega, Mannheim, Germany) with a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA, U.S.A.).
Immunoblot analysis
Immunoblots were performed according to standard methods using the following antibodies: monoclonal mouse anti-human PPARγ (E-8; Santa Cruz, Santa Cruz, CA, U.S.A.), monoclonal mouse anti-(human β-actin) (Sigma, St. Louis, MO, U.S.A.), monoclonal mouse anti-(human Aβ) (4G8 from Signet, Dedham, MA, U.S.A., and 6E10 from Chemicon, Temecula, CA, U.S.A.), monoclonal mouse anti-(human APP) (22C11; Chemicon, Temecula, CA, U.S.A.), polyclonal rabbit anti-(human APP) (Zymed, San Francisco, CA, U.S.A.), monoclonal mouse anti-(human BACE) (Chemicon, Temecula, CA, U.S.A.), and monoclonal mouse anti-ubiquitin (P4D1; Santa Cruz). Anti-mouse and anti-rabbit secondary antibodies were coupled to horseradish peroxidase (Amersham, Bucks., U.K.). Proteins were visualized with an enzyme-linked chemiluminescence detection kit according to the manufacturer's (Amersham) instructions. Chemiluminescence was monitored by exposure to film and the signals were analysed under non-saturating condition with an image densitometer (Bio-Rad, Hercules, CA, U.S.A.).
PCR
Total RNA was isolated with an RNA extraction kit from Qiagen. Semiquantitative assay for human APP mRNA expression was performed with AmpliTaq polymerase (PerkinElmer, Boston, MA, U.S.A.) and primers 5′-AGTGACAATGTGGATTCTGC-3′ (forward) and 5′-AGATACTTGTCAACGGCATC-3′ (reverse). Semi-quantitative PCR for human BACE was done using primers 5′-GGCAGCTGTCCAGCACATAC-3′ (forward) and 5′-AATGATCATGCTCCCTCCGA-3′ (reverse). Control reactions were performed with primers specific for human GAPDH (glyceraldehyde-3-phosphate dehydrogenase): (5′-AGCCACATCGCTCAGACACC-3′ and 5′-TGAGGCTGTTGTCATACTTCTC-3′). The identity of all the amplified fragments was confirmed by cloning and sequencing.
Proteasome activity
Cell extracts were prepared basically as described by Rodgers and Dean [15]. Briefly, HEKAPP+ cells were harvested by scraping them in 500 μl of water containing 1 mM DTT (dithiothreitol) and snap-frozen on solid CO2. After three cycles of freezing and thawing, the samples were centrifuged at 14000 g for 30 min and an equal amount of 2×buffer A (100 mM Tris/40 mM KCl/1 mM magnesium acetate/2 mM DTT, pH 7.8) was added to the supernatant. The samples were stored at −20 °C. Proteasome activity was measured using the 20 S proteasome activity assay kit (Chemicon, Chandlers Ford, Eastleigh, Hants., U.K.) according to the manufacturer's protocol, and quantified using a 380/460 filter set with a fluorescence spectrophotometer [FluoroMax-2; Instruments SA (now Horiba Jobin Yvon Ltd), Stanmore, Middx., U.K.].
Annexin V/PI (propidium iodide) assay
Annexin V/PI analysis of cell death was carried out according to the manufacturer's (BD PharMingen, San Diego, CA, U.S.A.) instructions. Incubation of subconfluent cells with 10 μM Aβ1–42 (Bachem, Bubendorf, Switzerland) and/or 1 mM H2O2 was performed for the indicated time periods. Aβ1–42 stock solution was stored at −20 °C in hexafluoropropan-2-ol. The solvent was completely evaporated and the peptide dissolved in sterile double-distilled water at 1 mg/ml just before treatments. Where indicated, cells were pretreated for 24 h with 100 nM β-secretase inhibitor II (Calbiochem, San Diego, CA, U.S.A.). The double staining was observed under a fluorescence microscope using a dual filter set for FITC and rhodamine.
RESULTS
Effect of PPARγ on Aβ production
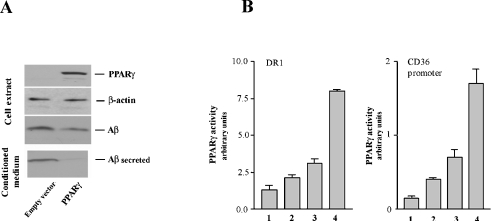
N2a cells stably transfected with human APP695 were transiently transfected to express PPARγ. The efficiency of transfections was monitored by immunoblotting (Figure 1A, upper panel). In line with other recently published work [14,15], overexpression of PPARγ resulted in a remarkable decrease in Aβ either in the cell extract or in the culture medium (Figure 1A, lower panels). The activity of the PPARγ construct was confirmed by luciferase assay on two distinct PPARγ response elements: DR1 [12] and a specific CD36 promoter region [11] (Figure 1B).
Figure 1. Effect of PPARγ overexpression on Aβ generation.
(A) N2a cells overexpressing human APP695 were transiently transfected with PPARγ-expressing vector. After 48 h transfection, cells were processed for immunoblot analysis. Aβ was immunoprecipitated from cell extracts and culture media and immunodetected with 4G8 antibody reactive to amino acid residues 17–24 of human Aβ. (B) The luciferase assay was performed as described in the Materials and methods section. Lanes are as follows: 1, empty vector; 2, empty vector+50 μM troglitazone; 3, PPARγ-expressing vector; 4, PPARγ-expressing vector+50 μM troglitazone. Troglitazone treatment was performed for 24 h. Results are means±S.D. for three independent experiments.
Effect of PPARγ on APP expression and secretase activities
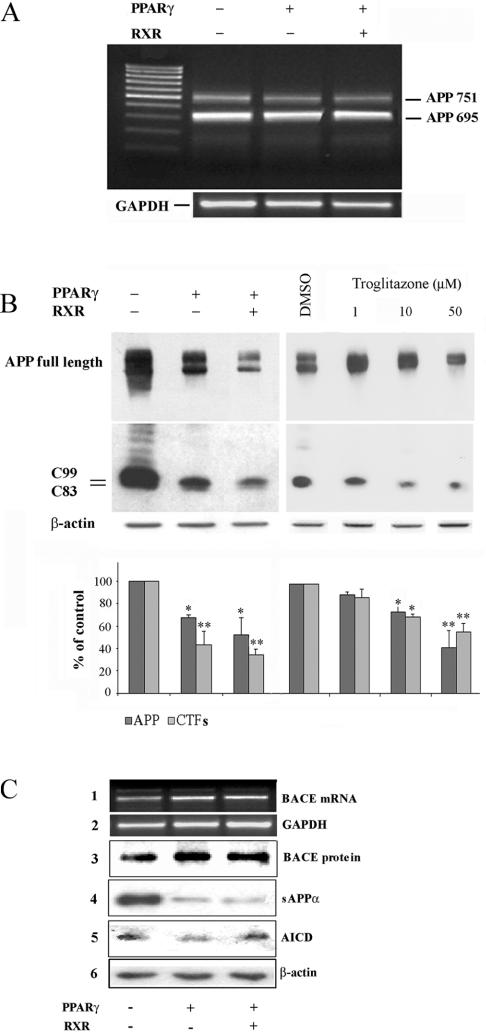
We performed RT-PCR (reverse-transcription PCR) and immunoblot analyses in order to test whether PPARγ could somehow interfere with the expression of APP. Figure 2(A) shows that, in HEKAPP+ cells, APP mRNA levels were not modulated by PPARγ, either in the presence or in the absence of its co-activator RXR. In contrast, the protein level of APP was significantly decreased by 35 and 50% in cells expressing PPARγ alone and PPARγ together with RXR respectively. A similar result was obtained by activating the endogenous PPARγ with troglitazone, a well established PPARγ activator drug [16], definitely implying a post-transcriptional event, PPARγ-mediated, affecting APP expression (Figure 2B, upper panels).
Figure 2. Effect of PPARγ on APP processing.
(A) RT-PCR analysis of APP expression in HEKAPP+ cells transiently transfected with PPARγ alone or together with RXR. Primer sequences for human APP are given in the Materials and methods section. APP751 bands represent the endogenous APP isoform. (B) HEKAPP+ cells transiently transfected with PPARγ alone and together with RXR (left panels) or treated with troglitazone for 24 h (right panels), were processed for total protein extraction. SDS/PAGE was performed with a gradient gel (4/10/15% polyacrylamide). After blotting of the proteins on nitrocellulose membrane, APP and C-terminal fragments (CTFs) were immunodetected using a polyclonal antibody (Zymed) recognizing the C-terminus of the human APP. The β-actin signal represents the internal loading control. Results are expressed as means±S.D. for three independent experiments. *P<0.05 and **P<0.01 versus the corresponding control. (C) HEKAPP+ cells transiently transfected with PPARγ and, where indicated, with RXR, were processed for total protein and RNA extraction. BACE expression was analysed by RT-PCR (panel 1) and by immunoblotting (panel 3); sAPPα in the cell-culture medium was detected by immunoblotting (6E10 antibody) (panel 4); a band apparently corresponding to the AICD fragment was detected by immunoblotting using the Zymed antibody (panel 5). GAPDH and β-actin signals (panels 2 and 6) represent the internal loading control.
APP is cleaved either by the α- or by the β-secretase, generating the APP C-terminal fragments (C83 or C99 respectively), remaining in the cell membrane. Figure 2(B) shows that, in HEKAPP+ cells, the decrease in APP full-length protein, induced either by PPARγ expression or troglitazone, is paralleled by a decrease in both the C83 and the C99 fragment. This result may exclude the notion of an inhibitory effect of PPARγ on the proteolytic activity of α- and β-secretases, which would likely increase the protein level of APP rather than decrease it. However, previous reports indicated that PPARγ agonists reduce Aβ secretion by lowering BACE expression and activity, although the direct evidence that BACE inhibition occurs in cells overexpressing PPARγ was not proven [15]. To investigate this issue further, we performed RT-PCR and immunoblot analyses of BACE, finding no effect of PPARγ on the expression of the β-secretase (Figure 2C, panels 1 and 3).
It could be argued that the simultaneous decrease in the APP protein level and Aβ secretion may represent the result of α-secretase activation. Although PPARγ was not able to increase the C83 fragment (Figure 2B), we further analysed the sAPPα (APP secreted by the cell) after the α-secretase cleavage. As expected, immunoprecipitation of sAPPα from the conditioned medium of HEKAPP+ cells expressing PPARγ was notably decreased, demonstrating that the activation of α-sectretase does not take place (Figure 2C, panel 4).
The C83 and C99 fragments, produced by α and β-secretase activity, are further cleaved by γ-secretase to generate the AICD (cytoplasmic APP intracellular domain). Our results, indicating a reduction of C83, C99, and APP full-length protein, already suggest that PPARγ does not affect the activity of γ-secretase. However, because it has been shown that agonists of PPARγ inhibit γ-secretase [17,18], we evaluated the generation of the AICD in our cell system. The result shown in Figure 2(C), panel 5, indicated that the immunodetected band, apparently corresponding to the AICD fragment, is not decreased by PPARγ expression, as previously confirmed also by others [14].
Overexpression of PPARγ increases APP ubiquitination
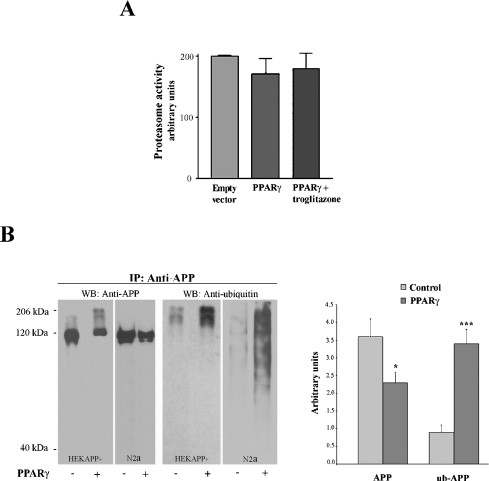
Several studies suggested that the proteasome could contribute to the pathogenesis of AD [19,20]. Thus we investigated the possibility that PPARγ expression stimulates an alternative, proteasome-dependent, processing of APP, consequently decreasing the production of Aβ. PPARγ was transfected into HEKAPP+ cells and optionally treated with troglitazone; nevertheless, the 20 S proteasome activity, measured with a commercially available kit (see the Materials and methods section), turned out not to be significantly modulated by PPARγ expression (Figure 3A). Because proteasome-dependent degradation pathways involve multiple steps, including ubiquitination of proteins targeted for proteolysis, we next investigated the effect of PPARγ expression on the formation of ubiquitinated APP. HEKAPP+ and N2a cell extracts were immunoprecipitated with a polyclonal anti-(human APP) antibody. Immunoprecipitated proteins were analysed by immunoblotting with anti-APP or anti-ubiquitin antibody. The results shown in Figure 3(B) indicated that, in PPARγ-transfected cells, the decrease of APP full-length protein is accompanied by a 3.4-fold induction of APP ubiquitination (P=0.001). The fact that the anti-APP antibody hardly recognized the ubiquitinated APP is probably due to the lack of affinity for the modified protein.
Figure 3. Effect of PPARγ on proteasome activity and APP ubiquitination.
(A) HEKAPP+ cells were transfected as indicated and the proteasome activity was measured as described in the Materials and methods section. The histogram shows the mean±S.D. for three independent experiments. (B) HEKAPP+ and N2a-overexpressing human APP695 were transfected with PPARγ for 48 h and then APP was immunoprecipitated from the cell extracts using the polyclonal Zymed anti-APP antibody. Immunoblot analysis was performed with 22C11 anti-APP or P4D1 anti-ubiquitin monoclonal antibodies. The histogram shows the mean±S.D. for two independent experiments. *P<0.05 and ***P=0.001 versus the corresponding control.
PPARγ protects from Aβ mediated H2O2-induced cell death
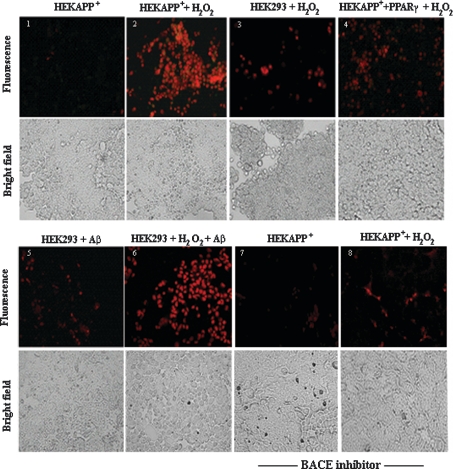
There is compelling evidence that Aβ peptide has cytotoxic effects. HEKAPP+ cells, despite the large amount of Aβ produced, show an apparent normal and healthy growth. However, if compared with HEK293 cells, in which APP is weakly expressed, HEKAPP+ were much more prone to H2O2-induced necrosis (Figure 4, panels 1–3). Interestingly, overexpression of PPARγ decreased the susceptibility of HEKAPP+ to H2O2 (Figure 4, panel 4), suggesting a role of Aβ as mediator of the H2O2-induced cell damage. To test our hypothesis, we exposed HEK293 cells to H2O2 and Aβ peptide, and found that the two compounds, added together to the culture medium, increased cell death in a synergistic manner (Figure 4, panels 3, 5 and 6). Moreover, and consistent with the above results, HEKAPP+ cells pretreated for 24 h with a specific BACE inhibitor [21,22] showed a strong protection against the damage induced by H2O2 (Figure 4, panels 7 and 8). The assay used for the cell-death analysis is based on the ability of annexinV (green fluorescence) to bind to the phosphatidylserine exposed on the surface of the cells undergoing apoptosis and the ability of propidium iodide (red fluorescence) to enter cells which have lost their membrane integrity [23]. It must be noted that, under the tested conditions, no sign or evidence of apoptosis was observed.
Figure 4. Aβ increases the cell necrosis induced by H2O2.
Cells were treated, where indicated, with 1 mM H2O2 and/or 10 μM Aβ1–42 for 24 h. Cells in panels 7 and 8 were additionally pretreated with BACE inhibitor (100 nM) for 24 h. Cell-death analysis was performed as described in the Materials and methods section. Necrotic cells show red fluorescence. Images were captured at ×20 magnification. The Figure is representative of three independent experiments all showing essentially similar results.
DISCUSSION
The present study provides evidence that PPARγ decreases Aβ secretion by increasing the rate of APP degradation. Previous reports indicated that proteasome inhibitors increase Aβ secretion [24,25]. This observation, together with the finding that PPARγ induces proteasome-dependent degradation of proteins such as cyclin D and oestrogen receptor-α [26], supported the hypothesis of an alternative, PPARγ-mediated, APP processing mechanism involving the proteasome system. Our experiments demonstrate that PPARγ overexpression does not directly stimulate the proteasome activity, but significantly (P=0.001) increases the ubiquitination level of APP.
In order to explain the recently observed PPARγ anti-amyloidogenic properties, contradictory mechanisms have been proposed. Sastre and colleagues [10] showed that the Aβ downregulation induced by PPARγ is a consequence of β-secretase inhibition; Camacho et al. [11] demonstrated that high levels of PPARγ decrease Aβ without altering either the β- or the γ-secretase activity, suggesting the existence of a not-well-identified extracellular clearance mechanism. In any event, our results clearly indicate that PPARγ decreases the cellular content of APP without affecting its mRNA level, thus implying a post-transcriptional event, unlikely to be linked to the inhibition of the secretase pathways. Importantly, the evidence that PPARγ stimulates the ubiquitination of APP supports the fact that the Aβ-lowering effect of PPARγ is due to the proteasome-mediated degradation of APP.
Another issue in the present study is the finding that PPARγ, by decreasing Aβ secretion, protects the cells against H2O2-mediated necrosis. Our results show, for the first time, that Aβ peptide makes the cell more susceptible to external stresses, without being, in itself, highly cytotoxic.
Additional detailed experiments are needed to elucidate the complete picture, firstly by investigating the mechanism by which Aβ mediates the cell damage induced by H2O2 and clarifying whether it is a phenomenon strictly related to oxidative events, or represents a more generic process. However, in the light of the present evidence, it can be assumed that the neuroprotective effects shown by certain NSAIDs, agonists of PPARγ [27], are not related to the inhibition of the Aβ-induced inflammatory process, but more likely to the capability of decreasing Aβ levels.
The fact that PPARγ agonists have been used for years in the treatment of Type II diabetes [7] raises the possibility that PPARγ could also soon become a drug target for the treatment of AD.
Acknowledgments
This work was supported by grants from COFIN 2004 (no. 2004068928_003), FIRB 2001 (National Co-ordinator G. Poli) and Genoa University (ex 60%). We thank Professor Peter Davies and Dr Riccardo Ruffini (Rapallo, Genoa, Italy) for advice and support.
References
- 1.Rogers J., Kirby L. C., Hempelman S. R., Berry D. L., McGeer P. L., Kaszniak A. W., Zalinski J., Cofield M., Mansukhani L., Willson P., et al. Clinical trial of indomethacin in Alzheimer's disease. Neurology. 1993;43:1609–1611. doi: 10.1212/wnl.43.8.1609. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama H., Barger S., Barnum S., Bradt B., Bauer J., Cole G. M., Cooper N. R., Eikelenboom P., Emmerling M., Fiebich B. L., et al. Inflammation and Alzheimer's disease. Neurobiol. Aging. 2000;21:383–421. doi: 10.1016/s0197-4580(00)00124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.in t' Veld B. A., Ruitenberg A., Hofman A., Launer L. J., van Duijn C. M., Stijnen T., Breteler M. M., Stricker B. H. Nonsteroidal antiinflammatory drugs and the risk of Alzheimer's disease. N. Engl. J. Med. 2001;345:1515–1521. doi: 10.1056/NEJMoa010178. [DOI] [PubMed] [Google Scholar]
- 4.Lim G. P., Yang F., Chu T., Chen P., Beech W., Teter B., Tran T., Ubeda O., Ashe K. H., Frautschy S. A., Cole G. M. Ibuprofen suppresses plaque pathology and inflammation in a mouse model for Alzheimer's disease. J. Neurosci. 2000;20:5709–5714. doi: 10.1523/JNEUROSCI.20-15-05709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim G. P., Yang F., Chu T., Gahtan E., Ubeda O., Beech W., Overmier J. B., Hsiao-Ashec K., Frautschy S. A., Cole G. M. Ibuprofen effects on Alzheimer pathology and open field activity in APPsw transgenic mice. Neurobiol. Aging. 2001;22:983–991. doi: 10.1016/s0197-4580(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 6.Weggen S., Eriksen J. L., Das P., Sagi S. A., Wang R., Pietrzik C. U., Findlay K. A., Smith T. E., Murphy M. P., Bulter T., et al. A subset of NSAIDs lower amyloidogenic Aβ42 independently of cyclooxygenase activity. Nature (London) 2001;414:212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 7.Willson T. M., Brown P. J., Sternbach D. D., Henke B. R. The PPARs: from orphan receptors to drug discovery. J. Med. Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- 8.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlmann T., Evans R. M. Nuclear receptors in Sicily: all in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- 10.Sastre M., Dewachter I., Landreth G. E., Willson T. M., Klockgether T., van Leuven F., Heneka M. T. Nonsteroidal anti-inflammatory drugs and peroxisome proliferator-activated receptor-γ agonists modulate immunostimulated processing of amyloid precursor protein through regulation of β-secretase. J. Neurosci. 2003;23:9796–9804. doi: 10.1523/JNEUROSCI.23-30-09796.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camacho I. E., Serneels L., Spittaels K., Merchiers P., Dominguez D., De Strooper B. Peroxisome proliferator-activated receptor γ induces a clearance mechanism for the amyloid-β peptide. J. Neurosci. 2004;24:10908–10917. doi: 10.1523/JNEUROSCI.3987-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagi S. A., Weggen S., Eriksen J., Golde T. E., Koo E. H. The non-cyclooxygenase targets of non-steroidal anti-inflammatory drugs, lipoxygenases, peroxisome proliferator-activated receptor, inhibitor of κB kinase, and NFκB, do not reduce amyloid β42 production. J. Biol. Chem. 2003;278:31825–31830. doi: 10.1074/jbc.M303588200. [DOI] [PubMed] [Google Scholar]
- 13.Ricciarelli R., Zingg J. M., Azzi A. Vitamin E reduces the uptake of oxidized LDL by inhibiting CD36 scavenger receptor expression in cultured aortic smooth muscle cells. Circulation. 2000;102:82–87. doi: 10.1161/01.cir.102.1.82. [DOI] [PubMed] [Google Scholar]
- 14.Juge-Aubry C., Pernin A., Favez T., Burger A. G., Wahli W., Meier C. A., Desvergne B. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5′-flanking region. J. Biol. Chem. 1997;272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- 15.Rodgers K. J., Dean R. T. Assessment of proteasome activity in cell lysates and tissue homogenates using peptide substrates. Int. J. Biochem. Cell Biol. 2003;35:716–727. doi: 10.1016/s1357-2725(02)00391-6. [DOI] [PubMed] [Google Scholar]
- 16.Marx N., Duez H., Fruchart J. C., Staels B. Peroxisome proliferator-activated receptors and atherogenesis: regulators of gene expression in vascular cells. Circ. Res. 2004;94:1168–1178. doi: 10.1161/01.RES.0000127122.22685.0A. [DOI] [PubMed] [Google Scholar]
- 17.Weggen S., Eriksen J. L., Sagi S. A., Pietrzik C. U., Ozols V., Fauq A., Golde T. E., Koo E. H. Evidence that nonsteroidal anti-inflammatory drugs decrease amyloid β42 production by direct modulation of γ-secretase activity. J. Biol. Chem. 2003;278:31831–31837. doi: 10.1074/jbc.M303592200. [DOI] [PubMed] [Google Scholar]
- 18.Eriksen J. L., Sagi S. A., Smith T. E., Weggen S., Das P., McLendon D. C., Ozols V. V., Jessing K. W., Zavitz K. H., Koo E. H., Golde T. E. NSAIDs and enantiomers of flurbiprofen target γ-secretase and lower Aβ42 in vivo. J. Clin. Invest. 2003;112:440–449. doi: 10.1172/JCI18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Checler F., da Costa C. A., Ancolio K., Chevallier N., Lopez-Perez E., Marambaud P. Role of the proteasome in Alzheimer's disease. Biochim. Biophys Acta. 2000;1502:133–138. doi: 10.1016/s0925-4439(00)00039-9. [DOI] [PubMed] [Google Scholar]
- 20.Nunan J., Williamson N. A., Hill A. F., Sernee M. F., Masters C. L., Small D. H. Proteasome-mediated degradation of the C-terminus of the Alzheimer's disease β-amyloid protein precursor: effect of C-terminal truncation on production of β-amyloid protein. J. Neurosci. Res. 2003;74:378–385. doi: 10.1002/jnr.10646. [DOI] [PubMed] [Google Scholar]
- 21.Abbenante G., Kovacs D. M., Leung D. L., Craik D. J., Tanzi R. E., Fairlie D. P. Inhibitors of β-amyloid formation based on the β-secretase cleavage site. Biochem. Biophys. Res. Commun. 2000;268:133–135. doi: 10.1006/bbrc.2000.2098. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto R., Yoneda S., Hara H. Neuroprotective effects of β-secretase inhibitors against rat retinal ganglion cell death. Neurosci. Lett. 2004;370:61–64. doi: 10.1016/j.neulet.2004.07.087. [DOI] [PubMed] [Google Scholar]
- 23.Vermes I., Haanen C., Steffens-Nakken H., Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J. Immunol. Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 24.Marambaud P., Lopez-Perez E., Wilk S., Checler F. Constitutive and protein kinase C-regulated secretory cleavage of Alzheimer's β-amyloid precursor protein: different control of early and late events by the proteasome. J. Neurochem. 1997;69:2500–2505. doi: 10.1046/j.1471-4159.1997.69062500.x. [DOI] [PubMed] [Google Scholar]
- 25.Flood F., Murphy S., Cowburn R. F., Lannfelt L., Walker B., Johnston J. A. Proteasome-mediated effects on amyloid precursor protein processing at the γ-secretase site. Biochem. J. 2005;385:545–550. doi: 10.1042/BJ20041145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin C., Burghardt R., Smith R., Wormke M., Stewart J., Safe S. Peroxisome proliferator-activated receptor γ agonists induce proteasome-dependent degradation of cyclin D1 and estrogen receptor α in MCF-7 breast cancer cells. Cancer Res. 2003;63:958–964. [PubMed] [Google Scholar]
- 27.Combs C. K., Johnson D. E., Karlo J. C., Cannady S. B., Landreth G. E. Inflammatory mechanisms in Alzheimer's disease: inhibition of β-amyloid-stimulated proinflammatory responses and neurotoxicity by PPARγ agonists. J. Neurosci. 2000;20:558–567. doi: 10.1523/JNEUROSCI.20-02-00558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]