Abstract
The lbp gene, which encodes a laminin-binding protein (Lbp) of Streptococcus pyogenes, was found in all S. pyogenes M types. An Lbp-deficient mutant showed a significantly lower efficiency of adhesion to HEp-2 cells than did the wild-type strain. These results indicate that Lbp is one of the important S. pyogenes adhesins.
Streptococcus pyogenes (group A streptococcus) is a gram-positive pathogenic bacterium that causes pharyngitis, impetigo, scarlet fever, and streptococcal toxic shock-like syndrome (TSLS) (23). These diseases are initiated by the adhesion of S. pyogenes to epithelial cells in the upper respiratory tract or skin. In the process of adhesion, extracellular matrix proteins such as fibronectin (Fn) and laminin (Lm) serve as mediators between the bacteria and host cells (2, 3). Previous studies have demonstrated that Fn-binding proteins of S. pyogenes, including protein F1/SfbI (21, 26), protein F2 (9), SfbII (15), FBP54 (1), PFBP (19), and Fba (28), function as adhesins and invasins. S. pyogenes has been reported to bind to Lm (25); however, its Lm-binding protein has not been identified, although Hytönen et al. recently suggested that the Lm-binding activity of S. pyogenes is mediated by SpeB (8).
Several streptococcal strains possess Fn-binding proteins and other adhesins (10-13), which are present in membrane- or cell-associated form. These proteins are linked either to the bacterial cell wall with the C-terminal region harboring an LPXTG motif (5, 20) or to the cell membrane with the N-terminal region harboring an LXXC or XXGC motif (24). Recently, the complete genome sequence of S. pyogenes strain SF370 (M type 1) has been deposited in the GenBank database (accession number AE004092) (4). Using this genome database, we found a novel open reading frame (ORF) containing the XXGC motif in the N-terminal region. These typical sequences are known as the signal peptidase II cleavage site of cell surface lipoprotein (12, 16, 22). In this report, we identify a novel gene encoding a cell surface protein and characterize the role of this protein in bacterial adhesion.
ORF analysis of the complete genome sequence.
The bacterial strains and plasmids used in this study are described in Table 1. Based on the complete genome sequence of S. pyogenes, the putative ORF sequences were translated into amino acid sequences with the help of GeneWorks software (IntelliGenetics, Campbell, Calif.). When we searched for genes harboring an XXGC motif in the N terminus, one ORF was found and designated lbp (the gene encoding the Lm-binding protein of group A streptococci). The nucleotide sequences of strains SSI-9 (M1) and SSI-1 (M3) were then determined by using an ABI PRISM 310 DNA sequencer (PE Applied Biosystems, Foster City, Calif.), and sequencing reactions were performed by the Sanger dideoxy-chain termination method. The lbp gene was found to consist of 921 nucleotides and encode a protein of 306 amino acids (designated Lbp) with a calculated molecular mass of ∼34.1 kDa. A putative signal peptidase cleavage site was revealed between amino acids 16 and 17 in the N-terminal region by using a method described previously (29). An alignment analysis of the deduced amino acid sequences of Lbp from strains SSI-9 (M1), SF370 (M1) (4), and SSI-1 (M3) showed that Lbp is 100% conserved. The high degree of similarity (98%) seen between Lbp and the Lm-binding protein of S. agalactiae, Lmb (22), strongly indicates that Lbp possesses Lm-binding activity.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain and plasmid | Relevant characteristics | Reference |
|---|---|---|
| S. pyogenes strains | ||
| SSI-9 | M type 1, isolated from patient with TSLS | T. Murai and Y. Shimizu |
| SSI-1 | M type 3, isolated from patient with TSLS | T. Murai and Y. Shimizu |
| #42 | M type 12, isolated from patient with TSLS | H. Watanabe |
| TR-7 | Isogenic mutant of SSI-9; derivative of pYT1088; lbp::aphA3 Kmr | This study |
| SE strains | Clinical isolates in Japan | Saga Prefectural Institute of Public Health |
| MJ strains | Clinical isolates in Japan | Osaka Prefectural Institute of Public Health |
| Others | Laboratory collection | |
| Other streptococcal strains | ATCCa | |
| Plasmids | ||
| pGEX-6P-1 | Expression vector; Ampr | Amersham Pharmacia Biotech |
| pSF151 | Suicide vector for insertional mutagenesis; Kmr | 27 |
| pYT1088 | pSF151 with lbp from SSI-9; Kmr | This study |
| pYT1097 | pGEX-6P-1 with lbp from SSI-9; Ampr | This study |
ATCC, American Type Culture Collection.
Distribution of lbp among streptococci.
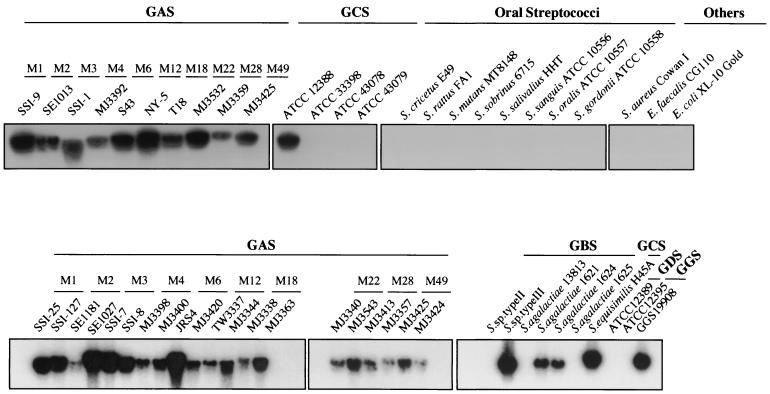
The distribution of lbp among a variety of streptococcal species was examined by Southern hybridization using lbp as a probe. Chromosomal DNA samples were digested with EcoRI and subjected to hybridization. Lbp was detected in strains M1, -2, -3, -4, -6, -12, -18, -22, -28, and -49 of S. pyogenes, as well as in some group B, C, D, and G strains (Fig. 1). Furthermore, an lbp-specific PCR analysis of various strains demonstrated that lbp is found in all M type strains of S. pyogenes (data not shown); however, lbp was not detected in oral streptococci.
FIG. 1.
Chromosomal DNAs from group A (GAS), B (GBS), C (GCS), D (GDS), and G (GGS) and oral streptococci were purified with a Puregene DNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.). DNA was digested with EcoRI and subjected to Southern hybridization analysis. The lbp gene was employed as a probe.
Lbp is a novel Lm-binding protein of S. pyogenes.
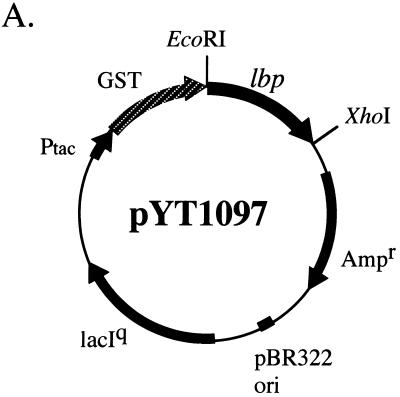
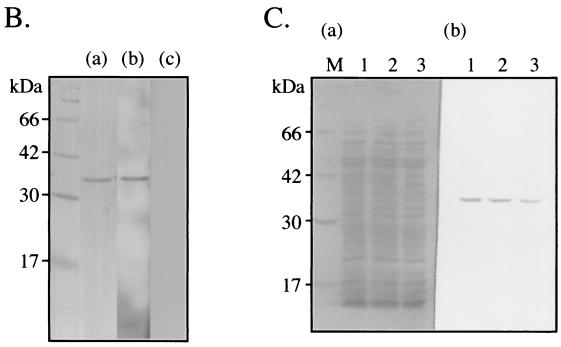
To determine whether Lbp possesses Lm-binding ability, recombinant Lbp (rLbp) was purified from the cell lysate of Escherichia coli BL21 harboring pYT1097 (Fig. 2A) by using glutathione Sepharose 4B affinity chromatography, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2B). rLbp, glutathione S-transferase (GST), and 8 M urea extracts of S. pyogenes were subjected to SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane, after which the membrane was incubated with 100 μg of biotinylated human Lm (Life Technology, Rockville, Md.) per ml. Biotinylated human Lm was prepared with an ECL protein biotinylation module (Amersham Pharmacia Biotech, Uppsala, Sweden). Lbp reacted with human Lm but not with GST, streptavidin only (Fig. 2B and 3B), Fn, or immunoglobulins (data not shown).
FIG. 2.
(A) Construction of the Lbp expression plasmid vector. The fragment containing codons 17 to 306 of the lbp gene was amplified from the SSI-9 (M1) genome as a template and inserted into pGEX-6P-1 (Amersham Pharmacia Biotech), which was then named pYT1097. The recombinant protein was lacking a signal peptide in the N-terminal region. (B) rLbp was purified by single-step affinity chromatography and immobilized on a PVDF membrane. (a) Coomassie brilliant blue staining. (b) Biotinylated human Lm solution (100 μg/ml) was added to the membrane. The reaction was developed with horseradish peroxidase (HRP)-labeled streptavidin. (c) Only HRP-labeled streptavidin was added. (C) Western blot analysis with rabbit anti-Lbp serum and urea extracts of S. pyogenes. Proteins were extracted from M1 (SSI-9), M3 (SSI-1), and M12 (#42) with 8 M urea (lanes 1 to 3, respectively). Samples were subjected to SDS-PAGE and then transferred to a PVDF membrane. (a) Coomassie brilliant blue staining. (b) Western blotting using rabbit anti-Lbp serum. Lane M contained molecular size markers.
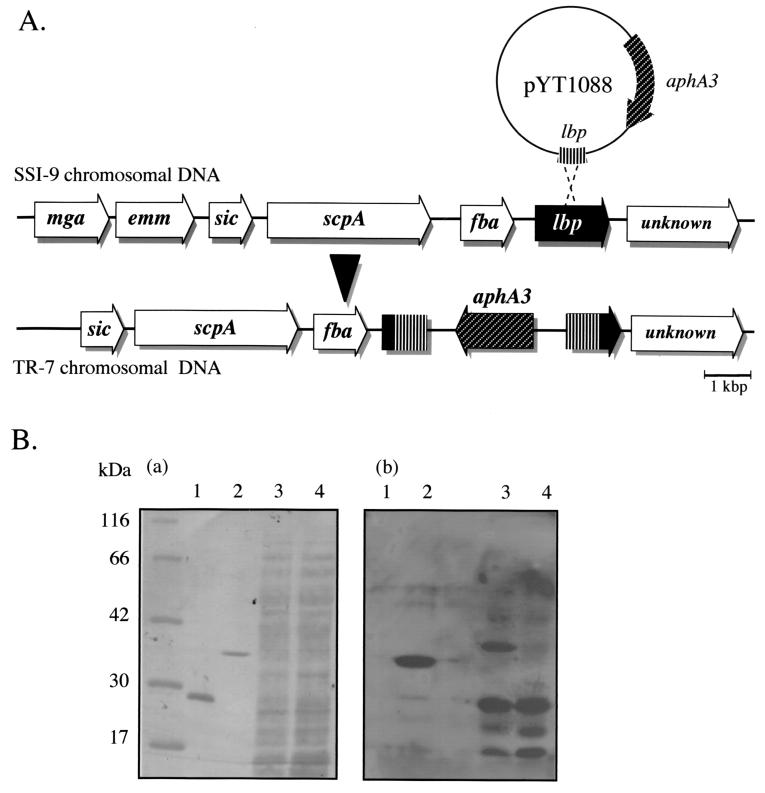
FIG. 3.
(A) Targeted mutagenesis of the lbp gene in S. pyogenes SSI-9. pYT1088 contains an internal fragment of lbp and a kanamycin resistance-encoding gene (aphA3), whereas mutant TR-7 was produced by single-crossover recombination. (B) Lm-binding assay with GST (lane 1), rLbp (lane 2), and 8 M urea extracts of strains SSI-9 (wild type, lane 3) and TR-7 (Δlbp, lane 4). Samples were separated by SDS-PAGE and transferred to a PVDF membrane. (a) Coomassie brilliant blue staining. (b) Biotinylated human Lm solution (100 μg/ml) and HRP-labeled streptavidin.
To investigate the function of Lbp, mutant strain TR-7 (Δlbp) was prepared as described below. The PCR product of an internal portion of lbp was ligated into a pGEM-T Easy vector (Promega, Madison, Wis.) and then digested with EcoRI, after which the fragments were cloned into vector pSF151 (27). The resultant plasmid, pYT1088, was transformed into strain SSI-9 by electroporation (Fig. 3A). Introduction of pYT1088 resulted in mutant strain TR-7 (Δlbp), which did not react with rabbit anti-Lbp antibody (data not shown). The 8 M urea extracts of strains SSI-9 (wild type) and TR-7 (Δlbp) were found to contain a 28-kDa Lm-binding protein (Fig. 3B). A recent report by Hytönen et al. (8) has indicated that SpeB is not only secreted from but also binds to bacterial cell surfaces and that the 28-kDa mature SpeB protein may mediate Lm binding. Furthermore, another 34-kDa Lm-binding protein (Lbp) was found in the extract of strain SSI-9 (Fig. 3B), while other, unknown, components (20 and 15 kDa) were shown to bind to Lm. These results clearly indicated that Lbp is a novel Lm-binding protein of S. pyogenes. Since 8 M urea extraction is a useful method by which to extract bacterial cell surface-associated proteins (6), cellular extracts of S. pyogenes in 8 M urea were used for Western blot analysis. Based on the results demonstrated in Fig. 2C and 3B, we speculated that Lbp is expressed on the cell surface of S. pyogenes. This organism expresses a variety of cell surface proteins with a specific ligand. Therefore, it may be hypothesized that Lbp eventually accelerates the infection of S. pyogenes by its binding to Lm in the underlying tissues once the organism adheres to and invades host cells.
Role of Lbp in cell adhesion of S. pyogenes.
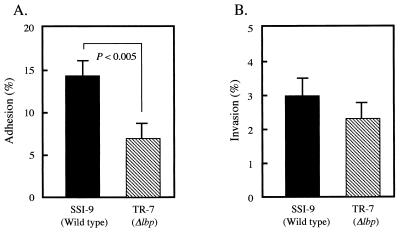
To investigate the role of Lbp of S. pyogenes, the adhesion and invasion efficiencies of Lbp-deficient mutant strain TR-7 were compared with those of parent strain SSI-9. Streptococcal adherence and invasion assays were performed as previously described (14). The TR-7 mutant demonstrated a significantly lower level of adhesion than wild-type strain SSI-9 (Fig. 4A; P < 0.005); however, the TR-7 mutant invaded HEp-2 cells as efficiently as did strain SSI-9 (Fig. 4B). These results suggest that Lbp works as an adhesin but not as an invasin.
FIG. 4.
Effects of Lbp on bacterial adhesion to and invasion of HEp-2 cells. Bacteria were suspended in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and then used to infect HEp-2 monolayers. HEp-2 cells were grown in a 24-well plate at a density of 5 × 104 per well. Approximately 107 bacteria were added to each well (multiplicity of infection, 1:200), and the plate was incubated for 3 h at 37°C. To determine bacterial adhesion, cells were washed with DMEM, lysed with 1 ml of sterile distilled water, and then plated to determine the number that were adhered to or invaded by S. pyogenes. (A) The percentage of adhesion or invasion was calculated as follows: (no. of CFU adhered to or invaded/total no. of CFU in inoculum) × 100. (B) Prior to bacterial invasion, cells were washed and incubated for 1 h in DMEM containing gentamicin (100 μg/ml) and penicillin (10 U/ml). Cells were washed, lysed, and plated for counting of those invaded by S. pyogenes. The percentage of invasion was calculated as follows: (no. of CFU invaded/total no. of CFU in inoculum) × 100. The results shown are the means ± the standard errors of the means of six wells (three experiments were performed in triplicate). Statistical analysis was performed with a nonparametric Mann-Whitney U test. All conclusions were based on a significance level of P < 0.005.
The mga, emm, sic, and scpA genes are known as members of the mga regulon. The fba and lbp genes are located downstream of the scpA gene in type M1 S. pyogenes. We investigated the transcriptional level of the Δmga mutant by reverse transcription-PCR analysis; however, transcriptional levels of mga-, emm-, sic-, scpA-, and fba-specific mRNAs were not detected in the Δmga mutant, although the lbp gene was not influenced (28). Furthermore, we found putative promoter sequences (−35 and −10), a ribosome-binding recognition site, and two pairs of inverted repeats as putative transcriptional terminators in the nucleotide sequence between the fba gene and the lbp gene. These findings suggested that the fba gene is a member of the mga regulon and that the expression of lbp mRNA is not controlled by the Mga regulator.
Common and highly conserved cell surface proteins for a protective antigen are highly regarded as an ideal vaccine against infection by S. pyogenes. In this regard, Fn-binding proteins were considered good candidates as a universal vaccine; however, a previous report showed that protein F1 was not present in the M1 and M3 strains among many M types of S. pyogenes (18). Our own recent study (28) has revealed that another Fn-binding protein, Fba, is expressed in M1, -2, and -4 and several other types but not in M3. M1 and M3 are the most common serotypes isolated from patients with TSLS and severe invasive diseases (17). There are more than 90 serotypes of the M protein, and protective antibodies are type specific (7). The results of the present study suggest the possibility that Lbp functions as a protective antigen against M1 and M3, as well as several other major serotypes of S. pyogenes. If Lbp could be an effective antigen, it might be useful as a universal vaccine against infection by a wide variety of S. pyogenes M types.
Nucleotide sequence accession number.
The sequence of the lbp gene was deposited in DDBJ/EMBL/GenBank under accession number AB040535.
Acknowledgments
This work was supported by grants from the Japan Society for Promotion of Science and the Ministry of Health, Labor and Welfare of Japan.
Editor: E. I. Tuomanen
REFERENCES
- 1.Courtney, H. S., Y. Li, J. B. Dale, and D. L. Hasty. 1994. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cue, D., P. E. Dombek, H. Lam, and P. P. Cleary. 1998. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect. Immun. 66:4593-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cue, D., S. O. Southern, P. J. Southern, J. Prabhakar, W. Lorelli, J. M. Smallheer, S. A. Mousa, and P. P. Cleary. 2000. A nonpeptide integrin antagonist can inhibit epithelial cell ingestion of Streptococcus pyogenes by blocking formation of integrin α5β1-fibronectin-M1 protein complexes. Proc. Natl. Acad. Sci. USA 97:2858-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. Mclaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from Gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 6.Hamada, S., T. Horikoshi, T. Minami, S. Kawabata, J. Hiraoka, T. Fujiwara, and T. Ooshima. 1991. Oral passive immunization against dental caries in rats by use of hen egg yolk antibodies specific for cell-associated glucosyltransferase of Streptococcus mutans. Infect. Immun. 59:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoe, N., D. K. Nakashima, D. Grigsby, X. Pan, S. J. Dou, S. Naidich, M. Garcia, E. Kahn, D. Bergmire-Sweat, and J. M. Musser. 1999. Rapid molecular genetic subtyping of serotype M1 group A streptococcus strains. Emerg. Infect. Dis. 5:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hytönen, J., S. Haataja, D. Gerlach, A. Podobielski, and J. Finne. 2001. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol. Microbiol. 39:512-519. [DOI] [PubMed] [Google Scholar]
- 9.Jaffe, J., S. Natanson-Yaron. M. G. Caparon, and E. Hanski. 1996. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Mol. Microbiol. 21:373-384. [DOI] [PubMed] [Google Scholar]
- 10.Jenkinson, H. F., and R. J. Lamont. 1997. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 8:175-200. [DOI] [PubMed] [Google Scholar]
- 11.Jenkinson, H. F. 1994. Cell surface protein receptors in oral streptococci. FEMS Microbiol. Lett. 121:133-140. [DOI] [PubMed] [Google Scholar]
- 12.Jenkinson, H. F. 1992. Adherence, coaggregation, and hydrophobicity of Streptococcus gordonii associated with expression of cell surface lipoproteins. Infect. Immun. 60:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joh, D., P. Speziale, S. Gurusiddappa, J. Manor, and M. Höök. 1998. Multiple specificities of staphylococcal and streptococcal fibronectin-binding microbial surface components recognizing adhesive matrix molecules. Eur. J. Biochem. 258:897-905. [DOI] [PubMed] [Google Scholar]
- 14.Kawabata, S., H. Kuwata, I. Nakagawa, S. Morimatsu, K. Sano, and S. Hamada. 1999. Capsular hyaluronic acid of group A streptococci hampers their invasion into human pharyngeal epithelial cells. Microb. Pathog. 27:71-80. [DOI] [PubMed] [Google Scholar]
- 15.Kreikemeyer, B., S. Talay, and G. S. Chhatwal. 1995. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Mol. Microbiol. 17:137-145. [DOI] [PubMed] [Google Scholar]
- 16.Lowe, A. M., P. A. Lambert, and A. W. Smith. 1995. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect. Immun. 63:703-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natanson, S., S. Sela, A. E. Moses, J. M. Musser, M. G. Caparon, and E. Hanski. 1995. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J. Infect. Dis. 171:871-878. [DOI] [PubMed] [Google Scholar]
- 19.Rocha, C. L., and V. A. Fischetti. 1999. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect. Immun. 67:2720-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 21.Sela, S., A. Aviv, A. Tovi, I. Burstein, M. G. Caparon, and E. Hanski. 1993. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10:1049-1055. [DOI] [PubMed] [Google Scholar]
- 22.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lütticken, and A. Podbielski. 1999. Lmb, a protein with similarities to LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens, D. L. 1992. Invasive group A streptococcus infections. Clin. Infect. Dis. 14:2-13. [DOI] [PubMed] [Google Scholar]
- 24.Sutcliffe, I., and R. R. B. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switalski, L. M., P. Speziale, M. Höök, T. Wadströme, and R. Timpl. 1984. Binding of Streptococcus pyogenes to laminin. J. Biol. Chem. 259:3734-3738. [PubMed] [Google Scholar]
- 26.Talay, S. R., P. Valentin-Weigand, K. N. Timmis, and G. S. Chhatwal. 1994. Domain structure and conserved epitopes of Sfb protein, the fibronectin-binding adhesin of Streptococcus pyogenes. Mol. Microbiol. 13:531-539. [DOI] [PubMed] [Google Scholar]
- 27.Tao, L., D. J. LeBlanc, and J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 28.Terao, Y., S. Kawabata, E. Kunitomo, J. Murakami, I. Nakagawa, and S. Hamada. 2001. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Mol. Microbiol. 42:75-86. [DOI] [PubMed]
- 29.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]