Abstract
Transfection technology for malaria parasites provides a valuable tool for analyzing gene function and correlating genotype with phenotype. Transfection models are even more valuable when appropriate animal models are available in addition to complete in vitro systems to be able to fully analyze parasite-host interactions. Here we describe the development of such a model by using the nonhuman primate malaria Plasmodium knowlesi. Blood-stage parasites were adapted to long-term in vitro culture. In vitro-adapted parasites could readapt to in vivo growth and regain wild-type characteristics after a single passage through an intact rhesus monkey. P. knowlesi parasites, either in vitro adapted or in vivo derived, were successfully transfected to generate circumsporozoite protein (CSP) knockout parasites by double-crossover mechanisms. In vitro-transfected and cloned CSP knockout parasites were derived in a time span of only 18 days. Microscopic evaluation of developing oocysts from mosquitoes that had fed on CSP knockout parasites confirmed the impairment of sporozoite formation observed in P. berghei CSP knockout parasites. The P. knowlesi model currently is the only malaria system that combines rapid and precise double-crossover genetic manipulation procedures with complete in vitro as well as in vivo possibilities. This allows for full analysis of P. knowlesi genotype-phenotype relationships and host-parasite interactions in a system closely related to humans.
The development of transfection technology for blood-stage malaria parasites (16, 23–25, 28, 29) is of great importance in the postgenomic era. It provides a direct way in which to correlate genotype with phenotype, and this enhances the further understanding of parasite biology. This will facilitate rational design of new vaccines and drugs, which are urgently needed to fight the malaria epidemic that kills annually between 1.5 and 2.7 million people, mainly young children, in Sub-Saharan Africa alone (7).
Four species of Plasmodium are natural to humans (9), and two of them, Plasmodium falciparum and Plasmodium vivax, are the most prevalent and important in terms of disease. Phylogenetically, P. falciparum, the more deadly form of the two, forms a separate clade with Plasmodium reichenowi, which causes chimpanzee malaria, and P. vivax clusters with simian malarias (11). The nonhuman primate malaria, caused by Plasmodium knowlesi, a natural parasite of Macaca fascicularis, has a relatively broad host range extending to humans, where it causes a mild disease (8). The parasite is closely related to P. vivax (11), and many genes identified in P. vivax have homologues in P. knowlesi. To date, transfection techniques developed for malaria parasite blood stages (26) include episomal transfection and targeted integration with linear constructs for the rodent parasite Plasmodium berghei (24, 25), episomal transfection and targeted integration with circular DNA for the human parasite P. falciparum (28, 29), and episomal transfection for the nonhuman primate malaria parasites P. knowlesi (23) and Plasmodium cynomolgi (16). Targeted integration experiments in P. falciparum, exploiting the results from the genome project (4, 12), are time-consuming, requiring several cycles of drug pressure to select for integration events based on a single crossover (28). In addition, this system offers only restricted in vivo possibilities in scarce New World primate sytems. The P. berghei system offers a rapid targeted integration regime through double crossover (24). It has, however, only limited in vitro possibilities for blood-stage parasites due to reticulocyte restriction, precluding in vitro transfection and selection procedures and thus, for example, the use of selectable markers that are toxic to the host.
P. knowlesi offers an especially powerful experimental system, since both the natural host, M. fascicularis, and an experimental host, Macaca mulatta, are phylogenetically close to humans. Genetic manipulation of this parasite species (23) offers the unique possibility to study parasite-host interactions in a system that is highly predictive for the human situation (14, 15). A complete in vitro transfection and selection system for this parasite would greatly enhance the experimental possibilities, since some analyses could be carried out without any requirement for primates. Furthermore, where host-parasite interactions are being studied, in vitro selection of the cloned parasite of the correct genotype can be assured before primate use is required. Publications on in vitro cultivation of P. knowlesi blood-stage parasites are scarce, and the reported technology is cumbersome (5, 22, 27). Culture medium was refreshed at least twice daily (5), red blood cells were added five times per week leading to up to 40% erythrocyte concentrations, and multiplication rates of <2.5 per 24-h lifecycle were reported (5, 27). These labor-intensive culture conditions do not allow the generation of enough parasites of good quality to perform transfection experiments.
We set out to develop a complete in vitro system for P. knowlesi to allow long-term in vitro culture and an extremely rapid procedure for in vitro transfection, selection, and cloning of transfected parasites. In addition, we further developed the in vivo and in vitro transfection technology for P. knowlesi to include targeted integration with linear constructs. This was demonstrated by targeting the circumsporozoite protein (CSP) locus and phenotypical analysis of the CSP knockout. The P. knowlesi system now is the only malaria system that combines rapid genetic manipulation procedures with complete in vitro as well as in vivo possibilities, allowing full analysis of genotype-phenotype and parasite-host relationships in a host closely related to humans.
MATERIALS AND METHODS
Parasites and animals.
P. knowlesi Nuri (21) and H strain (8) cryopreserved stocks were used to initiate blood-stage infection in rhesus monkeys by intravenous inoculation of 105 parasites. In vitro cultures were started with the same parasite strains.
Adult rhesus monkeys (M. mulatta) of either sex, each weighing over 4 kg, were used in these studies. They were infected with cryopreserved stocks of P. knowlesi, in vitro-adapted blood-stage P. knowlesi parasites, or blood from another rhesus monkey. All experimental animal work in these studies was carried out under protocols approved by the independent Institutional Animal Care and Use Committee and performed according to Dutch and European laws.
Long-term in vitro culture of P. knowlesi.
In vitro cultures were initiated with either cryopreserved or fresh P. knowlesi parasites isolated from an infected rhesus monkey. Rhesus red blood cells were used to culture P. knowlesi. White blood cells were removed by Plasmodipur filtration (Eurodiagnostica, Apeldoorn, The Netherlands), and red cells were stored for up to 2 weeks in RPMI at 4°C. Parasites were grown in static cultures at 36.5°C under reduced oxygen conditions (5% CO2, 5% O2, 90% N2) in RPMI 1640, supplemented with 20% pooled, heat-inactivated rhesus serum and rhesus erythrocytes at a 2.5% hematocrit. During the adaptation period medium was changed every 24 h, and fresh erythrocytes were added every 4 days to a maximum hematocrit of 5%. Once they had been established in culture, parasites were maintained under the same conditions except for medium changes every 48 h and subculturing when parasitemias exceeded 5%. Parasitemia was determined by microscopic examination of Giemsa-stained thick and thin films prepared from the cultured material at regular intervals. The culture-adapted parasites were subsequently adapted to growth in RPMI 1640 supplemented with 20% pooled, heat-inactivated human A or AB serum, by using a similar strategy. Parasites were cryopreserved at the young ring stage of development by standard protocols (19). Cloning of culture adapted P. knowlesi was performed by limiting dilution.
Transfection constructs.
All transfection constructs contained a heterologous selection cassette based on mutagenized Toxoplasma gondii dihydrofolate reductase/thymidine synthase gene (dhfr/ts) conferring pyrimethamine resistance, flanked by P. berghei dhfr/ts flanking sequences (17).
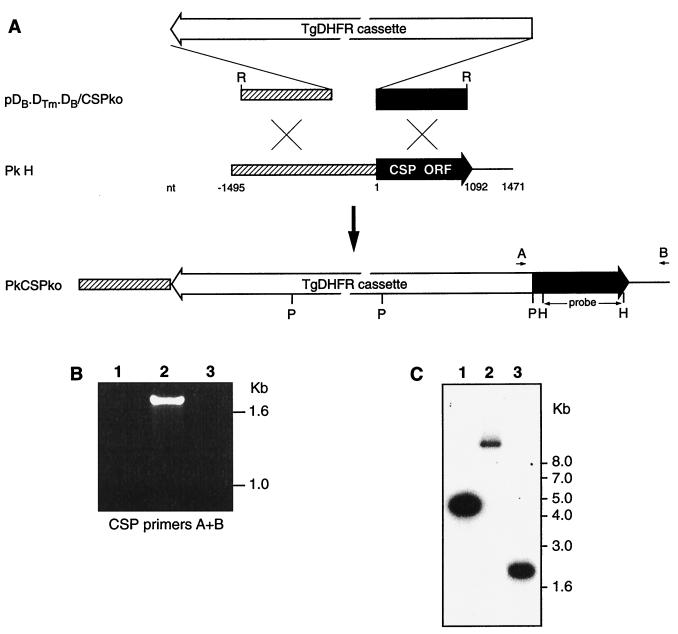
Sequences for P. knowlesi CSP locus were retreived from GenBank entries (accession numbers K00822 and M19749). To prepare a CSP knockout construct, 5′ and 3′ regions from CSP were amplified by PCR. These regions were as follows: 5′ nucleotides (nt) −1495 to −560 and 3′ nt 1 to 1089 (for numbering, nt 1 is the start of open reading frame [ORF]). Through a series of cloning steps plasmid pDB.DTm.DB/CSPko was derived (Fig. 1A). After purification on Plasmid Maxi columns (Qiagen), the vector backbone was excised from the construct by EcoRI digestion, and the linear DNA was used for transfection of P. knowlesi. Recombinant DNA manipulations and analyses were performed according to standard procedures (20).
FIG. 1.
DNA constructs and analysis of integration into P. knowlesi CSP locus after transfection. (A) Linear DNA construct designed for integration into the P. knowlesi CSP locus. The 5-kb selectable marker cassette (TgDHFR cassette) contains P. berghei dhfr/ts flanking regions controlling expression of mutagenized T. gondii dhfr/ts. Relevant restriction sites are indicated: R, EcoRI; P, PstI; H, HindIII; B, BamHI. (C) The location of the PCR primers A and B and the specific probe used for Southern blotting are shown. ORF-containing sequences are marked with an open arrow, and sequences used for targeted integration are indicated. (B) PCR analysis of transfected parasites. Lanes 1 to 3 show PCR with CSP integration-specific primers A and B. Lanes: 1, P. knowlesi H-strain DNA; 2, PkCSPko clone DNA; 3, pDB.DTm.DB/CSPko vector DNA. PkCSPko and the transfection construct were all positive for PCR with the two T. gondii dhfr/ts-specific primers (not shown). (C) Southern blot analysis of transfected parasites. PstI-digested DNA from P. knowlesi H strain (lane 2), PkCSPko clone (lane 3), and the transfection vector pDB.DTm.DB/CSPko (lane 1) was used to prepare a Southern blot. The blot was probed with a P. knowlesi CSP-specific probe (see panel A).
Transfection procedures and selection of transfected parasites.
Transfection of mature blood-stage schizonts of P. knowlesi H-strain derived from a donor rhesus monkey and in vivo selection of transfected parasites by using oral dosing of pyrimethamine were performed as described previously (23), except that pyrimethamine was given at 1 mg/kg of body weight daily for five consecutive days and thereafter every other day until the end of the experiment. For transfection of culture-adapted parasites, up to 2 ml (packed cell volume) of red cells containing between 0.5 × 109 and 1 × 109 mature schizont-infected red cells was washed with incomplete Cytomix (29), mixed with 100 μl of transfection construct containing 50 μg of DNA, and electroporated (Bio-Rad GenePulser) in three 700-μl aliquots at a pulse of either 1.5, 2, or 2.5 kV, with 25-μF capacitance and 200-Ω resistance (the time constant ranged from 0.7 to 1.1 ms). Samples electroporated under the same conditions were pooled, incubated on ice for 5 min, and used to initiate 20-ml in vitro cultures at a 10% hematocrit. Culture medium was refreshed daily for 3 days to remove parasite material resulting from dead parasites and lysed erythrocytes. After that, the medium was replenished every other day. Pyrimethamine selection (25 ng/ml [final concentration] from a stock solution in 0.5% lactic acid in phosphate-buffered saline) was started 24 h after culture inoculation and maintained throughout the culture period.
DNA analysis.
Total parasite DNA was isolated (Gentra Systems, Inc., Minneapolis, Minn.) directly from in vitro cultures or after Plasmodipur filtration of P. knowlesi-infected rhesus monkey blood according to the manufacturer’s instructions. Parasite DNA was analyzed through plasmid rescue by electroporation into E. coli, PCR, and Southern blotting according to standard procedures (20). The PCR primers used for confirming integration into the CSP locus were A (5′-GTGTCTATATTACCAACTC-3′) and B (5′-GTCAAAAAAGGGTCAGTCAAAAAGGG-3′) (Fig. 1A).
Transmission studies.
Rhesus monkeys were infected as described above, and Anopheles stephensi mosquitoes were allowed to feed for 10 min on the shaven chests of sedated monkeys 6 to 8 days after infection, when the parasitemia was between 0.5 and 3%. Unfed mosquitoes were removed, and engorged mosquitoes were maintained at 26°C and 80% relative humidity. From day 6 onward, midguts were dissected and examined by light microscopy (400× magnification) to monitor oocyst development and sporozoite formation within oocysts.
RESULTS
Long-term in vitro culture of P. knowlesi.
To improve the versatility of the P. knowlesi transfection system (23), in vitro protocols are required to allow the generation of genetically transformed parasites without the need for primates. Transformed parasites of the desired genotype can then be used in vivo to study biological questions at the parasite-host interface. Development of an in vitro transfection and selection protocol required establishment of stable, long-term in vitro culture of parasites. In vivo we have observed different growth characteristics between the H strain and the Nuri strain. Nuri strain reproducibly gives fulminating parasitemias with an average 15-fold multiplication rate that rises to >20% ca. 1 week after infection. Some rhesus monkeys infected with H strain showed similar parasite development but with a four- to fivefold multiplication rate. Others showed a rapid rise to ca. 5% parasitemia that subsequently developed into a chronic infection. P. knowlesi H strain and Nuri strain, originating from cryopreserved stocks or directly from an infected rhesus monkey, were both successfully adapted to in vitro culture by using RPMI 1640 supplemented with 20% rhesus serum and an incubation temperature of 36 to 37°C. After an initial lag phase of at least 3 weeks in which parasites were hardly detectable in the cultures, adapted parasites emerged that now had been in continuous culture for >18 months (Fig. 2). Routinely, three- to fourfold multiplication rates per 24 h were obtained for the H strain and slightly higher (four- to fivefold) for the Nuri strain parasites in static cultures, and parasitemias of up to 20% could easily be obtained. Culture-adapted H-strain parasites were subsequently adapted to growth in medium containing 20% human serum, in which similar growth rates were observed. The use of temperatures of 39°C (the normal temperature of rhesus monkeys) during cultivation slowed down the parasite growth, and temperatures of 36 to 37°C appeared to be the optimal growth temperature. Culturing parasites under shaking conditions or in a rollerbottle system did not dramatically improve growth rates, but rollerbottles now routinely supplied 1010 parasites from a 200-ml culture in a 2-liter flask. Finally, culture-adapted parasites were cloned by limiting dilution (Table 1), demonstrating the feasibility of the procedure required to obtain clonal parasite populations after transfection. The P. knowlesi H strain (either wild type or culture adapted) was used in subsequent transfection experiments.
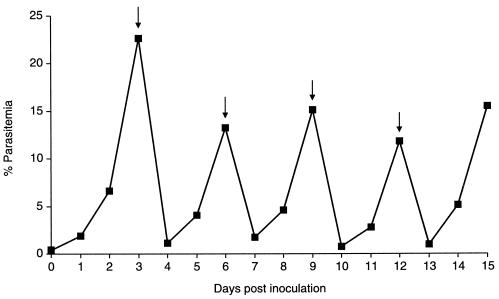
FIG. 2.
In vitro growth characteristics of P. knowlesi H strain. Parasitemia development over a 15-day period. Parasites were cultured as described in Materials and Methods, and after 3 months the parasitemia level was determined daily and expressed as a percentage of infected red blood cells. Arrows indicate the days on which the culture was diluted.
TABLE 1.
Cloning of in vitro culture-adapted P. knowlesi
| Seeding densitya | % Positive wellsb | % Probability of clonesc | No. of clones obtained |
|---|---|---|---|
| 100 | 100 | 0 | 0 |
| 20 | 100 | 0 | 0 |
| 4 | 100 | 0 | 0 |
| 0.8 | 75 | 46.2 | 0 |
| 0.16 | 21 | 91.7 | 5 |
| 0.032 | 4 | 98.4 | 1 |
Average number of parasites used to start cultures in 24 wells of a 96-well plate.
Cultures were maintained for 30 days before being scored as negative.
That is, the probability that parasites are derived from a single infected erythrocyte, as calculated according to the method of Gritzmacher and Reese (13).
Culture-adapted parasites can be readily readapted to in vivo growth.
To determine whether P. knowlesi H strain that had been maintained in culture for 11 months was still able to grow in rhesus monkeys, a rhesus monkey was infected with 105 parasites. Patent parasitemia was observed after 12 days, and a chronic low-level parasitemia developed with a peak parasitemia of 0.2%. On day 17, 1 ml of blood from this monkey was transferred to a second rhesus monkey, and this monkey developed a fulminating parasitemia from day 6 onward, reaching 19% parasitemia on day 10. Two further infections were initiated by using stocks from the first passage, resulting in peak parasitemias of 5% on day 8 and 1.2% on day 10, respectively. After the peak, the parasitemias declined in both rhesus. Two rhesus monkeys were infected with a stock from the second passage, and patency started on day 4, resulting in 4.2% parasitemia on day 7 and 6.3% parasitemia on day 9, respectively. At that time, the animals were bled to provide parasites for further transfection experiments. These results indicate that the in vivo H-strain phenotype of either a fulminant infection or a chronic infection after a ca. 5% peak parasitemia is obtained after one passage through an immunologically intact rhesus monkey. When these parasites were cultured in vitro again they immediately developed like in vitro-adapted parasites, without any adaptation period.
Transfected P. knowlesi can be readily selected in vitro.
By using previously determined transfection conditions (23), P. knowlesi schizonts, derived either from an infected rhesus monkey or from an in vitro culture, were transfected with plasmids containing the T. gondii dhfr/ts selection cassette. After transfection, parasites were maintained in 20-ml cultures, and pyrimethamine selection was applied from day 1 onward. The only transfection condition that consistently resulted in selection of transfected parasites was at 2.5 kV, 200 Ω, and 25 μF. Pyrimethamine-resistant parasites emerged in culture after 21 days for parasites derived from the monkey and after 8 days for in vitro-adapted parasites. Plasmid rescue from the resistant pools of parasites demonstrated the presence of plasmid DNA in an unrearranged form (not shown).
Targeted integration through double crossover can be obtained in vivo as well as entirely in vitro.
Previously, we reported transfection of P. knowlesi with plasmids that were maintained as episomes (23). To determine whether targeted integration in this system is feasible with linear constructs, as has been reported for P. berghei (24), an integration construct for the P. knowlesi CSP locus was prepared. The construct had 0.93 kb of P. knowlesi CSP 5′-untranslated region (UTR) sequence located 5′ of the selection cassette and 1.0 kb of P. knowlesi CSP ORF sequence located 3′ of the selection cassette (Fig. 1A). Parasites derived from a rhesus monkey were transfected with linearized constructs and selected in vivo as described above. In parallel, the CSP knockout construct was used to transfect in vitro cultured schizonts, and selection was performed in vitro. As with the episomal transfections, 2.5 kV consistently resulted in selection of pyrimethamine-resistant parasites. In vivo and in vitro, pyrimethamine-resistant parasites were detected on day 8. Plasmid rescue experiments from these parasites consistently failed to produce ampicillin-resistant E. coli, suggesting the absense of (recircularized) episomes within the transfected parasite populations. Cloning by limiting dilution of in vitro-selected transfectants was started on day 8, and clones were observed in thin films by day 18. Multiplication rates for these transfected parasites were about four times per 24 h in vivo and about three times per 24 h in vitro. PCR on in vitro-selected and cloned PkCSPko genomic DNA with primers A and B (Fig. 1 A) (amplifying a 1.7-kb P. berghei dhfr/ts 5′-UTR-CSP ORF region only present when targeted integration has occurred) showed integration into the CSP locus (Fig. 1B, lanes 1 to 3). This was also confirmed by Southern blotting with a 0.89-kb HindIII fragment (Fig. 1A) of the CSP ORF as a probe. As shown in Fig. 1C, lanes 1 to 3, the 10-kb wild-type PstI fragment from the CSP locus (lane 2) is converted to a 2.5-kb fragment in PkCSPko (lane 3) due to the introduction of PstI sites contained in the transfection construct.
CSP knockout P. knowlesi does not produce sporozoites in oocysts.
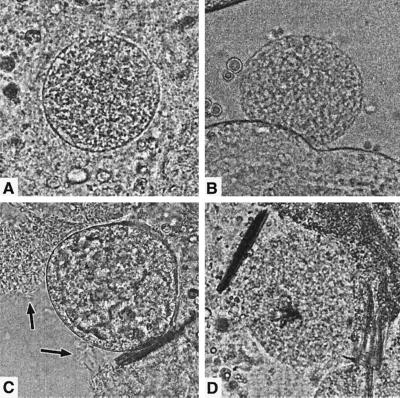
To examine the phenotype of CSP knockout P. knowlesi, rhesus monkeys were infected with either P. knowlesi H strain or the PkCSPko blood-stage parasites. The development of parasitemia in both monkeys was similar, showing a rapid rise to ca. 10% parasitemia on days 8 and 9 postinfection. A. stephensi mosquitoes were allowed to feed on day 8 on the wild-type infected monkey (0.9% parasitemia) and on days 6 and 7 on the PkCSPko-infected monkey (0.5 and 3% parasitemias, respectively). Fed mosquitoes were isolated and kept at 26°C to allow oocyst and sporozoite development. From day 6 postfeeding onward, mosquitoes were dissected and midguts were examined for the presence of oocysts and sporozoites. Day 7 feeding of the PkCSPko resulted in the highest transmission rates (100%), with ca. 50 oocysts per midgut. The wild-type feeding resulted in 83% infection and ca. 26 oocysts per midgut. Wild-type oocysts produced sporozoites in the hemocoel from day 8 postfeeding onward, as determined by lightmicroscopic examination of midguts (Fig. 3A and C). Spontaneously ruptured oocysts showed release of sporozoites, and rupture by applying gentle force to the coverslip also released sporozoites from the oocysts. As expected, due to rupture and release of sporozoites, the number of oocysts dramatically declined from days 8 to 11 postfeeding. Salivary gland sporozoites were not detected, since P. knowlesi sporozoites are not able to invade the salivary glands of A. stephensi (10). In contrast, in the PkCSPko oocysts no sporozoites developed, and this remained so until the experiment was stopped at a point when the great majority of the wild-type oocysts had matured and ruptured (Fig. 3B and D). The number of oocysts did not decline from days 8 to 12 as observed with wild-type oocysts. PkCSPko oocyst development otherwise appeared to be similar to that of wild-type oocysts, as observed by light microscopy (Fig. 3A and B), resulting in similar-sized oocysts at day 11.
FIG. 3.
Microscopic evaluation of wild-type and CSP knockout P. knowlesi oocyst development in A. stephensi midguts. (A) Intact wild-type oocyst 9 days postfeeding. (B) Intact CSP knockout oocyst 11 days postfeeding. (C) Wild-type oocyst releasing sporozoites (marked with arrows) 8 days postfeeding. (D) Force-induced rupture of CSP knockout oocyst 11 days postfeeding. Sporozoite release was never observed in CSP knockout oocysts.
DISCUSSION
The postgenomic analysis of the sequences now being obtained for a range of malaria-causing species will require versatile tools for genetic manipulation. Transfection permits targeted genomic manipulation of malaria parasites to characterize structural and functional relationships. Analysis of transfected parasites in a natural host-parasite setting is not possible with transfected human malaria parasites. In this respect P. knowlesi, a natural parasite of M. fascicularis (9), is a system with great potential because (i) the parasite is phylogenetically closely related to human malarial parasites, such as P. vivax (11) (in fact, the parasite can also infect humans [8]); (ii) animal hosts are available for experimentation (6, 14); (iii) animal hosts are primates, enabling analysis of transfectant phenotypes in systems closely related to humans (15); and (iv) we have developed in vivo transfection systems (23), allowing functional genome analysis.
We have now further improved and extended the P. knowlesi system in several ways to provide more widely available access to genetic manipulation of this parasite species. First, two P. knowlesi strains, Nuri and H, were adapted to long-term in vitro growth by using simple protocols. This is important since it obviates the need for primates as parasite donors to provide parasites for genetic manipulation. Second, by using protocols developed for in vivo selection of transfected parasites, transfected parasites were selected in vitro based on the T. gondii dhfr/ts gene providing resistance to pyrimethamine. This also obviates the need for primates as transfected parasite recipients during the initial selection for transfectants and allows development of new selectable markers, including those that would otherwise be toxic to the host, exploiting the benefits of complete in vitro selection.
Third, to determine whether gene targeting through double crossover was possible in P. knowlesi or whether the lengthy P. falciparum procedures needed to be adopted, transfection experiments with linear DNA constructs designed to target the CSP locus were performed. These experiments, performed in vivo as well as in vitro, showed that targeted integration could be obtained with linear constructs and entirely in vitro. Recently, by using similarly designed constructs, the P. knowlesi thrombospondin-related adhesive protein gene was also efficiently disrupted (H. Ozwara et al., unpublished results). The in vitro procedure allows for deriving cloned parasites within 18 days of transfection, demonstrating the relative speed of this procedure compared to in vitro P. falciparum procedures, and allowing high-throughput analysis of gene functions.
Recently, the P. knowlesi genome sequence to a threefold coverage has become available (http://www.sanger.ac.uk/Projects/P_knowlesi/). This greatly expands the possibilities for transfection studies since homologues from P. falciparum genes and from genes available from the expanding P. vivax gene sequence tag database (http://www.ncbi.nlm.nih.gov/Malaria/plasmodiumblcus.html) can now easily be identified in P. knowlesi.
Finally, we have shown that parasites that have been growing in vitro for a long period of time can readily readapt to in vivo growth, displaying wild-type H-strain growth characteristics after a single passage through an intact rhesus monkey. This observation is extremely important since in vitro-selected transfected parasites will be vital tools for studying host-parasite interactions. In vitro culture might lead to alteration of expression of SICAvar genes (1, 2, 3) and thus to loss of avoiding splenic clearence by sequestration, which can influence infectivity in nonsplenectomized rhesus monkeys. One passage through an intact rhesus is apparently sufficient to select for parasites that are readily able to infect rhesus monkeys and that can still be cultured in vitro.
In this study in a nonhuman primate malaria of the P. vivax type, we confirmed the earlier observations in a rodent malaria of the essential nature of CSP in sporozoite development (18). In P. knowlesi CSP is also critical for sporozoite formation in developing oocysts in mosquito midguts, and the lack of CSP expression results in absence of sporozoite formation. Unlike the situation in P. berghei, oocysts of the CSP knockout have similar morphology to wild-type oocysts, and the phenotype only becomes clear at the time of sporozoite release. This difference is mainly due to the fact that in P. knowlesi H-strain oocysts sporozoites are not visible “like rays from the sun” as described for P. berghei (18), and thus both wild-type and CSP knockout oocysts show an undefined morphology.
The P. knowlesi system now provides a unique malaria transfection system that allows for fast and simple in vitro genetic manipulations and combines this with the opportunity to perform in vivo studies in a nonhuman primate that is closely related to humans. It is the only malaria system described to date that is amenable to genetic manipulation that combines complete in vitro possibilities with studies in vivo in an animal model that is closely related to humans. This fact makes P. knowlesi highly suitable for the development of new antimalarial drugs and vaccines, as well as for studying basic biological questions in the postgenomic era at the parasite-host interface.
Acknowledgments
Clemens Kocken and Hastings Ozwara contributed equally to this work.
We thank John Barnwell for providing P. knowlesi H strain parasites and stimulating discussions and Wijnand Eling for provision of mosquitoes. We also thank Xander van der Linde, Geert-Jan van Gemert, and Willem Collignon for excellent technical assistance and members of the Animal Science Department of the BPRC for expert animal care.
This work was supported by EC DG XII contracts CT 97-9104 and CT99-10004, by ALW (805-33.332P and 809.35.004) of The Netherlands Organization for Scientific Research, and by PAD (9802.076.0) of the Dutch Health Research and Development Council. H.O. was funded in part by The Netherlands Foundation for the Advancement of Tropical Research (WOTRO).
Editor: J. M. Mansfield
REFERENCES
- 1.al-Khedery, B., J. W. Barnwell, and M. R. Galinski. 1999. Antigenic variation in malaria: a 3′ genomic alteration associated with the expression of a P. knowlesi variant antigen. Mol. Cell 3:131–141. [DOI] [PubMed] [Google Scholar]
- 2.Barnwell, J. W., R. J. Howard, H. G. Coon, and L. H. Miller. 1983. Splenic requirement for antigenic variation and expression of the variant antigen on the erythrocyte membrane in cloned Plasmodium knowlesi malaria. Infect. Immun. 40:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnwell, J. W., R. J. Howard, and L. H. Miller. 1982. Altered expression of Plasmodium knowlesi variant antigen on the erythrocyte membrane in splenectomized rhesus monkeys. J. Immunol. 128:224–226. [PubMed] [Google Scholar]
- 4.Bowman, S., D. Lawson, D. Basham, D. Brown, T. Chillingworth, C. M. Churcher, A. Craig, R. M. Davies, K. Devlin, T. Feltwell, S. Gentles, R. Gwilliam, N. Hamlin, D. Harris, S. Holroyd, T. Hornsby, P. Horrocks, K. Jagels, B. Jassal, S. Kyes, J. McLean, S. Moule, K. Mungall, L. Murphy, B. G. Barrell, et al. 1999. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature 400:532–538. [DOI] [PubMed] [Google Scholar]
- 5.Butcher, G. A. 1979. Factors affecting the in vitro culture of Plasmodium falciparum and Plasmodium knowlesi. Bull W. H. O. 57:17–26. [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher, G. A. 1996. Models for malaria: nature knows best. Parasitol. Today 12:378–382. [DOI] [PubMed] [Google Scholar]
- 7.Butler, D., J. Maurice, and C. O’Brien. 1997. Time to put malaria control on the global agenda. Nature 386:535–536. [DOI] [PubMed] [Google Scholar]
- 8.Chin, W., P. G. Contacos, G. R. Coatney, and H. R. Kimball. 1965. A naturally acquired quotidian-type malaria in man transferable to monkey. Science 149:865. [DOI] [PubMed] [Google Scholar]
- 9.Coatney, G. R., W. E. Collins, W. McWarren, and P. G. Contacos. 1971. The primate malarias. U.S. Government Printing Office, Washington, D.C.
- 10.Collins, W. E., P. G. Contacos, and E. G. Guinn. 1967. Studies on the transmission of simian malarias. II. Transmission of the H strain of Plasmodium knowlesi by Anopheles balabacensis balabacensis. J. Parasitol. 53:841–844. [PubMed] [Google Scholar]
- 11.Escalante, A. A., and F. J. Ayala. 1995. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc. Natl. Acad. Sci. USA 92:5793–5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner, M. J., H. Tettelin, D. J. Carucci, L. M. Cummings, L. Aravind, E. V. Koonin, S. Shallom, T. Mason, K. Yu, C. Fujii, J. Pederson, K. Shen, J. Jing, C. Aston, Z. Lai, D. C. Schwartz, M. Pertea, S. Salzberg, L. Zhou, G. G. Sutton, R. Clayton, O. White, H. O. Smith, C. M. Fraser, S. L. Hoffman, et al. 1998. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum Science 282:1126–1132. [DOI] [PubMed] [Google Scholar]
- 13.Gritzmacher, C. A., and R. T. Reese. 1984. Reversal of knob formation on Plasmodium falciparum-infected erythrocytes. Science 226:65–67. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy, R. C., M. H. Shearer, and W. Hildebrand. 1997. Nonhuman primate models to evaluate vaccine safety and immunogenicity. Vaccine 15:903–908. [DOI] [PubMed] [Google Scholar]
- 15.King, F. A., C. J. Yarbrough, D. C. Anderson, T. P. Gordon, and K. G. Gould. 1988. Primates. Science 240:1475–1482. [DOI] [PubMed] [Google Scholar]
- 16.Kocken, C. H., A. van der Wel, and A. W. Thomas. 1999. Plasmodium cynomolgi: transfection of blood-stage parasites using heterologous DNA constructs. Exp. Parasitol. 93:58–60. [DOI] [PubMed] [Google Scholar]
- 17.Kocken, C. H., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. van Gemert, X. van der Linde, L. H. Bannister, C. Janse, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119–15124. [DOI] [PubMed] [Google Scholar]
- 18.Menard, R., A. A. Sultan, C. Cortes, R. Altszuler, M. R. van Dijk, C. J. Janse, A. P. Waters, R. S. Nussenzweig, and V. Nussenzweig. 1997. Circumsporozoite protein is required for development of malaria sporozoites in mosquitoes. Nature 385:336–340. [DOI] [PubMed] [Google Scholar]
- 19.Rowe, A. W., E. Eyster, and A. Kellner. 1968. Liquid nitrogen preservation of red blood cells for transfusion: a low glycerol-rapid freeze procedure. Cryobiology 5:119–128. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Singh, J., A. P. Ray, and C. P. Nair. 1953. Isolation of a new strain of Plasmodium knowlesi. Nature 172:122. [DOI] [PubMed] [Google Scholar]
- 22.Trigg, P. I. 1967. Methods of in vitro culture of malaria parasites. Parasitology 57:3P–4P [PubMed] [Google Scholar]
- 23.van der Wel, A. M., A. M. Tomas, C. H. Kocken, P. Malhotra, C. J. Janse, A. P. Waters, and A. W. Thomas. 1997. Transfection of the primate malaria parasite Plasmodium knowlesi using entirely heterologous constructs. J. Exp. Med. 185:1499–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijk, M. R., C. J. Janse, and A. P. Waters. 1996. Expression of a Plasmodium gene introduced into subtelomeric regions of Plasmodium berghei chromosomes. Science 271:662–665. [DOI] [PubMed] [Google Scholar]
- 25.van Dijk, M. R., A. P. Waters, and C. J. Janse. 1995. Stable transfection of malaria parasite blood stages. Science 268:1358–1362. [DOI] [PubMed] [Google Scholar]
- 26.Waters, A. P., A. W. Thomas, M. R. van Dijk, and C. J. Janse. 1997. Transfection of malaria parasites. Methods 13:134–147. [DOI] [PubMed] [Google Scholar]
- 27.Wickham, J. M., E. D. Dennis, and G. H. Mitchell. 1980. Long term cultivation of a simian malaria parasite (Plasmodium knowlesi) in a semi-automated apparatus. Trans. R. Soc. Trop. Med. Hyg. 74:789–792. [DOI] [PubMed] [Google Scholar]
- 28.Wu, Y., L. A. Kirkman, and T. E. Wellems. 1996. Transformation of Plasmodium falciparum malaria parasites by homologous integration of plasmids that confer resistance to pyrimethamine. Proc. Natl. Acad. Sci. USA 93:1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, Y., C. D. Sifri, H. H. Lei, X. Z. Su, and T. E. Wellems. 1995. Transfection of Plasmodium falciparum within human red blood cells. Proc. Natl. Acad. Sci. USA 92:973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]