Abstract
The gonococcal transferrin receptor complex comprises two iron-regulated proteins, TbpA and TbpB. TbpA is essential for transferrin-iron uptake and is a TonB-dependent integral outer membrane protein. TbpB is thought to increase the efficiency of iron uptake from transferrin and is lipid modified and surface exposed. To evaluate the structure-function relationships in one of the components of the receptor, TbpA, we created constructs that fused individual putative loops of TbpA with amino-terminal affinity tags. The recombinant proteins were then overexpressed in Escherichia coli, and the fusions were recovered predominately from inclusion bodies. Inclusion body proteins were solubilized, and the epitope fusions were renatured by slow dialysis. To assess transferrin binding capabilities, the constructs were tested in a solid-phase dot blot assay followed by confirmatory quantitative chemiluminescent enzyme-linked immunosorbent assays. The constructs with only loop 5 and with loops 4 and 5 demonstrated dose-dependent specific ligand binding in spite of being out of the context of the intact receptor. The immunogenicities of individual TbpA-specific epitopes were investigated by generating rabbit polyclonal antisera against the fusion proteins. Most of the fusion proteins were immunogenic under these conditions, and the resulting sera recognized full-length TbpA in immunoblots. These results suggest that individual epitopes of TbpA are both immunogenic and functional with respect to ligand binding capabilities, and the vaccine implications of these findings are discussed.
The ability of a microbial pathogen to promote the disease state in a host is closely linked to its ability to adequately obtain essential nutrients from the surrounding host milieu (8). As iron is an essential cofactor required for the growth and replication of most bacteria (6), it is crucial for pathogens to efficiently compete with the host for its iron stores (8). Bacteria have developed diverse methods for iron acquisition from the surrounding environment and its subsequent internalization. In the mammalian host, most iron stores are found in intracellular pools consisting of heme compounds, ferritin, and metalloprotein complexes (41), while extracellular sources are sequestered by iron binding proteins, such as transferrin (in serum) and lactoferrin (at mucosal surfaces and in neutrophils). To utilize these extracellular sources, many bacteria synthesize and secrete low-molecular-weight iron-chelating siderophores (27).
There is no compelling evidence that the obligate human pathogen Neisseria gonorrhoeae produces classical siderophores (2); however, the bacterium is capable of utilizing iron bound to human transferrin, human lactoferrin, hemoglobin, and heme (25, 26). N. gonorrhoeae expresses an energy-dependent, iron-repressible receptor that binds host transferrin at the bacterial cell surface (3, 24). This receptor is expressed by all clinical gonococcal isolates, whereas only approximately 50% of isolated strains are capable of lactoferrin-iron utilization (25, 26). Furthermore, the transferrin receptor is not subject to high-frequency phase or antigenic variation (15, 26), is relatively well conserved (11), and is required for initiation of experimental urethral infection in human males (16).
The gonococcal transferrin receptor is made up of an integral outer membrane protein (TbpA) and a loosely associated surface lipoprotein (TbpB), which work together to bind and internalize iron from transferrin (13). TbpA, the larger of the two proteins, has sequence homology with the TonB-dependent family of outer membrane protein receptors and is essential for transferrin-iron uptake (15). TbpA is highly conserved among the pathogenic neisseriae (11), while TbpB is surface exposed, lipid modified, more variable, and not absolutely required for transferrin-iron acquisition (1, 12). For these reasons, we have focused on TbpA as a particularly attractive vaccine candidate.
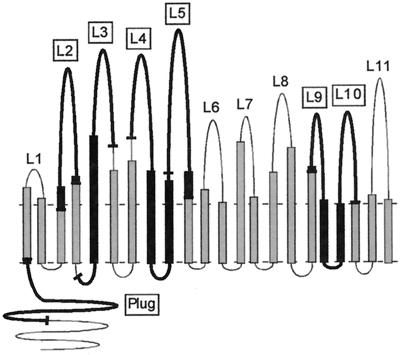
We used our proposed TbpA topology model (5) to guide the design of TbpA-specific epitope fusions, which are the focus of the present study. The hypothetical topology model was constructed using computer predictions for β-turns and amphipathic β-strands. Sequence alignments were subsequently superimposed, which identified significant patterns of diversity among the pathogenic Neisseria spp. (11). Variability in sequence was predicted to correspond to surface exposure, while regions of conservation were proposed to be constrained by function or structure (11). Pairwise alignments between TbpA and FepA, the Escherichia coli ferric enterobactin receptor, identified 22 putative amphipathic β-strands, which together are thought to constitute a β-barrel. Outside of the barrel and connecting the β-strands are 11 putatively surface-exposed loops. Likewise, comparisons with FepA allowed us to propose that the periplasmic opening of this barrel is blocked by an amino-terminal “plug” or “hatch,” as seen in the crystal structures of the siderophore receptors (7, 21). This domain is suggested to conditionally occlude the pore from the periplasmic face until ligand is bound and TonB-derived energy is sensed by the receptor (7, 21), at which point ligand entry is allowed by a shift in the position of the plug within the barrel.
The purpose of the present study was to determine if predicted surface-exposed epitopes of gonococcal TbpA are functional and immunogenic. To explore this, we expressed individual putative loops of TbpA with fusion partners to facilitate purification. These recombinant proteins were overexpressed in E. coli and isolated primarily from inclusion bodies. Solubilized and renatured fusion proteins were tested for transferrin binding by solid-phase colorimetric and chemiluminescent transferrin binding assays. By virtue of the fusion tag, the TbpA-specific loops were purified to a limited extent with an S- protein affinity matrix. The loop fusions were ultimately used as immunogens in rabbits in order to generate epitope-specific antisera.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. E. coli strains were routinely cultured on Luria-Bertani (LB) agar or in LB broth (23) containing appropriate antibiotics. For immunoblot analysis of whole-cell lysates, gonococci were maintained on plates containing Bacto GC medium base (Difco) with Kellogg's supplement I (1%) (19) and 12 μM Fe(NO3)3. The plates were incubated at 37°C in a 5% CO2 atmosphere. To induce iron stress, gonococci were grown in liquid CDM (a chemically defined medium) (43), which was pretreated with Chelex-100 (Bio-Rad) to remove residual iron, and grown at 35°C in a 5% CO2 atmosphere with vigorous shaking. The growth conditions for large-scale isolation of total gonococcal membranes for the purpose of solid-phase transferrin binding assay controls were as previously described (15).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB (rB−mB−) gal dcm (DE3) | Novagen |
| BL21(DE3) pLysS | F−ompT hsdSB (rB−mB−) gal dcm (DE3) pLysS (Cmr) | Novagen |
| NovaBlue | endA1 hsdR17(rk12−mk12+) supE44 thi-1 recA1 gyrA96 relA1 lac [F" proA+B+laclqZΔM15::Tn10 (Tcr)] | Novagen |
| N. gonorrhoeae | ||
| FA19 | Wild-type strain | 26 |
| FA6839 | Lbp− [lbpB::Ω (Strr)]; derivative of FA19 | 4 |
| MCV602 | TbpA− Lbp− [tbpA::mTn3 (Cmr) lbpB::Ω (Strr)]; derivative of FA19 | C. Kenney and C. N. Cornelissen, unpublished data |
| Plasmids | ||
| pET-34b (+) | Kanr | Novagen |
| pUNCH412 | HindIII-to-XbaI fragment of tbpA from pUNCH411 cloned into pET-11 | 14 |
| pVCU301 | PCR product encoding L5 sequence cloned into pET-34b (+) | This study |
| pVCU302 | PCR product encoding L4-L5 sequence cloned into pET-34b (+) | This study |
| pVCU303 | PCR product encoding amino-terminal plug region cloned into pET-34b (+) | This study |
| pVCU304 | PCR product encoding L2 sequence cloned into pET-34b (+) | This study |
| pVCU305 | PCR product encoding L3 sequence cloned into pET-34b (+) | This study |
| pVCU306 | PCR product encoding L9-L10 sequence cloned into pET-34b (+) | This study |
| pVCU307 | PCR product encoding LacZ control cloned into pET-34b (+) | This study |
Standard recombinant DNA techniques and DNA sequencing.
Isolation of plasmid DNA, digestion with restriction endonucleases, and ligations were carried out according to manufacturers' recommendations. For amplification of fragments encoding TbpA-specific epitopes, PCR conditions were as suggested by the supplier of the ligation-independent cloning expression vector, pET-34LIC (Novagen), with annealing conditions optimized for each primer pair. The sequence of inserts in the pET-34 recombinants was identical to the corresponding regions of tbpA from gonococcal strain FA19, with the exception of the construct consisting of loops 9 and 10 (L9-L10) construct, pVCU306 (Table 1), which contained two single-base changes. The first exchanged a glutamic acid for a glycine, and the second was a silent mutation. Due to the conservative nature of the base changes, no corrective mutagenesis was undertaken.
Cloning domains that encode individual epitopes of TbpA.
Six regions of tbpA, most encoding putative loops of TbpA (Fig. 1), were cloned individually into the expression vector pET-34LIC. The oligonucleotides oVCU-3 and oVCU-4 (Table 2) were used to PCR amplify the tbpA region that encodes the putative L5 (Fig. 1). These primers, like the others listed in Table 2, contain a 3′ region that anneals to tbpA and a 5′ segment that anneals to the vector. After PCR amplification, the product was treated with T4 DNA polymerase and dATP according to the manufacturer's recommendations. In this way, 13- to 14-base “sticky ends” which were compatible with the ends of the commercially prepared vector were created. Annealed vector and template mixtures were transformed directly into E. coli strain NovaBlue (Novagen) (Table 1) according to the manufacturer's recommendations; transformants were selected in the presence of kanamycin. Recombinant plasmid pVCU301 (Table 1) expressed a translational fusion between amino-terminal fusion tags and the domain encoding putative L5 of TbpA (Fig. 1). Plasmids pVCU302 through pVCU306 (Table 1) were created similarly, using the tbpA-specific oligonucleotides listed in Table 2. A negative control plasmid, pVCU307 (Table 1), in which the lacZ gene was translationally fused to the fusion tags in pET-34LIC, was constructed. The primers for this reaction were supplied by the manufacturer of the vector (Novagen). Because the CBD (cellulose-binding domain)-LacZ fusion protein was poorly expressed by recombinant E. coli (data not shown), subsequent experiments included CBD, expressed by pET-34 without an insert, as a negative control. The plasmid constructs (Table 1) were transformed into E. coli strain BL21(DE3) (Novagen) for expression of fusion proteins. This strain expresses an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 polymerase, which recognizes the promoter upstream of the fusion tags in the expression vector.
FIG. 1.
Schematic representation of proposed topology model for gonococcal TbpA (5). The horizontal dashed lines represent the boundaries of the gonococcal outer membrane. Twenty-two amphipathic β-strands are predicted to cross the outer membrane to form a β-barrel. Putative surface-exposed loops (numbered L1 to L11) are located above the membrane plane. Epitopes that were overexpressed as fusion proteins in the present study are indicated by thick black lines, the end points of which are shown by short bars.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Amplifyingb |
|---|---|---|
| oVCU-2 | 5"-GACGACGACAAGATGCAGCAGACGCACTGTTCCGCC-3" | L4-L5 |
| oVCU-3 | 5"-GACGACGACAAGATGCAGGATTATTATTATCAAAGTGC-3" | L5 |
| oVCU-4 | 5"-GAGGAGAAGCCCGGTTTAGCTTTTGCCGTTGATGCTGCG-3" | L4-L5 |
| oVCU-8 | 5"-GACGACGACAAGATGGCGGCATTGGGCGGGACGAGG-3" | Plug region |
| oVCU-9 | 5"-GAGGAGAAGCCCGGTTTACTGCCTGCCTTCCCCGATAAC-3" | Plug region |
| oVCU-10 | 5"-GACGACGACAAGATGCGCACCGGGCGGCACGCGGGG-3" | L2 |
| oVCU-11 | 5"-GAGGAGAAGCCCGGTTTACGGATCGGCGAGGAAGCGGTT-3" | L2 |
| oVCU-12 | 5"-GACGACGACAAGATGGAAAACAAACGGCACTACATC-3" | L3 |
| oVCU-13 | 5"-GAGGAGAAGCCCGGTTTACGCGCCCACCGGCGCATTGTT-3" | L3 |
| oVCU-14 | 5"-GACGACGACAAGATGGACATCAAAAAACGCGCAGAC-3" | L9-L10 |
| oVCU-15 | 5"-GAGGAGAAGCCCGGTTTAAGGGCGGGTACGGCGCGC-3" | L9-L10 |
Heterologous protein expression and refolding.
Cultures of recombinant E. coli strains were prepared by growing cells overnight in LB medium containing kanamycin (25 μg ml−1), ampicillin (50 μg ml−1), or kanamycin (30 μg ml−1) with chloramphenicol (12.5 μg ml−1). After 1:25 dilution in fresh medium containing the appropriate antibiotic, the cells were grown at 35°C to an A600 of 0.7, which corresponds to approximately 3 × 108 to 5 × 108 cells ml−1. IPTG was added to a concentration of 1 mM, and the cells were grown for 4 h. Whole-cell lysates were prepared from samples collected before and after IPTG induction. Whole-cell proteins were resolved by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using 12% acrylamide gels (20).
For protein fractionation, induced cultures were harvested by centrifugation (6,500 × g; 4°C; 15 min) and resuspended in inclusion body wash buffer (20 mM Tris-HCl [pH 7.5], 10 mM EDTA, and 1% Triton X-100) (Novagen). Lysozyme (Sigma Aldrich) was added to a concentration of 100 μg ml−1, and the resuspended pellets were incubated at 30°C for 15 min. Samples were sonicated on ice with a Branson sonifier. MgSO4, DNase (Promega), and RNase A (Sigma Aldrich) were added to the lysates at final concentrations of 12 mM, 1 U ml−1, and 10 μg ml−1, respectively, and the mixtures were incubated at room temperature for 20 min. The insoluble fraction was collected by centrifugation (10,000 × g; 4°C; 10 min), and the inclusion body pellets were washed three times in the above-described wash buffer. Inclusion body pellets were resuspended in solubilization buffer (50 mM 3-[cyclohexylamino]-1-propanesulfonic acid [pH 11.0], 0.3% Sarkosyl, and 1 mM dithiothreitol) (Novagen) to a final concentration of 10 to 20 mg ml−1, based on inclusion body wet weight. The resuspensions were incubated at room temperature with shaking to disperse the pellet. Resuspensions were either spun down at 3,200 × g for 15 min at room temperature in a Beckman GS-6KR tabletop centrifuge or spun down at 100,000 × g for 30 min in a Beckman L-60 ultracentrifuge. Soluble, resuspended inclusion body preparations were dialyzed overnight against buffer consisting of 20 mM Tris-HCl (pH 8.5), 100 mM NaCl, and 0.1 mM dithiothreitol.
Solid-phase transferrin binding assays.
A screening dot blot analysis was performed to assess whether any of the epitope fusions were capable of binding transferrin. Dialyzed inclusion body preparations were processed as indicated above. Serial dilutions were spotted onto nitrocellulose (Schleicher and Schuell) and probed with peroxidase-labeled human transferrin (HRP-Tf; Jackson ImmunoResearch) at a concentration of 1 μg ml−1. Binding was detected by addition of ECL reagent (Amersham Pharmacia). In subsequent experiments, blots were prepared by spotting 50 to 100 μl of resuspended or dialyzed inclusion body preparation onto Sequi-Blot polyvinylidene difluoride (Bio-Rad) and were allowed to bind for 1 h. The polyvinylidene difluoride membrane was blocked with 5% skim milk for 1 h, followed by incubation with serial dilutions of HRP-Tf with or without the addition of 20-fold excess unlabeled saturated human transferrin (Calbiochem). The blots were washed three times for 5 min each time with low-salt Tris-buffered saline, pH 7.5. Binding was detected by the addition of ECL Plus reagent (Amersham Pharmacia). Five micrograms of a total membrane preparation (generated as previously described [15]) from gonococcal strain FA19 was used as a positive control.
Chemiluminescent transferrin binding assay.
High-bind 96-well microtiter plates (Greiner) were used for solid-phase chemiluminescent binding assays. Dialyzed inclusion body preparations from epitope fusion strains, from a strain with vector alone (negative control), or from a recombinant E. coli strain expressing full-length recombinant TbpA (rTbpA; positive control [14]) were diluted to 50 μg ml−1 (with the exception of the construct consisting of loops 4 and 5 (L4-L5) which was diluted to 125 μg ml−1) in 100 mM carbonate buffer (pH 9.8). Fifty microliters of each of these protein preparations was applied per well and allowed to bind overnight at 4°C. Unbound proteins were removed, and the wells were washed with phosphate-buffered saline (PBS), pH 7.4, containing 0.1% Tween 20. The wells were then blocked with 200 μl of PBS containing 2% I-Block (Tropix) and 0.05% Tween 20 for 1 h at room temperature. This was followed by additional washes as indicated above. Serial dilutions of HRP-Tf were added with or without the competitor, human transferrin, and allowed to incubate with shaking at room temperature for 1 h. The plates were washed as indicated above and developed by addition of 150 μl of SuperSignal ELISA Femto Maximum Sensitivity Substrate (Pierce) per well with incubation at room temperature for 1 min. Luminescence was detected by a Tropix TR717 microplate luminometer (PE Applied Biosystems). All assays were performed in triplicate.
Purification of epitope fusions.
The epitope fusion proteins were purified with S protein-agarose (Novagen) according to the manufacturer's instructions. Briefly, resuspended or dialyzed inclusion body preparations were added to S protein slurry and incubated at 4°C overnight with gentle rocking. Following incubation, the entire volume was spun down at 500 × g for 10 min, and the unbound fraction was removed. The matrix was then washed briefly with bind-wash buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.1% Triton X-100) and spun down as described above. The epitope fusions were eluted by addition of 3 M MgCl2 to the matrix. This mixture was allowed to incubate for 1 h at room temperature, and the eluate was collected. The elutions were then dialyzed in Slide-A-Lyzer cassettes (3,500-MW cutoff [Pierce]) against PBS and concentrated by evaporation. The starting material and subsequent purified product were analyzed by SDS-PAGE on 12% acrylamide gels and transferred to nitrocellulose for immunoblot analysis.
Production of epitope-specific antisera.
Two New Zealand White Elite female rabbits were immunized (Covance Research Products, Denver, Pa.) with each recombinant protein preparation by intradermal and subcutaneous routes. One injection of 250 μg of the dialyzed inclusion body preparation in Freund's complete adjuvant was followed by five subsequent injections of 125 μg each in Freund's incomplete adjuvant. These five boosts occurred on days 21, 42, 63, 84, and 105 after the initial immunization. The rabbits were exsanguinated 18 weeks and 5 days (131 days total) after the first immunization. Sera from individual rabbits were maintained separately and screened individually.
Evaluating the immunogenicity of short epitopes of TbpA.
Whole-cell proteins from iron-stressed gonococcal strains FA6839 (TbpA+ Lbp− [Table 1]) and MCV602 (TbpA− Lbp− [Table 1]) were separated by SDS-PAGE and transferred to nitrocellulose (36). These lysates were probed with individual production bleed sera (10 weeks and 4 days after the first immunization) by immunoblotting as previously described (15). For these experiments, antiserum raised against full-length gonococcal TbpA (15) was used as a positive control. Bound polyclonal antibodies were detected by addition of a secondary goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (Sigma Aldrich) followed by development with nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate).
RESULTS
Choice of TbpA-specific epitopes for overexpression.
To assess the function and immunogenicity of putative loops of gonococcal TbpA, we created epitope fusions with two amino-terminal fusion tags, the CBD from Clostridium cellulovorans and the S tag (Novagen). We focused our initial efforts on designing constructs in which L5 or L4-L5 (Fig. 1) were translationally fused to the affinity tags. This focus was due to several compelling findings. First, to test our hypothetical topology model of gonococcal TbpA, we previously generated polyclonal anti-peptide sera in mice that were directed at putative surface-exposed, antigenic regions of gonococcal TbpA (5, 11). Out of the seven anti-peptide sera generated, only four recognized full-length TbpA in immunblots, and the one that was specific for putative L5 sequence reacted with whole iron-stressed gonococci in a dot blot assay (11). This result suggested that at least part of the putative L5 was surface exposed. In another study, we individually deleted loops 4 and 5 from TbpA (5) and found that with the loss of either of these predicted loops, the resultant mutants were incapable of binding transferrin in solid- or liquid-phase transferrin binding assays. Together, these findings suggested that L5 was in fact surface exposed and that both L4 and L5 were inherently important for ligand binding. Finally, we did a pairwise comparison (5) between TbpA and the E. coli TonB-dependent receptor, FepA, whose crystal structure had recently been elucidated (7). From this comparison, we found that putative L5 fell within the same region as the FepA ligand binding domain (11, 28).
We used various criteria to design the other four fusion constructs (Fig. 1). We concentrated on regions of TbpA that had predicted hydrophilicity, antigenicity, and surface exposure, because these were the most likely to be involved in ligand interaction. First, we chose regions that contained most of the peptide sequences used in the previous immunizations (11); however, we added length. Whereas the peptides ranged from 11 to 14 residues, the TbpA sequences in the fusion constructs ranged from 62 to 140 amino acids in length. Additionally, we previously evaluated the sequence diversity among TbpA proteins from strains of pathogenic neisseriae, as well as from other species, including Haemophilus influenzae, Actinobacillus pleuropneumoniae, Pasteurella haemolytica, and Moraxella catarrhalis (11). From this analysis, we identified four hypervariable domains within the TbpA sequence and suggested that these areas were surface accessible (11). Accordingly, L2 represents a stretch of TbpA that is predicted to be hydrophilic and encodes a hypervariable domain (11). L2 also contains one of the three pairs of cysteine residues found within the protein that may form a surface-exposed disulfide bond (7, 11, 21). The L3 sequence also contains a hypervariable domain and is predicted to be on the surface (11). We also focused on the other end of the protein and designed a construct consisting of loops 9 and 10 (L9-L10), whose sequence is somewhat divergent among the neisseriae and which is predicted to be surface accessible (11). Finally, we chose to use sequence from the putative plug region, which has homology with the FepA globular plug domain (7). The TbpA-specific epitopes chosen for subsequent analysis are graphically depicted in Fig. 1.
Expression and localization of epitope fusion proteins.
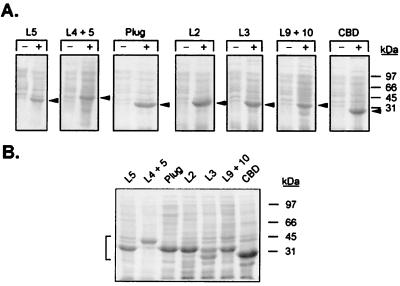
The fusion clones were produced by PCR amplification of tbpA-specific regions, followed by cloning into the expression vector, pET-34 (Table 1), to allow production of recombinant proteins in E. coli. Recombinant plasmids (Table 1) were used to transform E. coli strain BL21(DE3). The recombinants were grown in liquid culture and induced with IPTG, which resulted in expression of proteins ranging in mass from approximately 33 to 43 kDa after 4 h (Fig. 2A). To determine if the recombinant proteins were expressed in soluble or insoluble form, the induced cells were disrupted by sonication and solubilized in Triton X-100, and subsequent fractions were analyzed by SDS-PAGE. All of the overexpressed proteins, with the exception of the plug construct (which was localized to the cytosolic fraction), were enriched in inclusion body fractions (Fig. 2B and data not shown).
FIG. 2.
Overexpression of epitope fusion proteins. (A) Coomassie blue-stained gels containing whole-cell lysates of E. coli strains expressing fusion proteins as indicated at the top. Lanes CBD contain whole-cell lysates from E. coli expressing the fusion partner alone. −, uninduced lysates; +, lysates induced with IPTG for 4 h. The arrowheads denote the position of the overexpressed fusion protein in each lane. (B) Coomassie blue-stained gel of inclusion body preparations separated by SDS-PAGE. The lanes are labeled according to the overexpressed fusion proteins they contain. The positions of the overexpressed proteins are indicated by the bracket on the left. For both panels, positions of molecular mass standards are indicated at the right. The images were acquired using a Hewlett-Packard Scan Jet 4C and annotated using Adobe PhotoShop version 4.0.
Transferrin binding by L5 and L4-L5.
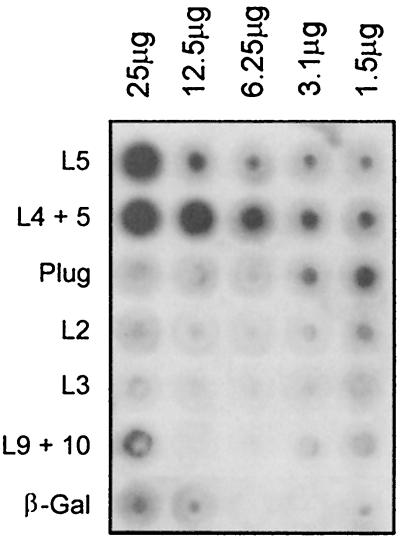
After overexpressing all of the epitope fusions, we assessed their ligand binding functions in a solid-phase dot blot assay. As shown in Fig. 3, the L5 fusion expressed by BL21(DE3)(pVCU301) bound HRP-Tf, as did the L4-L5 fusion expressed by BL21(DE3)(pVCU302), although the latter appeared to do so with more intensity. The other fusion constructs demonstrated virtually no binding of HRP-Tf above background levels attributable to the negative control (β-galactosidase [Fig. 3]). The L9-L10 fusion expressed by BL21(DE3)(pVCU306) demonstrated low-level binding at the highest concentration tested but showed no activity with lower concentrations. This preliminary evidence suggested that L5 and L4-L5, out of the context of the intact receptor, retained transferrin binding capabilities.
FIG. 3.
Solid-phase transferrin binding assay of dialyzed inclusion body preparations. Twofold dilutions (as indicated at the top) of dialyzed inclusion body preparations were spotted onto nitrocellulose and probed with HRP-Tf (1 μg/ml). The rows contain dialyzed inclusion body preparations and are labeled (left) according to the overexpressed protein they contain. The image was acquired using a Hewlett-Packard Scan Jet 4C and annotated using Adobe PhotoShop version 4.0.
Specificity of transferrin binding by L5 and L4-L5.
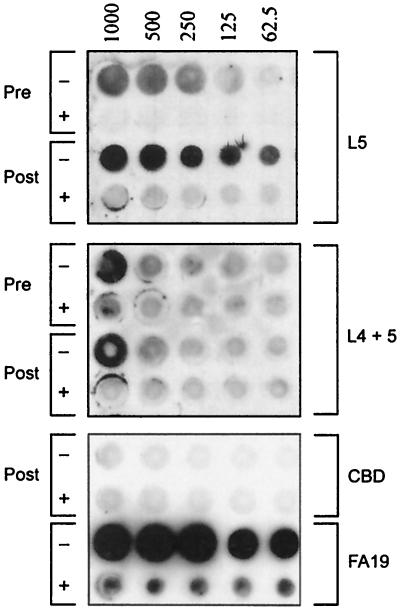
To determine if this binding phenomenon was dose dependent and specific, we analyzed the L5 and L4-L5 fusions in solubilized and in renatured forms by solid-phase dot blot analysis. Dialyzed inclusion body preparations expressed by BL21(DE3) containing vector alone (CBD) were used as a negative control (Fig. 4). Total membrane preparations from gonococcal strain FA19 were used as a positive control (Fig. 4). In the case of L5, the solubilized inclusion body preparations demonstrated concentration-dependent binding that was specific for human transferrin (Fig. 4). In the presence of excess unlabeled human transferrin, binding of HRP-Tf was eliminated or greatly diminished. The dialyzed, renatured inclusion body preparations demonstrated somewhat-increased ligand binding relative to the undialyzed preparations, which was likewise dose dependent and specific. The L4-L5 construct exhibited some binding activity that was dose dependent and specific (Fig. 4), but the results were more variable and less reproducible with both the solubilized and renatured forms. In total, these results indicated that L5, and possibly L4 as well, contained specific transferrin binding epitopes that are potentially critical for ligand binding to full-length TbpA.
FIG. 4.
Transferrin binding by L5 and L4-L5 in a solid-phase dot blot assay. The rows contain either resuspended inclusion body preparations before renaturing dialysis (Pre) or after dialysis (Post). The rows are labeled on the right according to the overexpressed proteins they contain. The rows labeled FA19 contain total membrane preparations from the wild-type gonococcal strain. The blot was probed with HRP-Tf alone (−) or with HRP-Tf in the presence of 20-fold excess saturated transferrin as a competitor (+). HRP-Tf dilutions are indicated above the blot in nanograms ml−1. The image was scanned using a Hewlett-Packard Scan Jet 4C and annotated using Adobe PhotoShop version 4.0.
Ligand binding by L5 and L4-L5 in a chemiluminescent solid-phase binding assay.
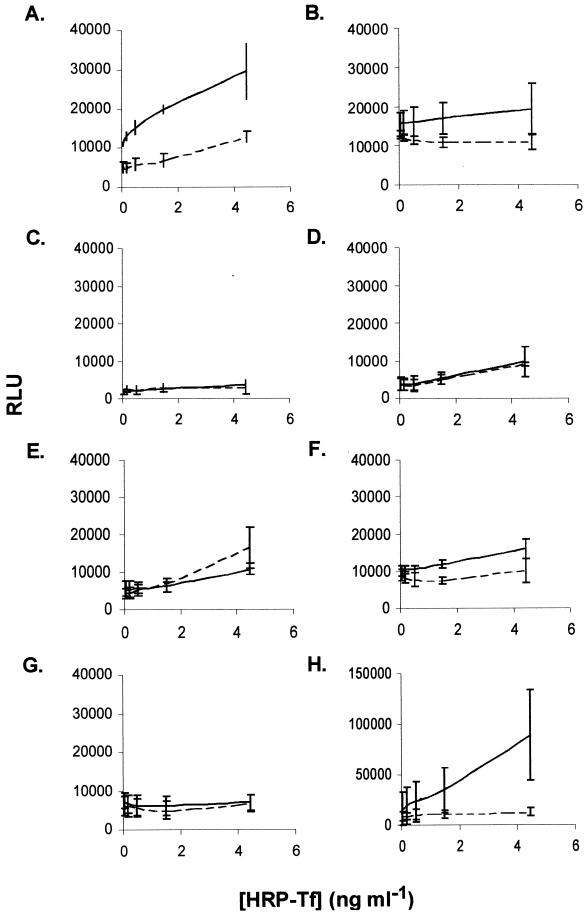
The dot blot assays allowed us to demonstrate specific binding, but not in a quantitative fashion. We subsequently developed and utilized a chemiluminescent solid-phase assay to measure the transferrin binding activity of each epitope fusion. Dialyzed inclusion body preparations were bound to microtiter plates and processed as described in Materials and Methods. The results from three independent assays are shown in Fig. 5 and were consistent with the results of the screening dot blot shown in Fig. 3. The L5 fusion (Fig. 5A) displayed specific, dose-dependent ligand binding; however, the L4-L5 preparation (Fig. 5B) was more variable and required the addition of 2.5-fold more protein. The other epitope fusions (Fig. 5C to F) did not display any significant binding compared to the negative control, in which we used inclusion body preparations containing CBD alone (the fusion partner) (Fig. 5G). As a positive control, we used inclusion body preparations containing overexpressed, full-length rTbpA (Fig. 5H).
FIG. 5.
Transferrin binding by L5 and L4-L5 in a solid-phase chemiluminescent binding assay. Dialyzed inclusion body preparations were probed with either HRP-Tf (solid line) or HRP-Tf in the presence of excess saturated transferrin as a competitor (dashed line). The results are plotted as relative luminescence units (RLU) as a function of the concentration of HRP-Tf added. The individual panels represent total binding to the fusion proteins L5 (A), L4-L5 (B), plug (C), L2 (D), L3 (E), and L9-L10 (F), CBD alone (G), and full-length rTbpA (H). Error bars represent standard deviations.
Specificity of ligand binding by L5 and L4-L5 in a chemiluminescent solid-phase assay.
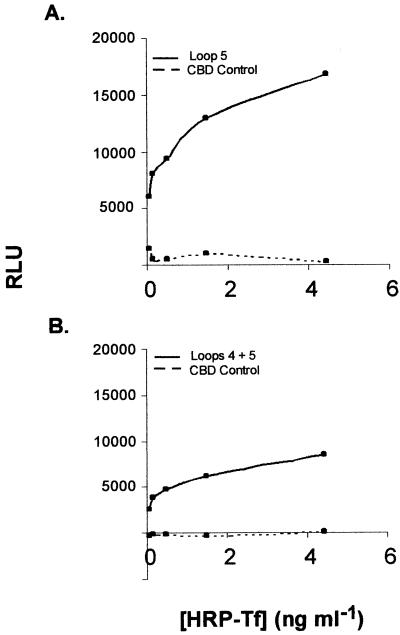
Specific transferrin binding in the chemiluminescent assay was calculated by subtracting the nonspecific binding (detected in the presence of competitor) from total binding for each of the two fusion constructs that bound ligand, L5 and L4-L5 (Fig. 5A and B). The specific binding attributable to each of these fusion proteins is displayed graphically in Fig. 6. The L5 construct demonstrated saturable, specific transferrin binding with a capacity that was between two- and threefold higher than that of the L4-L5 fusion.
FIG. 6.
Specific transferrin binding to L5 and L4-L5 fusions in solid-phase chemiluminescent transferrin binding assay. (A) Specific transferrin binding to L5 fusion (Loop 5) compared to CBD alone (CBD control). (B) Specific transferrin binding to L4-L5 fusion (Loops 4 + 5) compared to CBD alone. RLU, relative luminescence units.
Purification of epitope fusions using S protein-agarose.
We initially planned to purify the epitope fusions via their affinity tags for use in subsequent immunizations. We achieved limited purification of all fusions using the S-tag fusion partner. The purified products also contained some contaminating proteins, as analyzed by SDS-PAGE, and some breakdown products that reacted with anti-CBD tag polyclonal sera (data not shown). However, the yield of purified protein (ranging from 0.04 to 1.96 mg of purified protein ml−1 per 250 ml of induced culture) was major deterrent. We therefore decided to use dialyzed inclusion body preparations as immunogens, since contaminating E. coli-reactive antibodies could be absorbed out postproduction.
Immunogenicity of TbpA-specific epitopes.
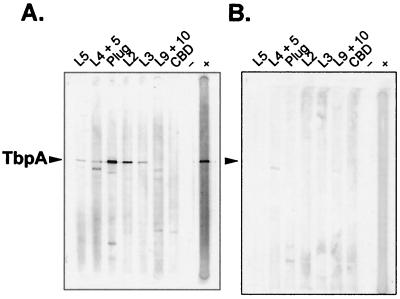
To determine if the sera generated against defined epitopes of gonococcal TbpA recognized denatured full-length TbpA, we performed immunoblots probing whole-cell lysates of strains FA6839 (TbpA+ Lbp− [Table 1]) and MCV602 (TbpA− Lbp−) with the epitope-specific sera. We used Lbp mutants to eliminate cross-reactivity between the anti-TbpA sera and gonococcal LbpA (15). All of the fusion sera reacted with TbpA to different degrees, with the exception of the anti-L9-L10 serum and the anti-CBD serum (Fig. 7A). The plug and L2 constructs appeared to have generated more-vigorous immune responses than did the other fusions. The sera reacted minimally with other proteins expressed by the isogenic TbpA− mutant (Fig. 7B), indicating that each was specific for its target antigen, TbpA. All of the sera were capable of recognizing their respective immunizing epitope fusions in immunoblots (data not shown).
FIG. 7.
Immunoblots showing specificities of antisera raised against TbpA-specific epitopes. (A) Immunoblot of whole-cell lysates from gonococcal strain FA6839 (TbpA+ Lbp−). (B) Immunoblot of whole-cell lysates from gonococcal strain MCV602 (TbpA− Lbp−). For both blots, the lanes were probed with antisera raised against TbpA-specific epitopes as indicated at the top. The arrowheads indicate the positions of gonococcal TbpA. Lanes −, negative control to which no primary antibody was added; lanes +, positive control, which was probed with an antiserum against holo-TbpA (15). The image was acquired using a Hewlett-Packard Scan Jet 4C and annotated using Adobe PhotoShop version 4.0.
DISCUSSION
In this study, we assessed the possibility that specific regions of gonococcal TbpA participate in transferrin binding and that these limited domains could elicit a defined, epitope-specific immune response. Our recently described TbpA topology model (5) aided us in predicting putatively immunogenic ligand binding domains. The regions chosen for the present study had the following characteristics in common. With the exception of the plug construct, all fusions included regions of TbpA predicted to form extracellular loops. These regions were rich in antigenic and sequence variability and thus are likely to be surface exposed and potentially involved in ligand binding. Several fusions contained paired cysteine residues, which might serve to stabilize or constrain the loop structure. All fusion constructs contained regions of TbpA with high antigenicity index values (18). Finally, all of the fusions contained peptide sequences for which we previously generated peptide-specific antisera (11). Peptide sequence located within putative L5 (in the fusion protein expressed by pVCU301) was previously shown to generate antibodies that react with intact gonococci, implicating this region as a surface-exposed epitope (5, 11). Thus, we focused our early efforts on determining whether L5 was also capable of ligand binding.
Diverse methods have been used to define the ligand binding domains of TbpB proteins expressed by several species, with various degrees of success. For example, we (12) constructed amino- and carboxy-terminal deletions of gonococcal TbpB which were translationally fused to the α-peptide and overexpressed in E. coli. These truncated TbpB proteins were then tested for transferrin binding capability in ligand binding Western blots. This method identified an amino-terminal domain of approximately 400 residues within gonococcal TbpB that retained transferrin binding capabilities. Vonder Haar et al. identified a similarly sized stable transferrin binding domain in meningococcal TbpB (40). Strutzberg et al. (34) used TnphoA mutagenesis and a peptide spot approach to map functional regions of TfbA (a TbpB homologue) of A. pleuropneumoniae to three different regions of the amino-terminal half of the protein. The group subsequently generated multiple antigenic peptides containing these binding domains and raised sera against them. Ultimately, they found that the peptide constructs inhibited binding of transferrin to recombinant TfbA but the anti-peptide antibodies did not (33). In another approach, Renault-Montgenie et al. identified binding domains within meningococcal TbpB with both a peptide spot approach and a translational fusion expression system (29). With these methods, they isolated and characterized binding sites in both the amino and carboxy termini of the TbpB protein.
In contrast to the wealth of TbpB ligand-binding studies, the present study is the first to attribute transferrin binding to individual domains of TbpA expressed by any species. These TbpA-specific epitopes were capable of saturable, specific binding of human transferrin, which was surprising, since the excised domains of TbpA were out of the context of the intact receptor. Moreover, we could reconstitute ligand binding to TbpA fragments isolated from inclusion body preparations. Production of recombinant proteins in inclusion body form is generally avoided because of the difficulty in manipulating proteins to their functional states (see references 22 and 32 for reviews). We found, however, that renatured inclusion body preparations containing L5 bound transferrin in a dose-dependent and specific manner. This coincides well with the results of our previous deletion mutagenesis study, which showed that deletion of L5 abolished transferrin binding and transferrin-iron uptake (5). The deletion analysis also implicated L4 as a critical determinant for transferrin binding (5). In the present study, the fusion protein consisting of the L4-L5 construct displayed specific binding, but it was much more variable than the activity attributable to L5 alone. This variability may be due to several compromising factors, such as the increased length of the L4-L5 construct (140 versus 69 residues), the increased number of putative transmembrane domains in the L4-L5 construct (two versus zero), or the additional pair of cysteine residues in the L4-L5 construct. Any or all of these differences might have influenced the success of our efforts to refold the epitopes into a more native conformation. The deletion study also indicated that removal of L8 decreased transferrin binding to TbpA (5), suggesting that loops or domains other than L4 and L5 are likely to be important for optimal ligand binding as well. While the present study did not implicate any other putative loops in ligand binding, we cannot rule out the possibility of low-affinity interactions attributable to other loops or of binding phenomena that could not be reconstituted under the conditions utilized here. Since the epitopes examined in this study were expressed out of the context of the intact receptor, conformational determinants that influence or mediate ligand binding would have been lost, and this may have contributed to our inability to attribute binding functions to other putative loops.
Comparison of gonococcal TbpA sequences demonstrated that the L5 domain contains much sequence variability, including one of four hypervariable domains, interspersed with discrete pockets of conservation (11). In spite of its ligand binding capability, this region was not defined by linear sequence conservation, which would appear to contradict the theme of conserved domains acting as functional components. This observation raises the possibility that conserved functional domains could be hidden within variable sequence as a way to camouflage vulnerable epitopes from the immune system. Could this be similar to the immune evasion ploy used by viruses, as described in the “canyon hypothesis”? This theory suggests that the receptor attachment sites of a virus lie within a domain lined with conserved residues and that this domain is surrounded by hypervariable regions that are capable of binding neutralizing antibodies (31). The TbpA situation could also be similar to the strategy used by the foot-and-mouth disease virus, in which a flexible, disordered loop covers the structural component of the receptor attachment site and protects it from the immune system until the presence of the receptor is sensed. This produces a conformational change in the loop, which exposes the conserved interior (10, 31). Further characterization of L5 in and out of the context of the entire receptor is needed before we can discern the true relevance of the juxtaposition of conservation and diversity observed in this bacterial system.
The TbpA-specific fusion constructs generated in this study were used as immunogens for the generation of epitope-specific antisera. We found that all except one fusion protein elicited antibodies that recognized full-length TbpA on Western blots. It is unclear why the L9-L10 fusion did not elicit a response that was reactive against full-length TbpA; however, it should be noted that the anti-peptide sera directed against L10 generated in our previous study likewise did not recognize full-length TbpA (11). The sequence contained in the L9-L10 fusion is relatively invariant, contains none of the defined hypervariable domains, and is predicted to include two conserved transmembrane strands (11). It is possible that this domain is simply not as immunogenic as predicted; however, the serum elicited against the L9-L10 fusion did recognize the immunizing antigen. We postulated that by adding length, the loop fusions would elicit a more vigorous immune response in animals than did the preceding peptide immunogens. However, the additional length in this particular construct did not appear to enhance the immune response in the present study.
TbpA has been proposed as a vaccine candidate, most recently by West et al. for immunoprophylaxis against Neisseria meningitidis (42). Earlier, Danve et al. had suggested that TbpA-specific antibodies were not bactericidal against meningococci (17); however, West et al. demonstrated that sera generated against recombinant TbpA, while less bactericidal, were more cross-protective in an intraperitoneal mouse model than those generated against recombinant TbpB (42). Thus, the more conserved of the Tbp proteins was capable of eliciting a cross-reactive, protective immune response. Antigenically variable epitopes have been recognized as immunodominant in neisserial porins (35, 37), in neisserial FrpB (38) (now called FetA; [9]), and in meningococcal TbpB (30); however, these immunodominant epitopes have also been shown to elicit strain-specific antibody responses that did not confer broadly cross-reactive biological activities (30, 35, 37, 38). Thus, in the search for a cross-reactive response, we may be more successful identifying immunorecessive epitopes that could be “unmasked” by removal of the immunodominant, antigenically variable residues. Van der Voort et al. (39) found that deleting specific loops of meningococcal porin skewed the immune response toward constant, immunorecessive epitopes against which bactericidal antibodies were generated. Because the targets were more conserved among strains, these authors suggested that the response could be more cross protective (39). In principle, the Tbp proteins may be similar in that if we could molecularly excise the constant, immunorecessive domains and fuse the epitopes with an appropriate adjuvant, we might be able to generate a cross-reactive immune response that is broadly protective. In light of these possibilities, future studies will be directed at characterizing the immune responses elicited against the TbpA-specific epitopes described in the present study, including determination of bactericidal or opsonophagocytic functions, as well as cross-reactivity and interference with transferrin-iron internalization.
Acknowledgments
We acknowledge the Nucleic Acids Core facility at Virginia Commonwealth University for performing automated DNA sequencing. We are also grateful to Gour Biswas and Fred Sparling for providing chromosomal DNA from gonococcal strain FA6839, with which Chris Kenney constructed MCV602, the Lbp strain used to test antibody specificity.
This work was supported by Public Health Service grants AI39523 and AI47141 from the National Institute of Allergy and Infectious Disease and by a grant from the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust.
Editor: J. T. Barbieri
REFERENCES
- 1.Anderson, J. E., P. F. Sparling, and C. N. Cornelissen. 1994. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol. 176:3162-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald, F. S., and I. W. DeVoe. 1980. Iron acquisition by Neisseria meningitidis in vitro. Infect. Immun. 27:322-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archibald, F. S., and I. W. DeVoe. 1979. Removal of iron from human transferrin by Neisseria meningitidis. FEMS Microbiol. Lett. 6:159-162. [Google Scholar]
- 4.Biswas, G. D., J. E. Anderson, C.-J. Chen, C. N. Cornelissen, and P. F. Sparling. 1999. Identification and functional characterization of the Neisseria gonorrhoeae lbpB gene product. Infect. Immun. 67:455-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton, I. C., M. K. Yost, J. E. Anderson, and C. N. Cornelissen. 2000. Identification of discrete domains within gonococcal transferrin-binding protein A that are necessary for ligand binding and iron uptake functions. Infect. Immun. 68:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briat, J.-F. 1992. Iron assimilation and storage in prokaryotes. J. Gen. Microbiol. 138:2475-2483. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 8.Bullen, J. J., H. J. Rogers, and E. Griffiths. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 80:1-35. [DOI] [PubMed] [Google Scholar]
- 9.Carson, S. D. B., P. E. Klebba, S. M. C. Newton, and P. F. Sparling. 1999. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J. Bacteriol. 181:2895-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman, M. S., and M. G. Rossmann. 1993. Comparison of surface properties of picornaviruses: strategies for hiding the receptor site from immune surveillance. Virology 195:745-756. [DOI] [PubMed] [Google Scholar]
- 11.Cornelissen, C. N., J. E. Anderson, I. C. Boulton, and P. F. Sparling. 2000. Antigenic and sequence diversity in gonococcal transferrin-binding protein A (TbpA). Infect. Immun. 68:4725-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelissen, C. N., J. E. Anderson, and P. F. Sparling. 1997. Characterization of the diversity and the transferrin-binding domain of gonococcal transferrin-binding protein 2. Infect. Immun. 65:822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornelissen, C. N., J. E. Anderson, and P. F. Sparling. 1997. Energy-dependent changes in the gonococcal transferrin receptor. Mol. Microbiol. 26:25-35. [DOI] [PubMed] [Google Scholar]
- 14.Cornelissen, C. N., G. D. Biswas, and P. F. Sparling. 1993. Expression of gonococcal transferrin-binding protein 1 causes Escherichia coli to bind human transferrin. J. Bacteriol. 175:2448-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelissen, C. N., G. D. Biswas, J. Tsai, D. K. Paruchuri, S. A. Thompson, and P. F. Sparling. 1992. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J. Bacteriol. 174:5788-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelissen, C. N., M. Kelley, M. M. Hobbs, J. E. Anderson, J. G. Cannon, M. S. Cohen, and P. F. Sparling. 1998. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Mol. Microbiol. 27:611-616. [DOI] [PubMed] [Google Scholar]
- 17.Danve, B., L. Lissolo, M. Mignon, P. Dumas, S. Colombani, A. B. Schryvers, and M.-J. Quentin-Millet. 1993. Transferrin-binding proteins isolated from Neisseria meningitidis elicit protective and bactericidal antibodies in laboratory animals. Vaccine 11:1214-1220. [DOI] [PubMed] [Google Scholar]
- 18.Jameson, B. A., and H. Wolf. 1989. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput. Appl. Biosci. 4:181-186. [DOI] [PubMed] [Google Scholar]
- 19.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and C. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Locher, K. P., B. Rees, R. Koebnik, A. Mitschler, L. Moulinier, J. P. Rosenbusch, and D. Moras. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771-778. [DOI] [PubMed] [Google Scholar]
- 22.Makrides, S. C. 1996. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60:512-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.McKenna, W. R., P. A. Mickelsen, P. F. Sparling, and D. W. Dyer. 1988. Iron uptake from lactoferrin and transferrin by Neisseria gonorrhoeae. Infect. Immun. 56:785-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mickelsen, P. A., E. Blackman, and P. F. Sparling. 1982. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from lactoferrin. Infect. Immun. 35:915-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mickelsen, P. A., and P. F. Sparling. 1981. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from transferrin and iron compounds. Infect. Immun. 33:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neilands, J. B. 1992. Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-K30). Can. J. Microbiol. 38:728-733. [DOI] [PubMed] [Google Scholar]
- 28.Newton, S. M. C., J. S. Allen, Z. Cao, Z. Qi, X. Jiang, C. Sprencel, J. D. Igo, S. B. Foster, M. A. Payne, and P. E. Klebba. 1997. Double mutagenesis of a positive charge cluster in the ligand-binding site of the ferric enterobactin receptor, FepA. Proc. Natl. Acad. Sci. USA 94:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renault-Montgenie, G., D. Poncet, L. von Olleschik-Elbheim, T. Cournez, M. Mignon, M. A. Schmidt, and M.-J. Quentin-Millet. 1997. Identification of human transferrin-binding sites within meningococcal transferrin-binding protein B. J. Bacteriol. 179:6400-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rokbi, B., G. Renauld-Mongenie, M. Mignon, B. Danve, D. Poncet, C. Chabanel, D. A. Caugant, and M.-J. Quentin-Millet. 2000. Allelic diversity of the two transferrin binding protein B gene isotypes among a collection of Neisseria meningitidis strains representative of serogroup B disease: implication for the composition of a recombinant TbpB-based vaccine. Infect. Immun. 68:4938-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossmann, M. B. 1989. The canyon hypothesis. J. Biol. Chem. 264:14587-14590. [PubMed] [Google Scholar]
- 32.Rudolph, R., and H. Lilie. 1996. In vitro folding of inclusion body proteins. FASEB J. 10:49-56. [PubMed] [Google Scholar]
- 33.Strutzberg, K., B. Franz, and G. F. Gerlach. 1997. Interference of peptides and specific antibodies with the function of the Actinobacillus pleuropneumoniae transferrin-binding protein. Infect. Immun. 65:5346-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strutzberg, K., L. von Olleschik, B. Franz, C. Pyne, M. A. Schmidt, and G.-F. Gerlach. 1995. Mapping of functional regions on the transferrin-binding protein (TfbA) of Actinobacillus pleuropneumoniae. Infect. Immun. 63:3846-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toropainen, M., L. Saarinen, P. van der Ley, B. Buipers, and H. Kayhty. 2001. Murine monoclonal antibodies to PorA of Neisseria meningitidis show reduced protective activity in vivo against B:15:P1.7,16 subtype variants in an infant rat infection model. Microb. Pathog. 30:139-148. [DOI] [PubMed] [Google Scholar]
- 36.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Ley, P., J. E. Heckels, M. Virji, P. Hoogerhout, and J. T. Poolman. 1991. Topology of outer membrane porins in pathogenic Neisseria spp. Infect. Immun. 59:2963-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Ley, P., J. van der Biezen, R. Sutmuller, P. Hoogerhout, and J. T. Poolman. 1996. Sequence variability of FrpB, a major iron-regulated outer-membrane protein in the pathogenic Neisseriae. Microbiology 142:3269-3274. [DOI] [PubMed] [Google Scholar]
- 39.van der Voort, E. R., H. van Dijken, B. Kuipers, J. van der Biezen, P. van der Ley, J. Meylis, I. Claassen, and J. Poolman. 1997. Human B- and T-cell responses after immunization with a hexavalent PorA meningococcal outer membrane vesicle vaccine. Infect. Immun. 65:5184-5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vonder Haar, R. A., M. Legrain, H. V. J. Kolbe, and E. Jacobs. 1994. Characterization of a highly structured domain in Tbp2 from Neisseria meningitidis involved in binding to human transferrin. J. Bacteriol. 176:6207-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg, E. D. 1978. Iron and infection. Microbiol. Rev. 42:45-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West, D., K. Reddin, M. Matheson, R. Heath, S. Funnell, M. Hudson, A. Robinson, and A. Gorringe. 2001. Recombinant Neisseria meningitidis transferrin binding protein A protects against experimental meningococcal infection. Infect. Immun. 69:1561-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West, S. E. H., and P. F. Sparling. 1987. Aerobactin utilization by Neisseria gonorrhoeae and cloning of a genomic DNA fragment that complements Escherichia coli fhuB mutations. J. Bacteriol. 169:3414-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]