Abstract
The Lyme disease spirochete, Borrelia burgdorferi, is capable of infecting a wide variety of vertebrates. This broad host range implies that B. burgdorferi possesses the ability to contravene the immune defenses of many potential hosts. B. burgdorferi produces multiple different Erp proteins on its outer membrane during mammalian infection. It was reported previously that one Erp protein can bind human factor H (J. Hellwage, T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppälä, and S. Meri, J. Biol. Chem. 276:8427–8435, 2001). In this paper we report that the ability to bind the complement inhibitor factor H is a general characteristic of Erp proteins. Furthermore, each Erp protein exhibits different relative affinities for the complement inhibitors of various potential animal hosts. The data suggest that the presence of multiple Erp proteins on the surface can allow a single B. burgdorferi bacterium to resist complement-mediated killing in any of the wide range of potential hosts that it might infect. Thus, Erp proteins likely contribute to the persistence of B. burgdorferi in nature and to the ability of this bacterium to cause Lyme disease in humans and other animals.
The causative agent of Lyme disease, Borrelia burgdorferi, is maintained in nature through infectious cycles involving Ixodes sp. tick vectors and a variety of vertebrate reservoir hosts. In addition, many species of mammals, including humans, are susceptible to incidental infection by this bacterium (64). The diversity of animals that B. burgdorferi is capable of infecting indicates that this bacterium has evolved mechanisms by which it can evade clearance by the immune systems of its numerous potential hosts. In fact, in the absence of antibiotic therapy, the spirochetes can be cultured from tissues of immunocompetent animals more than 1 year after infection (17, 23).
The alternative pathway of complement activation forms an important part of the innate immune systems of vertebrates. This pathway results in the deposition of factor C3b on the surfaces of invading microorganisms, leading to opsonization and killing of the organisms (33). Several successful pathogens have evolved mechanisms of resistance to complement-mediated killing, greatly increasing their virulence (34, 48). Consistent with this generalization, virulent B. burgdorferi strains are resistant to the direct bactericidal effects of complement from many of the animals that they are capable of infecting (10, 36, 38, 44, 61, 76). In humans and other animals, host cells protect themselves from the alternative pathway of complement-mediated killing by coating their surfaces with the inhibitory plasma protein factor H, which promotes deactivation of C3 convertases and degradation of C3b (33). Like many other pathogenic organisms, B. burgdorferi exploits the host’s self-defense mechanism by also binding factor H, thus inhibiting complement activation on the bacterial surface (3, 40, 41).
It was recently reported that a B. burgdorferi surface protein, OspE of strain N40, can specifically interact with human factor H (29). The N40 OspE protein is one member of the protein family collectively known as Erp (OspEF-related) proteins (45, 70, 73). All Lyme disease borreliae have genes that encode multiple homologous Erp proteins; some of these proteins have very similar amino acid sequences, while others exhibit only limited similarity. For example, the well-characterized B. burgdorferi type strain, B31, can carry 17 erp genes arranged in 10 separate loci, and the predicted Erp proteins have primary structures that are between 17 and 100% identical (13, 14, 68, 72). The B. burgdorferi erp genes are considered a family of genes because they have nearly identical promoter sequences, are transcriptionally coregulated, occupy allelic positions on members of the cp32 family of plasmids, and encode highly charged lipoproteins with well-conserved leader polypeptide sequences and other motifs (reviewed in reference 73). Erp proteins localize to the bacterial outer surface (22, 45) and are expressed during mammalian infection (73). Furthermore, multiple Erp proteins can be expressed simultaneously by an individual bacterium (28; El-Hage and Stevenson, unpublished results). While the maintenance of this large repertoire of seemingly redundant genes suggests that multiple Erp proteins play an important role in B. burgdorferi, until now there has been no convincing explanation for this phenomenon. In this report we show that most, if not all, Erp proteins bind the complement inhibitor factor H. Moreover, the affinities of each Erp protein for the complement inhibitors of different animal species are different, which probably contributes to the ability of the bacteria to infect a broad range of vertebrate hosts.
MATERIALS AND METHODS
Bacteria.
B. burgdorferi strain B31 is a wild-type bacterium that was originally isolated from a tick collected on Shelter Island, N.Y. (12). The subculture used in this study, B31-RML (55), is infectious for both mice and ticks and has been passaged fewer than five times in culture since reisolation from an infected mouse. Bacteria were grown in Barbour-Stoener-Kelly II medium (7) at 23°C until they reached the mid-exponential phase (approximately 107 bacteria per ml), and then the cultures were diluted 1:100 in fresh medium and grown to the mid-exponential phase at 35°C. A temperature shift from 23 to 35°C, which mimics the increase in temperature experienced by B. burgdorferi in a feeding tick, induces B. burgdorferi to produce many proteins known to be expressed during mammalian infection (65, 68, 71). A lysate of the bacteria was used for the immunoblot analyses of human and animal sera described below.
Recombinant Erp proteins.
Strain B31 erp genes were cloned into either pProEX-1 (Life Technologies, Gaithersburg, Md.), pET15b (Novagen, Madison, Wis.), or pUni/V5-His (Invitrogen, Carlsbad, Calif.) so that they were in the correct reading frame to encode a fusion protein with the plasmid-encoded polyhistidine polypeptide (22, 68). All fusion proteins were designed so that they lacked the N-terminal leader polypeptide and type II signal processing sequences, since elimination of such sequences generally allows greater protein recovery (18). Escherichia coli cells were transformed with each plasmid and induced to synthesize recombinant protein. Fusion proteins were purified from E. coli lysates by using His-Bind Resin column chromatography kits (Novagen). As previously reported (68), some protein preparations contained multimers or degradation products. Note that in strain B31 three genes, erpA, erpI, and erpN, encode identical proteins; since they are indistinguishable, these proteins are collectively designated ErpA/I/N (14, 22, 56, 69).
Factor H and sera.
Purified human factor H was purchased from Calbiochem (La Jolla, Calif.). Two preexisting human serum samples from anonymous donors were obtained from local physicians. Serum was obtained from strain F344/N rats (Rattus norvegicus), an outbred domestic dog (Canis familiaris), a domestic shorthaired cat (Felis domesticus), and a horse (Equus caballus). Mouse (Mus musculus) and rabbit (Oryctolagus cuniculus) sera were purchased from Sigma (St. Louis, Mo.), and fetal bovine (Bos taurus) serum was purchased from Hy-Clone (Logan, Utah). All human and animal serum samples were collected and used according to guidelines administered by the University of Kentucky Office of Research Integrity and Institutional Animal Care and Use Committee.
Immunoblot analysis.
Animal sera were examined by immunoblotting to determine whether an antiserum raised against human factor H contained antibodies that bound the factor H protein of each animal species. A 3-μl aliquot of serum was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), electrotransferred to a nitrocellulose membrane, and then blocked by incubation with 5% nonfat dry milk in Tris-buffered saline-Tween 20 (TBS-T) (20 mM Tris [pH 7.5], 150 mM NaCl, 0.05% [vol/vol] Tween 20). The membrane was incubated with goat anti-human factor H polyclonal antiserum (Calbiochem) diluted 1:8 in TBS-T. Then, the membrane was washed with TBS-T and incubated with donkey anti-goat horseradish peroxidase conjugate (Santa Cruz Biotechnology, Santa Cruz, Calif.), and bound antibodies were visualized by enhanced chemiluminescence (Amersham, Piscataway, N.J.) (56).
In addition, each serum sample was examined to ensure that it did not contain antibodies directed against B. burgdorferi proteins, such as the Erp proteins, which might obscure results obtained in the immunoaffinity analyses described below. A B. burgdorferi lysate was separated by SDS-PAGE and analyzed by immunoblotting with each human or animal serum. Membranes were incubated with protein A-horseradish peroxidase conjugate (Amersham), and bound antibodies were visualized by enhanced chemiluminescence. The resulting blots were examined for the presence of specific immunoblot bands indicative of exposure to B. burgdorferi (15).
Factor H affinity analysis.
A 0.1-μg portion of each recombinant Erp protein was subjected to SDS-PAGE and transferred to nitrocellulose membranes. Each membrane was blocked by incubation for at least 1 h with 5% nonfat dry milk in TBS-T, followed by incubation for 1 h at room temperature in either purified human factor H (0.5 μg/ml in TBS-T) or a serum sample from a human or animal source. The membranes were then washed with TBS-T and incubated for 1 h at room temperature in goat anti-human factor H polyclonal antiserum diluted 1:8 in TBS-T. Finally, the membranes were washed with TBS-T and incubated with either donkey anti-goat antibody-horseradish peroxidase or protein A-horseradish peroxidase conjugate, and bound antibodies were identified by enhanced chemiluminescence (16, 40, 57).
RESULTS
Affinity of Erp proteins for human factor H.
Hellwage and coworkers recently observed that one member of the Erp protein family, the strain N40 OspE protein, can bind human factor H (29). Since all B. burgdorferi bacteria encode multiple different Erp proteins, we queried whether factor H binding was a general characteristic of Erp proteins. Since it has been suggested that the Erp protein family may be subdivided into three groups (1), we included representatives of each group in this study.
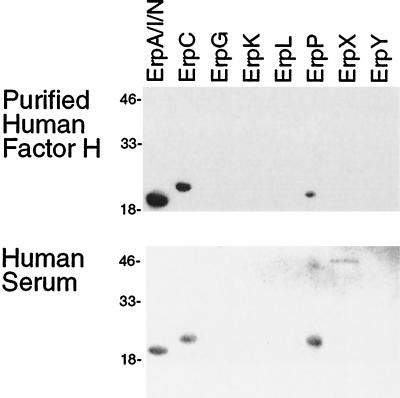
By immunoaffinity analysis, we determined that recombinant forms of the strain B31 ErpA/I/N, ErpC, and ErpP proteins could bind purified human factor H (Fig. 1 and Table 1). Recombinant ErpA/I/N had the greatest relative affinity, followed by ErpC, and ErpP had the lowest apparent affinity of the three proteins. There was no detectable binding of factor H by any of the other recombinant Erp proteins. No signals were detected with control preparations which lacked either factor H or anti-factor H antiserum (data not shown). Of the strain B31 Erp proteins, the three that bound purified human factor H exhibited the greatest levels of similarity to the strain N40 OspE protein; 85, 88, and 82% of the ErpA/I/N, ErpC, and ErpP amino acids are identical to amino acids of OspE, respectively (72). The primary sequences of the remaining strain B31 Erp proteins are significantly different from those of OspE, ErpA/I/N, ErpC, and ErpP (69).
FIG. 1.
Immunoaffinity blot analyses of recombinant Erp proteins performed with either purified human factor H or whole human serum. Some purified recombinant protein preparations contain multimers or breakdown products (68). The lanes contained different Erp proteins, as indicated at the top. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
TABLE 1.
Binding of factor H proteins of different mammals by Erp proteins
| Source of factor H | Binding by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ErpA/I/N | ErpC | ErpG | ErpK | ErpL | ErpP | ErpX | ErpY | |
| Human (purified) | +a | + | − | − | − | + | − | − |
| Human (serum) | + | + | − | − | − | + | + | − |
| Mouse | + | + | − | − | − | + | + | + |
| Rat | + | + | − | − | − | + | + | + |
| Rabbit | + | + | − | − | − | + | + | + |
| Horse | + | + | − | − | − | + | + | − |
| Cattle | + | + | + | − | + | + | − | − |
| Cat | + | + | + | − | − | + | + | + |
| Dog | + | + | + | − | − | + | + | + |
+, detectable binding; −, no detectable binding.
As human serum contains approximately 0.5 mg of factor H per ml (59), whole serum can also be used in an analysis of factor H binding (3, 40). Immunoaffinity blot analysis of recombinant Erp proteins performed with human serum samples revealed that in addition to the binding to the three proteins identified above, the anti-human factor H antiserum also detected binding to ErpX (Fig. 1). The same result was obtained with serum samples from two different individuals (data not shown). The primary sequence of ErpX is approximately 20% identical to the primary sequences of ErpA/I/N, ErpC, and ErpP, although all of these proteins contain a number of similar sequence motifs (68). The results which we obtained might represent interactions between ErpX and a glycosylated form of factor H that was not present in the purified sample described above. Alternatively, ErpX may have bound a related protein, such as factor H-like protein 1 (FHL-1), which is produced from the same gene through alternative splicing of the mRNA (26, 48, 78). Although FHL-1 apparently functions like factor H in modulating C3b degradation, it is a much smaller protein and contains a different carboxy terminus; thus, it may have affinity for some Erp proteins that do not bind well to factor H.
Erp protein binding of factor H proteins from other animals.
While the binding of human factor H by Erp proteins probably plays a role in human Lyme disease, humans are biological dead ends for B. burgdorferi, and our susceptibility to infection is coincidental to the ability of this bacterium to infect many different reservoir host species in nature. As one would expect, the amino acid and carbohydrate compositions of factor H are somewhat different in different animal species (2, 30, 42). Different animals’ factor H proteins also differ in their interactions with other proteins (31). Since B. burgdorferi can infect many types of animals, we hypothesized that this bacterium can also bind factor H proteins from potential host animals. Accordingly, we examined the abilities of the strain B31 Erp proteins to bind the complement inhibitors of several susceptible mammals, some of which also serve as reservoirs of B. burgdorferi in nature.
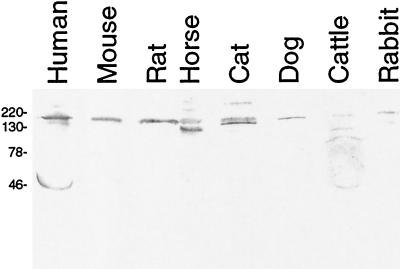
Prior to our study, it was not known whether the polyclonal goat anti-human factor H antibodies could recognize the factor H proteins of the animals examined (Calbiochem, personal communication). Immunoblot analysis indicated that the goat antibodies did in fact recognize at least one serum protein whose size was comparable to that of human factor H (Fig. 2). There did not appear to be any small factor H-related proteins in the sera of the nonhuman mammals tested, suggesting that the immunoaffinity analyses of these sera, as described below, examined interactions between Erp proteins and full-length factor H proteins. Differences in the affinities of the goat antiserum for the different factor H proteins prevented direct comparisons of the Erp-factor H binding affinities of different species. For example, the goat antiserum data revealed relatively weak binding of the bovine homolog, presumably reflecting factor H sequence similarities between these two ungulates. Each animal serum was also tested for the presence of antibodies that could bind B. burgdorferi proteins. All samples were negative (data not shown), indicating that they could be used in immunoaffinity analyses without complications resulting from antibodies that bound directly to the Erp proteins.
FIG. 2.
Immunoblot analysis of animal serum samples performed with goat anti-human factor H polyclonal antiserum. Human factor H has a molecular mass of 155 kDa, and FHL-1 has a molecular mass of 42 kDa (26, 78). The positions of molecular mass markers (in kilodaltons) are indicated on the left.
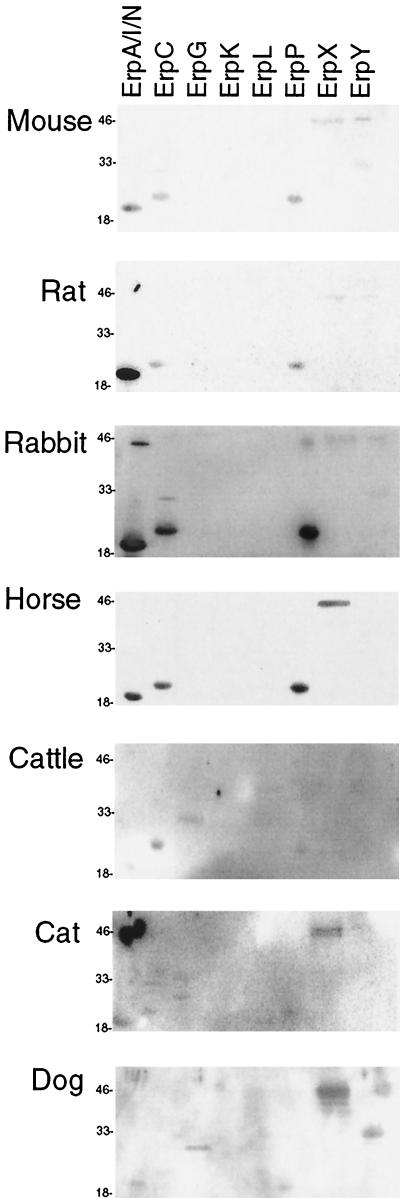
Rodents are often key reservoirs of B. burgdorferi in nature (64, 66). Domestic mice and rats are susceptible to B. burgdorferi infection and are often used in laboratory studies of Lyme disease (8, 9). Factor H in mouse serum and factor H in rat serum both bound to the same five Erp proteins, detectably interacting with ErpX and ErpY in addition to the three proteins that also bound human factor H (Fig. 3 and Table 1). There did, however, appear to be differences in the relative affinities of the factor H proteins of the two rodents tested.
FIG. 3.
Immunoaffinity blot analyses of recombinant Erp proteins performed with whole sera from various animals. Some purified recombinant protein preparations contain multimers or breakdown products (68). The lanes contained different Erp proteins, as indicated at the top. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
Rabbits are frequently used in laboratory studies of B. burgdorferi infection (24, 37) and can also serve as reservoirs of B. burgdorferi in the wild (4, 75). Other researchers have observed that virulent Lyme disease spirochetes bind rabbit factor H from serum (3). Consistent with this previous report, we found that rabbit factor H bound with approximately the same strength to ErpA/I/N, ErpC, and ErpP and bound significantly less well to ErpX and ErpY (Fig. 3 and Table 1).
Many domestic animals are susceptible to B. burgdorferi infection, often exhibiting symptoms similar to those of Lyme disease in humans (49, 51, 60). Domestic dogs have been used as models of Lyme disease in laboratory studies (5) and may also serve as reservoirs of B. burgdorferi in some settings (53). In agricultural animals, symptoms of Lyme disease, such as weight loss and decreased milk production, can have significant financial impact. Four Erp proteins bound factor H from horse serum, and these four proteins had similar apparent affinities (Fig. 3 and Table 1). Five Erp proteins could detectably bind the factor H in cattle serum, with ErpC exhibiting the greatest apparent affinity. Cat factor H was bound by six Erp proteins, with ErpX having the greatest apparent affinity. The same six Erp proteins bound dog factor H, with ErpX and ErpY having the greatest apparent affinities.
DISCUSSION
Ever since their discovery, why B. burgdorferi contain multiple different erp genes has been puzzling. However, the observation that all Lyme disease borreliae carry numerous erp genes led to the hypothesis that Erp proteins are likely to perform essential functions in these bacteria (14, 73). The data presented in this report, together with the data of Hellwage et al. (29), suggest that these proteins provide resistance to complement-mediated killing through their interactions with host factor H. Erp proteins are exposed on the bacterial outer surface (22, 45) and so are positioned to bind factor H. Expression of all erp genes carried by an individual bacterium appears to be regulated through a single pathway, as all of the proteins are expressed in response to the same environmental stimuli (6, 28, 68, 71) and can be simultaneously coexpressed by individual bacteria (28; El-Hage and Stevenson, unpublished results). The entire repertoire of Erp proteins encoded by the genes of a bacterium is synthesized during the initial stages of mammalian infection, a stage in the B. burgdorferi infection cycle when the bacteria encounter host serum and complement (28, 68). The bacteria also encounter complement while they are in the tick vector, both when the tick acquires an infection and when the bacteria are being transmitted to a new host. Consistent with this, analysis of B. burgdorferi gene expression during various stages of tick infection has demonstrated that erp genes are transcribed at all times at which the bacteria are exposed to host blood (27).
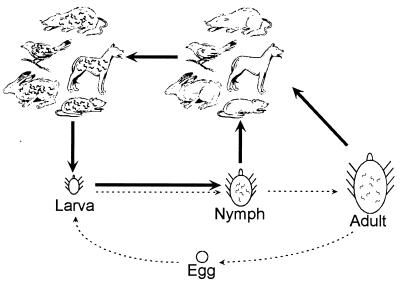
While many pathogenic organisms make themselves insensitive to complement-mediated killing by coating their surfaces with host factor H (34), the amino acid sequences and glycosylation patterns of factor H differ from animal to animal (2, 30, 42). Because of the variations, the host range of a pathogen can be severely restricted by which factor H proteins its receptors are able to bind. Our study indicated that Erp proteins bind factor H but they do so with different specificities for the complement inhibitor factors of various animals. An examination of the natural history of B. burgdorferi indicates why such an ability is important to this bacterium (Fig. 4) (35, 67). There are three postembryonic stages in the development of the vector Ixodes sp. ticks: larva, nymph, and adult. There is negligible transovarial transmission of B. burgdorferi (50, 63), so a larva is not infected upon hatching but must acquire B. burgdorferi through feeding on an infected host. Such a tick feeds only two more times in its life, once after each molt, during which B. burgdorferi can be transmitted to a new host. Therefore, to be biologically successful, a B. burgdorferi bacterium must be transmitted from a nymph or adult tick, successfully infect the host upon which that tick feeds, and persist in that host until a larval tick feeds on the host. Since Ixodes sp. ticks may feed on a variety of vertebrate hosts, a B. burgdorferi bacterium cannot be guaranteed of its next host’s species. Thus, the hypothesized need for multiple different Erp proteins must be present on a single bacterium, providing the bacterium with an ability to bind complement-inhibiting factors of a wide variety of potential hosts and ensuring survival of the bacterium and its progeny.
FIG. 4.
Diagram of the life cycles of B. burgdorferi and Ixodes sp. vector ticks. Tick larvae hatch and feed once; then they molt into nymphs, which also feed once, and then into adults, which mate, feed once, lay eggs, and then die (dashed arrows). Larvae acquire B. burgdorferi by feeding on infected hosts and then transmit the bacteria to new hosts during feeding in the nymph and adult stages (solid arrows). The tick hosts include a large variety of species of mammals, birds, and reptiles, many of which are susceptible to B. burgdorferi infection and serve as reservoir animals. In many geographic areas, adult Ixodes sp. ticks do not feed on the same animal hosts as the larvae or nymphs and so do not contribute greatly to the maintenance of B. burgdorferi in these locations (35, 67).
Although virulent isolates of B. burgdorferi are generally resistant to complement-mediated killing (3, 10, 36, 41, 61, 76), they may be susceptible to the alternative pathways of some vertebrates (10, 43, 44). This observation led to the hypothesis that the host range of a particular B. burgdorferi bacterium is restricted by its ability to defeat the alternative pathway of complement-mediated killing in susceptible hosts (43, 44, 46). Our data refine this hypothesis to include the ability of a bacterium’s Erp proteins to bind factor H as an important element in determining whether the bacterium can infect a particular host. There is often considerable variation in the Erp protein sequences of different B. burgdorferi isolates, and we predict that the differences can have significant effects on the host ranges of the isolates. Additional analyses of B. burgdorferi host ranges, sensitivity to complement, and Erp protein characteristics should continue to test and refine the hypotheses described above.
The location of erp genes on extrachromosomal elements, rather than on the bacterial chromosome, is consistent with the theory that genes which confer specific local advantages are often plasmid-borne, since they are more readily transferred between bacteria (19). Furthermore, there is strong evidence that the members of the cp32 family of plasmids which carry the erp genes are prophages (20, 21), which could allow very efficient shuffling of desirable erp genes among a population via bacteriophage transduction (54). Indeed, it is evident that exchange of erp genes does occur in nature (1, 69, 74). However, different B. burgdorferi strains isolated in the same geographic region may contain similar erp gene sequences (52, 73; unpublished results), suggesting that there is genetic stability in bacteria that share a host reservoir pool.
Recent studies found that human factor H bound to at least two different proteins in whole-cell lysates of B. burgdorferi (3, 40). Our data strongly suggest that these unidentified proteins are members of the Erp protein family. However, the previous studies were performed with bacteria other than strain B31, and since the erp gene sequences of different bacterial strains are often considerably different, it is unlikely that any of these factor H-binding proteins are identical to any of the factor H-binding proteins encoded by strain B31.
We noted that the recombinant ErpK protein examined in this study did not detectably bind a factor H protein and that some other Erp proteins exhibited relatively weak factor H binding. This may have been a consequence of the SDS-PAGE-based immunoaffinity analysis technique used; perhaps the recombinant proteins were not folded so that they were able to bind factor H, while the native proteins may have been able to bind factor H. There is also the strong possibility that ErpK and some of the other Erp proteins preferentially bind the factor H proteins of animals not examined in this study. B. burgdorferi is capable of infecting a wide variety of animals (11, 32, 47, 58), and ErpK may play a role in inactivating the alternative pathways of some of these hosts.
It has been noted that while some strains of B. burgdorferi are resistant to complement-mediated killing, mutants of the same strains may be sensitive to killing (39, 61). Since B. burgdorferi regulates synthesis of Erp proteins (73), it is possible that such mutants lack the ability to produce Erp proteins under the conditions tested. Indeed, many such mutants are defective in gene regulation and exhibit protein profiles different from those of their parent strains (39, 61). Additionally, B. burgdorferi is known to lose plasmids during cultivation, including the plasmids that encode Erp proteins (14), so it is also possible that the mutants lost plasmids encoding the Erp proteins that bind factor H for the type of serum being tested. Further analysis of such mutants should provide additional insight into the mechanisms by which B. burgdorferi resists killing via the alternative pathway.
Based on comparisons of the amino acid sequences, it was recently proposed that B. burgdorferi Erp proteins can be subdivided into three groups (1). Furthermore, it was also suggested that the groups should be given the following different names, suggesting that the groups have different functions: OspE for the proteins most like the strain N40 OspE protein; OspF for the proteins most like the strain N40 OspF protein; and Elp for the proteins less similar to either strain N40 protein (1). The results of our study indicate that members of all three groups can bind factor H. ErpA/I/N, ErpC, and ErpP are in the OspE group; ErpG, ErpL, and ErpY are in the OspF group; and ErpX is in the Elp group (1). Our data suggest that most, if not all, Erp proteins perform similar functions for the bacteria and that sequence variations are probably necessary consequences of the affinities of the proteins for different factor H proteins. Thus, we conclude that a name change is not necessary for the Erp proteins.
In addition to members of the erp multigene family, individual B. burgdorferi strains may contain members of several other families of paralogous genes; strain B31 contains members of at least 161 such families (13, 25). For example, it is known that B. burgdorferi expresses Mlp proteins during mammalian infection (62, 77), yet strain B31 contains genes for at least nine different paralogs of these proteins (13, 62). We wonder whether each Mlp paralog might permit advantageous interactions with a different potential host.
In conclusion, our studies demonstrated that B. burgdorferi Erp proteins bind the factor H proteins of several diverse mammalian species. Additionally, the relative affinities of each Erp protein for factor H proteins differed depending on the animal serum tested, with ErpA/I/N having the greatest relative affinity for rat factor H but ErpX having the greatest relative affinity for the cat homolog. These data suggest that Erp proteins contribute to the expansive host range of B. burgdorferi by making the bacteria insensitive to the alternative pathways of many different types of animals. Additional studies of the bacteriophage-encoded Erp proteins will undoubtedly reveal important information concerning these apparent virulence factors and their role in the pathogenesis of Lyme disease.
Acknowledgments
This research was supported by National Institutes of Health grant RO1-AI44254 to B. Stevenson.
We thank Ken Dickey, Mark Dobbs, Kathy Forrest, Jens Goebel, Mary Ann Kenneson, Jarlath Nally, Janet Rodgers, Tom Roszman, John Timoney, Creighton Trahan, and Penny Wildman for assistance in obtaining human and animal serum and Don Cohen, Mike Hubank, Patti Rosa, Tom Schwan, Tony Sinai, Mark Wooten, and Wolf Zückert for constructive discussions and comments on the manuscript.
Editor: D. L. Burns
REFERENCES
- 1.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, J. J., B. K. Hack, P. N. Cunningham, and R. J. Quigg. 2001. A protein with characteristics of factor H is present on rodent platelets and functions as the immune adherence receptor. J. Biol. Chem. 276:32129–32135. [DOI] [PubMed] [Google Scholar]
- 3.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. F., L. A. Magnarelli, R. B. LeFebvre, T. G. Andreadis, J. B. McAninch, G.-C. Perng, and R. C. Johnson. 1989. Antigenically variable Borrelia burgdorferi isolated from cottontail rabbits and Ixodes dentatus in rural and urban areas. J. Clin. Microbiol. 27:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appel, M. J. G., S. Allan, R. H. Jacobson, T. L. Lauderdale, Y. F. Chang, S. J. Shin, J. W. Thomford, R. J. Todhunter, and B. A. Summers. 1993. Experimental Lyme disease in dogs produces arthritis and persistent infection. J. Infect. Dis. 167:651–664. [DOI] [PubMed] [Google Scholar]
- 6.Babb, K., N. El-Hage, J. C. Miller, J. A. Carroll, and B. Stevenson. 2001. Distinct regulatory pathways control the synthesis of Borrelia burgdorferi infection-associated OspC and Erp surface proteins. Infect. Immun. 69:4146–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 8.Barthold, S. W., D. S. Beck, G. M. Hansen, G. A. Terwilliger, and K. D. Moody. 1990. Lyme borreliosis in selected strains and ages of laboratory mice. J. Infect. Dis. 162:133–138. [DOI] [PubMed] [Google Scholar]
- 9.Barthold, S. W., K. D. Moody, G. A. Terwilliger, R. O. Jacoby, and A. C. Steere. 1988. An animal model for Lyme arthritis. Ann. N. Y. Acad. Sci. 539:264–273. [DOI] [PubMed] [Google Scholar]
- 10.Brade, V., I. Kleber, and G. Acker. 1992. Differences of two Borrelia burgdorferi strains in complement activation and serum resistance. Immunobiology 185:453–465. [DOI] [PubMed] [Google Scholar]
- 11.Brown, R. N., and R. S. Lane. 1996. Reservoir competence of four chaparral-dwelling rodents for Borrelia burgdorferi in California. Am. J. Trop. Med. Hyg. 54:84–91. [DOI] [PubMed] [Google Scholar]
- 12.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. [DOI] [PubMed] [Google Scholar]
- 13.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516. [DOI] [PubMed] [Google Scholar]
- 14.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 1995. Recommendations for test performance and interpretation from the second national conference on serologic diagnosis of Lyme disease. Morb. Mortal. Wkly. Rep. 44:590–591. [PubMed] [Google Scholar]
- 16.Dave, S., A. Brooks-Walter, M. K. Pangburn, and L. S. McDaniel. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect. Immun. 69:3435–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Souza, M. S., A. L. Smith, D. S. Beck, G. A. Terwilliger, E. Fikrig, and S. W. Barthold. 1993. Long-term study of cell-mediated responses to Borrelia burgdorferi in the laboratory mouse. Infect. Immun. 61:1814–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn, J. J., B. N. Lade, and A. G. Barbour. 1990. Outer surface protein A (OspA) from the Lyme disease spirochete, Borrelia burgdorferi: high level expression and purification of a soluble recombinant form of OspA. Protein Expr. Purif. 1:159–168. [DOI] [PubMed] [Google Scholar]
- 19.Eberhard, W. G. 1989. Why do bacterial plasmids carry some genes and not others? Plasmid 21:167–174. [DOI] [PubMed] [Google Scholar]
- 20.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. F. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by φBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggers, C. H., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308–7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, R. D. Gilmore, Jr., M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821–830. [DOI] [PubMed] [Google Scholar]
- 23.El-Hage, N., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foley, D. M., R. J. Gayek, J. T. Skare, E. A. Wagar, C. I. Champion, D. R. Blanco, M. A. Lovett, and J. N. Miller. 1995. Rabbit model of Lyme borreliosis: erythema migrans, infection-derived immunity, and identification of Borrelia burgdorferi proteins associated with virulence and protective immunity. J. Clin. Investig. 96:965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. [DOI] [PubMed] [Google Scholar]
- 26.Friese, M. A., J. Hellwage, T. S. Jokiranta, S. Meri, H. H. Peter, H. Eibel, and P. F. Zipfel. 1999. FHL-1/reconectin and factor H: two human complement regulators which are encoded by the same gene are differently expressed and regulated. Mol. Immunol. 36:809–818. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799–808. [DOI] [PubMed] [Google Scholar]
- 28.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppälä, and S. Meri. 2001. The complement regulatory factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427–8435. [DOI] [PubMed] [Google Scholar]
- 30.Horstmann, R. D., and J. J. Müller-Eberhard. 1985. Isolation of rabbit C3, factor B, and factor H and comparison of their properties with those of the human analog. J. Immunol. 134:1094–1100. [PubMed] [Google Scholar]
- 31.Horstmann, R. D., M. K. Pangburn, and H. J. Müller-Eberhard. 1985. Species specificity of recognition by the alternative pathway of complement. J. Immunol. 134:1101–1104. [PubMed] [Google Scholar]
- 32.Humair, P., and L. Gern. 2000. The wild hidden face of Lyme borreliosis in Europe. Microbes Infect. 2:915–922. [DOI] [PubMed] [Google Scholar]
- 33.Janeway, C. A., P. Travers, M. Walport, and J. D. Capra. 1999. Immunobiology, 4th ed. Elsevier Science Ltd., New York, N.Y.
- 34.Joiner, K. A. 1988. Complement evasion by bacteria and parasites. Annu. Rev. Microbiol. 42:201–230. [DOI] [PubMed] [Google Scholar]
- 35.Keirans, J. E., H. J. Hutcheson, L. A. Durden, and J. S. H. Klompen. 1996. Ixodes (Ixodes) scapularis (Acari: Ixodidae): redescription of all active stages, distribution, hosts, geographical variation, and medical and veterinary importance. J. Med. Entomol. 33:297–318. [DOI] [PubMed] [Google Scholar]
- 36.Kochi, S. K., and R. C. Johnson. 1988. Role of immunoglobulin G in killing of Borrelia burgdorferi by the classical complement pathway. Infect. Immun. 56:314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornblatt, A. N., A. C. Steere, and D. G. Brownstein. 1984. Experimental Lyme disease in rabbits: spirochetes found in erythema migrans and blood. Infect. Immun. 46:220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraiczy, P., K.-P. Hunfeld, S. Breiner-Ruddock, R. Würzner, G. Acker, and V. Brade. 2000. Comparison of two laboratory methods for the determination of serum resistance in Borrelia burgdorferi isolates. Immunobiology 201:406–419. [DOI] [PubMed] [Google Scholar]
- 39.Kraiczy, P., K.-P. Hunfeld, S. Peters, R. Würzner, G. Acker, B. Wilske, and V. Brade. 2000. Borreliacidal activity of early Lyme disease sera against complement-resistant Borrelia afzelii FEM1 wild-type and an OspC-lacking FEM1 variant. J. Med. Microbiol. 49:917–928. [DOI] [PubMed] [Google Scholar]
- 40.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. Eur. J. Immunol. 31:1674–1684. [DOI] [PubMed] [Google Scholar]
- 41.Kraiczy, P., C. Skerka, M. Kirschfink, P. F. Zipfel, and V. Brade. 2001. Mechanism of complement resistance of pathogenic Borrelia burgdorferi isolates. Int. Immunopharmacol. 1:393–401. [DOI] [PubMed] [Google Scholar]
- 42.Kristensen, T., and B. F. Tack. 1986. Murine protein H is comprised of 20 repeating units, 61 amino acids in length. Proc. Natl. Acad. Sci. USA 83:3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo, M. M., R. S. Lane, and P. C. Giclas. 2000. A comparative study of mammalian and reptilian alternative pathway of complement-mediated killing of the Lyme disease spirochete (Borrelia burgdorferi). J. Parasitol. 86:1223–1228. [DOI] [PubMed] [Google Scholar]
- 44.Kurtenbach, K., H.-S. Sewell, N. H. Ogden, S. E. Randolph, and P. A. Nuttall. 1998. Serum complement sensitivity as a key factor in Lyme disease ecology. Infect. Immun. 66:1248–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam, T. T., T.-P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane, R. S., and G. B. Quistad. 1998. Borreliacidal factor in the blood of the western fence lizard. J. Parasitol. 84:29–34. [PubMed] [Google Scholar]
- 47.Levin, M., J. F. Levine, S. Yang, P. Howard, and C. S. Apperson. 1996. Reservoir competence of the southeastern five-lined skink (Eumeces inexpectatus) and the green anole (Anolis carolinensis) for Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 54:92–97. [DOI] [PubMed] [Google Scholar]
- 48.Lindahl, G., U. Sjšbring, and E. Johnsson. 2000. Human complement regulators: a major target for pathogenic microorganisms. Curr. Opin. Immunol. 12:44–51. [DOI] [PubMed] [Google Scholar]
- 49.Lissmann, B. A., E. M. Bosler, H. Camay, B. G. Ormiston, and J. L. Benach. 1984. Spirochete-associated arthritis (Lyme disease) in a dog. J. Am. Vet. Med. Assoc. 185:219–220. [PubMed] [Google Scholar]
- 50.Magnarelli, L. A., J. F. Anderson, and D. Fish. 1987. Transovarial transmission of Borrelia burgdorferi in Ixodes dammini (Acari: Ixodidae). J. Infect. Dis. 156:234–236. [DOI] [PubMed] [Google Scholar]
- 51.Magnarelli, L. A., J. F. Anderson, H. R. Levine, and S. A. Levy. 1990. Tick parasitism and antibodies to Borrelia burgdorferi in cats. J. Am. Vet. Med. Assoc. 197:63–66. [PubMed] [Google Scholar]
- 52.Marconi, R. T., S. Hohenberger, S. Jauris-Heipke, U. Schulte-Spechtel, C. P. LaVoie, D. Rößler, and B. Wilske. 1999. Genetic analysis of Borrelia garinii OspA serotype 4 strains associated with neuroborreliosis: evidence for extensive genetic homogeneity. J. Clin. Microbiol. 37:3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mather, T. N., D. Fish, and R. T. Coughlin. 1994. Competence of dogs as reservoirs for Lyme disease spirochetes (Borrelia burgdorferi). J. Am. Vet. Med. Assoc. 205:186–188. [PubMed] [Google Scholar]
- 54.Miao, E. A., and S. I. Miller. 1999. Bacteriophages in the evolution of pathogen-host interactions. Proc. Natl. Acad. Sci. USA 96:9452–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller, J. C., J. L. Bono, K. Babb, N. El-Hage, S. Casjens, and B. Stevenson. 2000. A second allele of eppA in Borrelia burgdorferi strain B31 is located on the previously undetected circular plasmid cp9–2. J. Bacteriol. 182:6254–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, J. C., N. El-Hage, K. Babb, and B. Stevenson. 2000. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J. Clin. Microbiol. 38:1569–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neeleman, C., S. P. M. Geelen, P. C. Aerts, M. R. Daha, T. E. Mollnes, J. J. Roord, G. Posthuma, H. van Dijk, and A. Fleer. 1999. Resistance to both complement activation and phagocytosis in type 3 pneumococci is mediated by the binding of complement regulatory factor H. Infect. Immun. 67:4517–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olsen, B., T. G. T. Jaenson, L. Noppa, J. Bunikis, and S. Bergström. 1993. A Lyme borreliosis cycle in seabirds and Ixodes uriae ticks. Nature 362:340–342. [DOI] [PubMed] [Google Scholar]
- 59.Pangburn, M. K. 2000. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology 49:149–157. [DOI] [PubMed] [Google Scholar]
- 60.Parker, J. L., and K. K. White. 1992. Lyme borreliosis in cattle and horses: a review of the literature. Cornell Vet. 82:253–274. [PubMed] [Google Scholar]
- 61.Patarakul, K., M. F. Cole, and C. A. N. Hughes. 1999. Complement resistance in Borrelia burgdorferi strain 297: outer membrane proteins prevent MAC formation at lysis susceptible sites. Microb. Pathog. 27:25–41. [DOI] [PubMed] [Google Scholar]
- 62.Porcella, S. F., C. A. Fitzpatrick, and J. L. Bono. 2000. Expression and immunological analysis of the plasmid-borne mlp genes of Borrelia burgdorferi strain B31. Infect. Immun. 68:4992–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoeler, G. B., and R. S. Lane. 1993. Efficiency of transovarial transmission of the Lyme disease spirochete, Borrelia burgdorferi, in the western blacklegged tick, Ixodes pacificus (Acari: Ixodidae). J. Med. Entomol. 30:80–86. [DOI] [PubMed] [Google Scholar]
- 64.Schwan, T. G., W. Burgdorfer, and P. A. Rosa. 1999. Borrelia, p.746–758. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 65.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith, R. P., P. W. Rand, E. H. Lacombe, S. R. Telford, S. M. Rich, J. Piesman, and A. Spielman. 1993. Norway rats as reservoir hosts for Lyme disease spirochetes on Monhegan Island, Maine. J. Infect. Dis. 168:687–691. [DOI] [PubMed] [Google Scholar]
- 67.Sonenshine, D. E. 1991. Biology of ticks, vol. 1. Oxford University Press, New York, N.Y.
- 68.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869–1879. [DOI] [PubMed] [Google Scholar]
- 70.Stevenson, B., S. Casjens, R. van Vugt, S. F. Porcella, K. Tilly, J. L. Bono, and P. Rosa. 1997. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J. Bacteriol. 179:4285–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stevenson, B., W. R. Zückert, and D. R. Akins. 2000. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol. 2:411–422. [PubMed] [Google Scholar]
- 74.Sung, S. Y., J. V. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 68:1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Telford, S. R., and A. Spielman. 1989. Enzootic transmission of the agent of Lyme disease in rabbits. Am. J. Trop. Med. Hyg. 41:482–490. [DOI] [PubMed] [Google Scholar]
- 76.van Dam, A. P., A. Oei, R. Jaspars, C. Fijen, B. Wilske, L. Spanjaard, and J. Dankert. 1997. Complement-mediated serum sensitivity among spirochetes that cause Lyme disease. Infect. Immun. 65:1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, X., T. G. Popova, K. E. Hagman, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 67:6008–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zipfel, P. F., T. S. Jokiranta, J. Hellwage, V. Koistinen, and S. Meri. 1999. The factor H protein family. Immunopharmacology 42:53–60. [DOI] [PubMed] [Google Scholar]