Abstract
Tuberculosis remains a leading killer worldwide, and new approaches for its treatment and prevention are urgently needed. This effort will benefit greatly from a better understanding of gene regulation in Mycobacterium tuberculosis, particularly with respect to this pathogen's response to its host environment. We examined the behavior of two promoters from the divergently transcribed M. tuberculosis genes acr/hspX/Rv2031c (α-crystallin homolog) and Rv2032/acg (acr-coregulated gene) by using a promoter-GFP fusion assay in Mycobacterium bovis BCG. We found that Rv2032 is a novel macrophage-induced gene whose expression is coregulated with that of acr. Relative levels of intracellular induction for both promoters were significantly affected by shallow standing versus shaking bacterial culture conditions prior to macrophage infection, and both promoters were strongly induced under low oxygen conditions. Deletion analyses showed that DNA sequences within a 43-bp region were required for expression of these promoters under all conditions. Multiple sequence alignment and database searches performed with PROBE indicated that Rv2032 is one of eight M. tuberculosis genes of previously unknown function that belong to an unusual superfamily of classical nitroreductases, which may have a role for bacteria within the host environment. These findings show that mycobacterial culture conditions can greatly influence the results and interpretation of subsequent gene regulation experiments. We propose that these differences might be exploited for dissection of the regulatory factors that affect mycobacterial gene expression within the host.
Mycobacterium tuberculosis, the etiologic agent of tuberculosis (TB), causes up to 2 million deaths per year and latently infects approximately one-third of the world's population (10). A worldwide rise in TB cases in the last decade and the emergence of multidrug-resistant strains led the World Health Organization to declare TB a global emergency (4, 36). Given this public health crisis and the significant gaps in our knowledge of the bacterium, a thorough study of the molecular basis of TB pathogenesis is required to develop better therapeutic and preventive methods.
The clinical course of TB infection is a poorly understood process, although it is known that the survival and replication of M. tuberculosis within unactivated macrophages are central to its ability to cause a sustained infection and disease (13). This intracellular survival is dependent on the specific regulation of M. tuberculosis genes in response to the intramacrophage environment. M. tuberculosis bacteria interact differently with lung epithelial cells and other newly infected macrophages following prior intracellular growth within other macrophages (3, 28, 29). These phenotypic alterations are likely due to changes in gene expression during the bacterium's time within the phagocytic environment. Two-dimensional gel-based proteomic analyses provide evidence for the extensive alteration of M. tuberculosis protein expression in response to these conditions (23, 28).
Recent studies have also begun to identify specific macrophage-induced M. tuberculosis genes that may be important in TB pathogenesis. Expression of the 16-kDa α-crystallin (Acr) protein is strongly induced and is required for M. tuberculosis growth within macrophages (57). acr expression is also induced in the extracellular milieu in response to nitric oxide, low oxygen, and stationary-phase conditions (9, 15, 19, 56). Members of the Acr family of heat shock proteins act as ATP-independent chaperones that are induced in response to environmental stresses in several microorganisms, although their specific function within macrophages is unknown. Three Mycobacterium marinum genes that have homologues of unknown function in M. tuberculosis are induced in cultured macrophages and frog granulomas (35). Additional studies using global screening approaches have identified a growing list of macrophage-induced genes, which have probable roles in nutrient utilization, protection from oxidative stress, and defense against xenobiotics (17, 26, 47).
Although these recent findings represent important advances in our knowledge of the genetics underlying the pathogenicity of M. tuberculosis, the regulatory networks that modulate M. tuberculosis gene expression in response to environmental stimuli within the macrophage are largely unknown. This gap in knowledge is further complicated by a generally poor understanding of gene regulation in M. tuberculosis.
We used the promoters of the macrophage-induced α-crystallin homolog gene, acr, and a divergently transcribed gene, Rv2032, in a reporter-based assay to evaluate the effects of growth conditions on bacterial gene expression within macrophages. We report here that Rv2032 is a novel macrophage-induced gene of M. tuberculosis and that its expression is coregulated with acr. Intracellular induction of both genes was affected by bacterial culture conditions prior to macrophage infection, and both promoters were strongly induced in response to low oxygen conditions. Multiple sequence alignment and database searching performed with PROBE (25, 33) indicated that Rv2032 is one of eight M. tuberculosis genes of previously unknown function which have products that belong to an unusual family of putative classical nitroreductases (CNRs) (50, 55). The implications of these findings are discussed.
MATERIALS AND METHODS
Growth of bacteria and macrophages.
Recombinant Mycobacterium bovis BCG (Pasteur strain, Trudeau Institute) seed-lot stocks were grown for 2 weeks at 37°C in mycomedium (Middlebrook 7H9 medium supplemented with 0.5% glycerol, 10% oleic acid-dextrose-catalase and 0.05% Tween 80) with kanamycin (25 μg/ml). These stocks were then frozen at ∼109 cells/ml in fresh mycomedium and stored in aliquots at −70°C until use. Fresh cultures were started for each experiment by inoculating 50 μl of M. bovis BCG from frozen seed-lots to 5 ml of mycomedium supplemented with kanamycin. All cultures were grown for the specified number of days in 25-cm2 tissue culture flasks, which were kept horizontal with the caps tightly screwed. The depth of the medium was 2 mm. Standing cultures were grown undisturbed, whereas shaking cultures were grown on a rocker platform (Reliable Scientific) at a speed setting of 30. Low-oxygen cultures were grown in vented-cap tissue culture flasks (25 cm2; Corning) with 1.3% oxygen and 5% CO2 in a Series II incubator (Forma Scientific, Inc). Low-oxygen conditions were achieved and maintained by the regulated addition of nitrogen to the incubator. Control experiments showed that 5% CO2 was required for growth of low-oxygen shaking BCG cultures, but had no effect on bacterial gene expression under high-oxygen (ambient) conditions (data not shown).
J774.16 murine macrophage cells were maintained by twice-weekly passages in tissue culture medium (Dulbecco's modified Eagle's medium [GIBCO] supplemented with 5% NCTC [GIBCO], 1% nonessential amino acids, 20% fetal bovine serum, and 1% glutamine) as described previously (30).
Cloning of mycobacterial promoters.
A 214-bp region upstream of and including the start codons of acr and the divergent open reading frame (ORF) for Rv2032 (7) was PCR amplified and cloned in pGFPoriM, which carries a promoterless gfpmut2 gene (8, 37). The DNA primer sequences used were GGATCCGTCCGGCATGATCAACCTCC (Rv2032) and GGATCCGGTGGCCATTTGATGCCTCC (acr). This DNA fragment contains 196 bp of intergenic sequence and was cloned in both orientations into a unique BamHI site immediately upstream of the gfpmut2 ORF. A 536-bp fragment containing the M. tuberculosis tuf gene promoter was cloned into pGFPoriM for use as a control. Preliminary studies had indicated that tuf promoter activity was unaffected inside macrophages (data not shown).
A series of promoter constructs with 5′ deletions of various lengths was made for the acr and acg promoters by using different 5′ primers for the PCR amplification step. Upstream primers for the acg promoter deletion series were as follows: 2032L3 (GGATCCGAAACGGATGCCTTTGATCC), 2032L2 (GGATCCAGGGCTAGGGACAGAAGTCC), 2032L1 (GGATCCGAAGCGCGGGCCATTTGTCC), and 2032SN (GGATCCCCCGTCGGTGATCCACTTGG). The upstream primers for the acr deletions included 2031L1 (GGATCCCTCCGCTGTTCGATATCACC), 2031L2 (GGATCCCCGATCTTTCTGAACGGCGG), 2031L3 (GGATCCGGTCAATGGTCCCCAAGTGG), 2031S1 (GGATCCGACAAATGGCCCGCGCTTCG), and 2031S2 (GGATCCCGGGGACTTCTGTCCCTAGC). Downstream primers in all cases were identical to those used for the full-length acg or acr promoter clones. All final promoter constructs were verified by DNA sequence analysis.
A single-copy integrative lacZ reporter vector (pLACint) was constructed by replacing a NarI-MunI DNA fragment that contained the oriM sequences in pJEM15 (46) (generously provided by B. Gicquel) with a 1.6-kbp cassette containing the attachment and integrating (attP/int) sequences from mycobacteriophage L5 (24). This attP/int cassette was amplified from pMH94 (kindly provided by G. Hatfull) by PCR by an overlap-extension procedure to specifically mutate an internal BamHI restriction site. This mutation was silent in that it did not affect the amino acid sequence. The final vector retains a unique BamHI cloning site at the start of the lacZ ORF that was used for insertion of acr (pACR:LACint) or gyrB (pGYRtb:LACint) promoter sequences from M. tuberculosis (37).
Measuring the growth phase of mycobacterial strains.
Multiple samples of each mycobacterial strain were grown under shaking and standing conditions to measure the growth phase of the bacterial cultures under the conditions mentioned above. At each time point, one new flask each of the shaking and standing samples was opened for sample removal and then discarded. The growth phase of the strains was determined by measuring the optical densities at 600 nm and by plating CFU in duplicate at each of two dilutions. In these assays, 5 days was approximately the mid-log phase, 7 days was approximately the late log phase, and 11 days was approximately the stationary phase for BCG. There was no appreciable difference in growth phase between strains grown shaking and those grown standing at the times measured.
Macrophage infection.
Recombinant BCG cultures were grown either shaking or standing for 5, 7, or 11 days prior to infection of macrophages. J774.16 cells were seeded at 4 × 105 cells/ml in six-well plates (Falcon) and grown overnight before infection with the recombinant BCG at a multiplicity of infection of 10 in tissue culture medium. An equal number of bacteria were incubated in tissue culture medium in six-well plates under identical conditions, to serve as the extracellular control. At 6 h postinfection (hpi), the inoculating medium was replaced with fresh tissue culture medium in the macrophage samples. At 24 or 48 hpi, the infected macrophages were washed three times with phosphate-buffered saline (PBS; 0.1 M NaCl, 0.01 M Na2HPO4, 0.001 M KH2PO4 [pH 7.6]) and lysed with a 0.5% (vol/vol) Triton X-100-0.5% (vol/vol) deoxycholate detergent mixture. Intracellular (IC) bacteria were harvested by centrifugation at 12,128 × g for 15 min. The bacteria from the extracellular (XC) control samples were also treated with the detergent mixture and harvested by centrifugation. Thereafter, all samples were kept covered on ice for flow cytometric analyses. Only a small percentage of bacteria within macrophages expressed green fluorescent protein (GFP) from the acr or acg promoters at 24 hpi, when assays were done with shaking cultures, so all data presented are from the 48-h time point.
Fluorescence assays.
Flow cytometric analysis of GFP-expressing bacteria was done in the FACScan system (Becton Dickinson Immunocytometry Systems), which was equipped with a 15-mW, 488-nm argon ion laser. Fluorescence emission was detected through a fluorescein isothiocyanate (FITC [530 ± 15 nm]) filter. Data were collected from 20,000 individual particles per sample, with gating for size using a forward scatter range of 35 to 55 relative units to exclude fluorescence from mycobacterial clumps. Microscopy was used in control experiments to confirm that the gated particles were predominantly single bacilli. Flow cytometric data were analyzed with the CellQuest (Becton Dickinson) software package.
β-Galactosidase activity was measured in a Cytofluor 4000 fluorometer (PerSeptive Biosystems) by using 5-acetylamino-fluorescein di-β-d-galactopyranoside (C2FDG; Molecular Probes) as described previously (37).
Microscopy.
Coverslips (22 by 22 cm2) were used in the six-well plates for microscopy experiments. Infected macrophages were washed three times with PBS and fixed for 30 min with 4% paraformaldehyde in PBS at room temperature before mounting onto slides with ProLong Antifade mounting medium (Molecular Probes). XC bacteria from control samples were spun onto slides at 1,000 rpm for 5 min in a cytospin centrifuge (Shandon). Slides were viewed with a Zeiss Axioskop microscope with an FITC filter (EX 450-490/EM 515 to 565 nm) at a magnification of ×400 or ×1,000. A DAGE MTI cooled charge-coupled device camera was used to record images with ScionImage 6.12c on a MacOS platform.
Sequence analysis.
Database mining and multiple sequence alignments were performed with PROBE (25, 33). Given a single protein sequence, PROBE performs a transitive BLAST search to identify a set of related sequences and proceeds to align only functionally constrained regions of the proteins, creating a “superlocal” multiple sequence alignment model consisting of individual motifs. The alignment model is then used to search the nonredundant protein database for additional sequences. PROBE iterates between refining the alignment model and searching the database until no additional sequences are recruited from the database.
PROBE calculates maximum a posteriori (MAP) scores for the model as well as for each motif. MAP scores are a measure of how well the alignment model describes the data relative to the unaligned random background. (MAP scores represent the log of the probability ratio.) A MAP score above zero indicates that the motif is more likely to belong to the alignment model than to the unaligned random background.
PROBE was initially used with default settings to identify and align two sets of sequences: one set was identified with the Rv2032 protein sequence (gi|2896769) as the query, and the second was identified by using the Thermus thermophilus NADH dehydrogenase (NOX) protein sequence (gi|3024211) as the query. These two sets of sequences were then combined into a single extended sequence set to form the family. PROBE was applied to this extended set for the final multiple sequence alignment of the family, setting the maximum expectation value at 1.0 and using 50 sampling iterations. PROBE requires that motifs occur in order in each protein sequence. The significant motifs identified by PROBE were then used with SCAN (32) to analyze the protein sequences for alternative motif order by analyzing each motif independently.
RESULTS
Differential regulation of acr and Rv2032/acg under shaking and standing growth conditions.
The acr gene is expressed at high levels in bacteria that have settled to the bottom of deep standing liquid cultures, presumably because of the lack of oxygen at the bottom of these tubes (9, 51, 52). In contrast, little acr expression occurs in mycobacteria grown with constant aeration in roller bottles (57).
We routinely grow very shallow cultures of mycobacteria undisturbed in tissue culture flasks, a condition in which acr expression has not been investigated. We compared acr promoter activity in these cultures, with (shaking) and without (standing) agitation, by using flow cytometry to measure GFP expressed from promoter-reporter fusion constructs in BCG. The promoter activity of the putative divergently transcribed gene Rv2032 was also measured at different stages of bacterial growth. A promoterless pGFPoriM construct and the constitutive EF-Tu promoter, tuf, were included as controls.
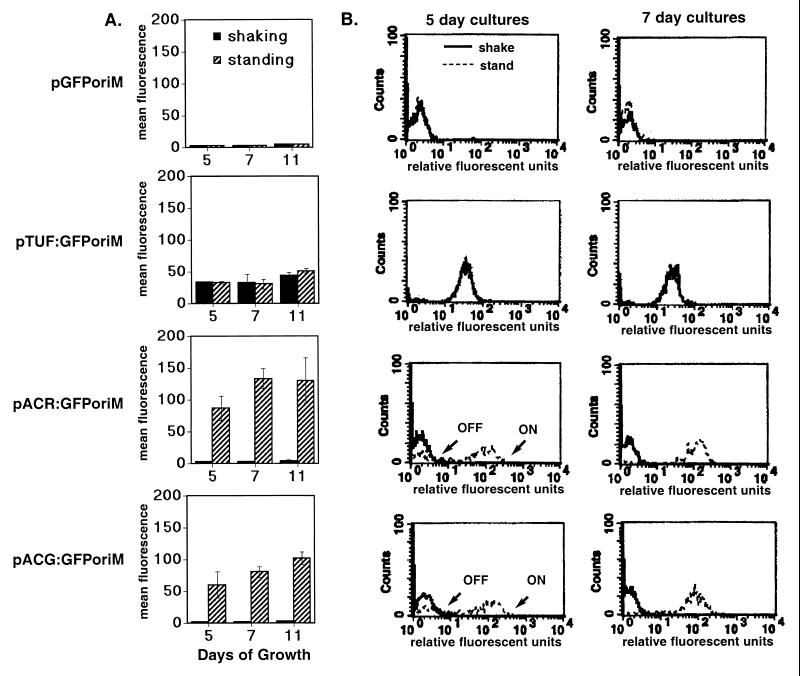
Very large differences in acr and Rv2032 promoter activity were observed between the standing and shaking cultures (Fig. 1). In contrast, expression levels for tuf were the same under both growth conditions. The acr promoter was strongly expressed in the standing cultures, but inactive in the shaking cultures. Maximal induction of the acr promoter was observed in the 7-day (late log) standing culture, which was ∼62-fold higher than in the shaking cultures (Fig. 1A). The Rv2032 promoter behaved similarly to acr (∼42-fold increase), and we have designated this gene acg (acr-coregulated gene). A key transition in activation of the acr and acg promoters was observed between 5 (mid-log)- and 7 (late log)-day standing cultures. The 5-day BCG(pACR:GFPoriM) and BCG(pACG:GFPoriM) cultures consisted of two populations of cells—one that showed promoter activity and another that did not (marked “on” and “off” [Fig. 1B]). In contrast, the entire bacterial population formed a single peak with a high level of promoter activity in the 7- and 11-day (early stationary) standing cultures. These results indicate that the promoters of acr and acg, but not tuf, were specifically up-regulated in standing versus shaking cultures.
FIG. 1.
Induction of acr and acg during growth in shaking and standing cultures. (A) The flow cytometry readings shown are the mean fluorescence per bacterium (in arbitrary fluorescence units) from the recombinant BCG grown under shaking (solid bars) or standing (striped bars) conditions for 5 (n = 3), 7 (n = 2), or 11 (n = 4) days. Error bars indicate standard deviations. (B) Representative flow profiles of recombinant BCG strains grown shaking (solid lines) or standing (broken lines) for 5 or 7 days. Relative fluorescence units are shown in a log scale on the x axis, and rightward peak migration correlates with increased fluorescence intensity in the sample bacteria. Counts shown on the y axis denote the number of bacteria counted at each fluorescence intensity. Relative peak position, rather than height, is most important when comparing GFP expression between samples. Dotted lines are not apparent in the pGFPoriM and pTUForiM samples, because the shaking and standing curves completely overlap. Note that the 5-day standing cultures (broken lines) for BCG(pACR:GFP) and BCG(pACG:GFP) have two subpopulations, which appear as distinct peaks labeled “off” and “on.”
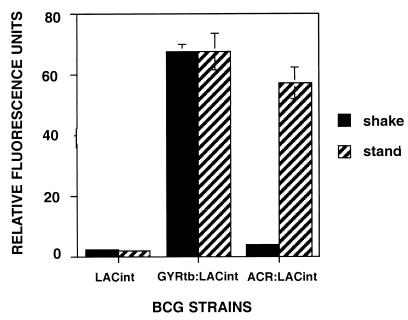
These experiments were done with plasmid-based promoter fusions with an estimated copy number of five plasmids per cell (42). Others have shown that some mycobacterial promoters regulate poorly on plasmids (42), so we confirmed this differential regulation for acr by using a single-copy lacZ reporter vector (pACR:LACint) integrated into the BCG chromosome. β-Galactosidase activity was measured with a fluorescence plate reader with C2FDG substrate in intact viable BCG, because this assay is more sensitive than GFP (37), which did not produce sufficient signal in the single-copy vector (data not shown). Reporter signal from the single-copy integrated construct was lower than that of the plasmid-based construct, but the ratios of differential expression between shallow standing and shaking conditions were similar in both systems (Fig. 2).
FIG. 2.
Comparison of acr promoter activity under shaking (solid) versus standing (striped) conditions with lacZ reporter fusions integrated into the BCG chromosome in single copy. Recombinant BCG bacteria contained a promoterless vector control (pLACint), an M. tuberculosis gyrB promoter-lacZ fusion (pGYRtb:LACint), or an acr promoter-lacZ fusion (pACR:LACint). Relative fluorescent units shown are per 106 bacteria and were measured in viable bacteria by using a Cytofluor fluorescence plate reader at an excitation/emission ratio of 485/530 after a 2-h incubation at 37°C with C2FDG substrate. The results shown are the mean of two experiments, and the bars denote standard deviations.
Growth conditions prior to infection affect the relative induction levels of acr and acg within macrophages.
A previous study reported strong induction of acr in M. tuberculosis within macrophages when bacteria from agitated cultures were used for infection (57). We compared the effects of shaking and standing culture conditions on the subsequent intracellular behavior of various promoters following macrophage infection. The four recombinant BCG strains carrying pGFPoriM-based constructs were used to infect macrophages and analyzed by flow cytometry at 48 hpi. Flow cytometry was chosen for these experiments, because it allows us to determine the mean fluorescent signal per bacterium in each sample. This measurement is independent of bacterial number and does not require normalization to adjust for size differences in the IC and XC populations that are sampled. Flow cytometry is also useful because it detects multiple populations if they are present, as shown in Fig. 1.
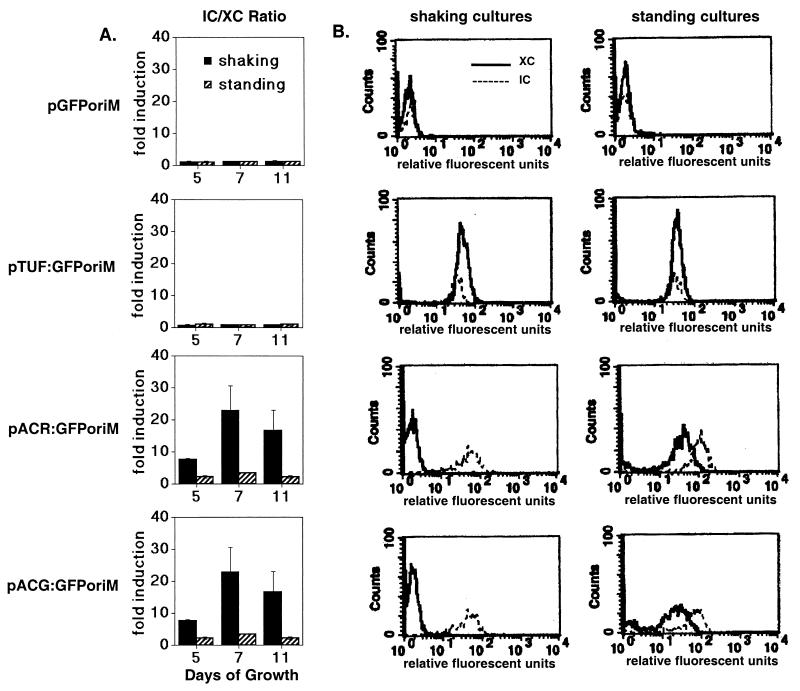
GFP expression from the acr and acg promoters increased up to 25-fold in bacteria within macrophages relative to XC bacteria when assays were done with shaking cultures, depending on the growth stage of the bacteria (Fig. 3). acr and acg promoter activity in bacteria from standing cultures also increased further following macrophage infection, but the maximal levels reached were only two to three times the level of extracellular bacteria. These lower relative levels of intracellular induction were observed with the standing cultures, because the promoters were already highly active in the shallow standing cultures prior to macrophage infection. The tuf promoter was expressed constitutively and showed no induction in any of the experiments (Fig. 3). These results show for the first time that the acg promoter, like acr, is highly active within macrophages. They also show that similar levels of acr and acg promoter activity can be achieved outside of macrophages under shallow standing culture conditions.
FIG. 3.
Relative expression of acr and acg within macrophages following prior growth of cultures under shaking or standing conditions. (A) Relative levels of promoter induction in BCG at 48 hpi of macrophages are shown for four recombinant BCG strains. Macrophages were infected with bacteria grown either shaking or standing prior to macrophage infection for 5 (n = 2), 7 (n = 2), or 11 (n = 4) days. Fold induction is calculated as mean fluorescence per bacterium of IC/mean fluorescence of XC bacteria. Error bars denote standard deviations. (B) Flow profiles for IC (solid lines) and XC (dotted lines) bacteria in a representative experiment with bacteria from 7-day shaking or standing cultures used to infect the macrophages. x and y axes are as described for Fig. 1, with the relative position of peaks on the x axis being more important than their height. Separation of the peaks reflects a difference in GFP production, whereas overlying peaks indicate the similarity of GFP levels within the bacteria of the sample populations.
Maximal intracellular induction of both promoters occurred in the 7-day late-log- to early-stationary-phase bacteria, while mid-log-phase (5 day) shaking cultures produced two populations (active and inactive) within macrophages. This biphasic pattern of acr and acg, but not tuf, expression within macrophages was nearly identical to that of the bacteria in 5-day standing cultures (Fig. 1B). The “active” group included 30 to 50% of the intracellular bacteria, which expressed GFP from either promoter at levels similar to the intracellular bacteria from 7-day cultures (not shown). acr and acg promoter expression remained at background levels in the remaining “inactive” group of intracellular bacteria.
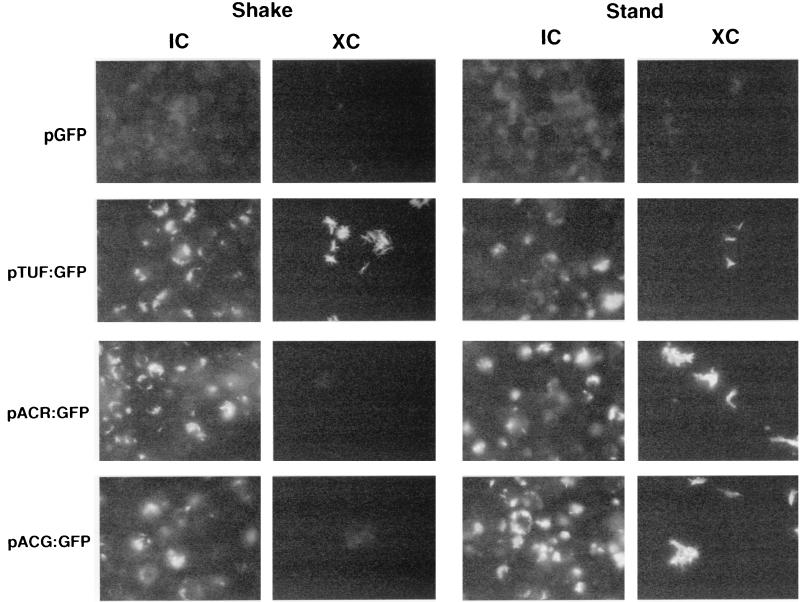
A potential drawback of flow cytometry is that the fluorescence measured from a bacterial population gated for single cells may not be representative of the whole sample, given that mycobacteria tend to clump and form cords in culture. We used fluorescence microscopy to visually assess the relative levels of acr and acg expression within the entire bacterial population. Promoter expression was examined in multiple fields by fluorescence microscopy at 48 hpi in an infection assay done with 5- or 11-day cultures. Bacteria carrying either pACR:GFPoriM or pACG:GFPoriM were brightly flourescent within macrophages, regardless of their prior growth conditions (Fig. 4). In contrast, XC bacteria fluoresced only if they had been pregrown under standing, but not shaking, conditions. BCG(pTUF:GFPoriM) was uniformly fluorescent in all of the IC and XC samples. Likewise, the vector-only control strain BCG(pGFPoriM) was nonfluorescent under all conditions.
FIG. 4.
Fluorescence microscopy of recombinant BCG strains within macrophages (IC) or in tissue culture media (XC) control samples. Representative micrographs shown are at 48 hpi of macrophages with 11-day-old cultures grown under shaking or standing conditions. Similar results were obtained with 7-day-old cultures. Fluorescence is due to GFP expression within the bacteria.
The acg promoter responds to low oxygen conditions.
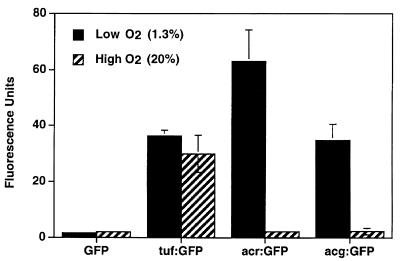
Previous studies showed that the acr promoter is strongly induced in response to low oxygen conditions (40, 57). We compared the activities of the acg, acr, and tuf promoters in the presence of ambient (∼20.9%) or low (1.3%) oxygen conditions in shaking cultures (Fig. 5). Relative fluorescence of recombinant BCG containing each promoter-GFP construct was measured by flow cytometry. Both the acg and acr promoters were strongly expressed under low oxygen conditions and inactive under high (ambient) oxygen conditions. Fluorescence associated with the acr promoter was nearly twice that observed with the acg construct. Expression of the tuf promoter was constitutive and was not affected by oxygen tension.
FIG. 5.
Response of the tuf, acr, and acg promoters to different oxygen conditions. Recombinant BCG bacteria containing the various promoter-GFP fusions were grown shaking for 7 days under low (solid bars) or high (striped bars) oxygen conditions, as described. The results shown are the mean relative fluorescence of three independent experiments. Error bars denote standard deviations. The absence of error bars indicates the standard deviation was <1.0.
Adjacent DNA sequences are required for expression of the divergent acr and acg promoters.
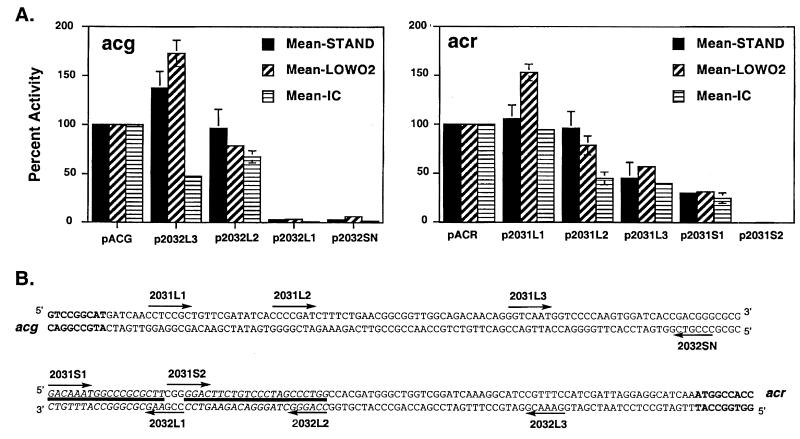
A series of DNA fragments were generated with successive 5′ deletions from the full-length acg and acr promoter fragments to identify cis-acting DNA sequences that may be involved in regulation (Fig. 6). In both cases, elimination of a short region at the 5′ end of the DNA fragment that contained the start codon of the divergently transcribed gene increased expression under low oxygen, but not intramacrophage, conditions (p2032L3 and p2031L1 in Fig. 6). In contrast, this deletion decreased acg promoter activity within macrophages ∼50%. Expression from the acg promoter was otherwise similar to that of the full-length DNA fragment under all conditions until a 22-bp region (between the 2032L2 to 2032L1 end points) was deleted, which included nucleotides (nt) 116 to 138 upstream of the putative acg start codon. Loss of this 22-bp region resulted in abrupt termination of expression from the acg promoter.
FIG. 6.
Deletion analysis of acg and acr promoters to identify potential cis regulatory sequences required for the response of these promoters to various conditions. (A) Percent activity for each shortened DNA promoter fragment is shown relative to the full-length (214 bp) acg (left) or acr (right) promoter fragments used throughout the study. Percent activity was calculated for each independent experiment as the ratio of mean fluorescence for the shortened fragment relative to the mean fluorescence of the full-length fragment × 100%. The values shown are the overall means of the percent activity calculated for each DNA fragment in several experiments. The different conditions (number of replicates in parentheses) are as follows: standing (n = 3), solid bars; low oxygen (n = 3), diagonal striped bars; and intramacrophage (n = 2), horizontal striped bars. Error bars denote standard deviations. The absence of error bars for values other than the full-length promoters indicates that the standard deviation was <5.0. (B) Sequence of full-length DNA promoter fragment used in these studies, with arrows marking the 5′ endpoint of each truncated fragment. acg and acr ORF sequences are shown in bold. Solid lines denote the regions of 18 to 22 bp for each promoter that were required for GFP expression under all three conditions tested. Deletion of additional upstream regions had effects for some, but not all of the conditions. The essentiality of downstream regions was not tested for either promoter.
Expression from the acr promoter was abolished under all conditions by deletion of an 18-bp region that extended from nt 81 to 99 upstream of the acr start codon. The essential regions in the two promoters are separated by 3 nt and may comprise a larger regulatory site that is important for expression of both promoters. Loss of an additional upstream region (nt −135 to −171 from the acr ORF) partially reduced expression of the acr promoter under standing and low oxygen conditions. Intramacrophage expression was instead influenced by deletion of a region immediately upstream of these sequences.
Rv2032/acg may encode a nitroreductase.
The product of the Rv2032/acg ORF has been annotated as a protein of unknown function with homology to two other M. tuberculosis proteins of unknown function (Rv3131 and Rv3127). BLAST analysis of the unfinished microbial genomes at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) showed that Rv2032 also had significant pairwise homology to several additional sequences from other mycobacterial (M. bovis and M. avium) and streptomycete species, but these were also of unknown function (1). Because Rv2032 contained no known protein domains described in the Pfam database (2), we decided to use PROBE, a more sophisticated sequence analysis technique to predict the functional nature of Rv2032. PROBE is a multiple sequence alignment and database search algorithm that dynamically identifies and aligns only functionally constrained regions of protein sequence patterns (33). The resulting “superlocal” alignment model typically consists of several colinear, conserved motifs that describe the protein domains common to that family (27).
PROBE identified a family of 74 protein sequences related to Rv2032. This family included known NAD(P)H oxidoreductases and nitroreductases, many of which are included in the Pfam CNR family (PF00881) (2). Seven additional M. tuberculosis proteins of unknown function and many hypothetical protein sequences from other organisms were also part of this family (Table 1). Three-dimensional structures have been determined for four of the oxido- and nitroreductase proteins in this family, and these were analyzed in the context of the PROBE motif models.
TABLE 1.
Superfamily proteinsa
| Species | Protein | Structure | Known function |
|---|---|---|---|
| Proteins with known function | |||
| Vibrio cholerae | Fra1 | Flavin oxidoreductase | |
| Vibrio fischeri | Fra1 | 1VFR | Flavin oxidoreductase |
| Vibrio cholerae | Frp | Flavin oxidoreductase | |
| Vibrio harveyi | Frp | 1BKJ | Flavin oxidoreductase |
| Enterobacter cloacae | NfnB | 1NEC | NAD(P)H nitroreductase |
| Escherichia coli | NfnB | NAD(P)H nitroreductase | |
| Salmonella enterica serovar Typhimurium | NfnB | NAD(P)H nitroreductase | |
| Escherichia coli | NfsA | NADPH nitroreductase | |
| Salmonella enterica serovar Typhimurium | NfsA | NADPH nitroreductase | |
| Bacillus subtilis | NfrA | Nitro/flavin reductase | |
| Helicobacter pylori | FrxA | NADPH-flavin oxidoreductase | |
| Helicobacter pylori | RdxA | NADPH nitroreductase | |
| Synechocystis sp. strain PCC 6803 | DrgA | Nitroreductase | |
| Thermus thermophilus | Nox | 1NOX | NADH oxidoreductase |
| M. tuberculosis proteins | |||
| Mycobacterium tuberculosis H37Rv | Rv0306 | ||
| Mycobacterium tuberculosis H37Rv | Rv1355c | ||
| Mycobacterium tuberculosis H37Rv | Rv2032 | ||
| Mycobacterium tuberculosis H37Rv | Rv2337c | ||
| Mycobacterium tuberculosis H37Rv | Rv3127 | ||
| Mycobacterium tuberculosis H37Rv | Rv3131 | ||
| Mycobacterium tuberculosis H37Rv | Rv3262 | ||
| Mycobacterium tuberculosis H37Rv | Rv3368c |
Family members not listed are hypothetical or function is assigned only by similarity. A full list including all family members is available on our web site.
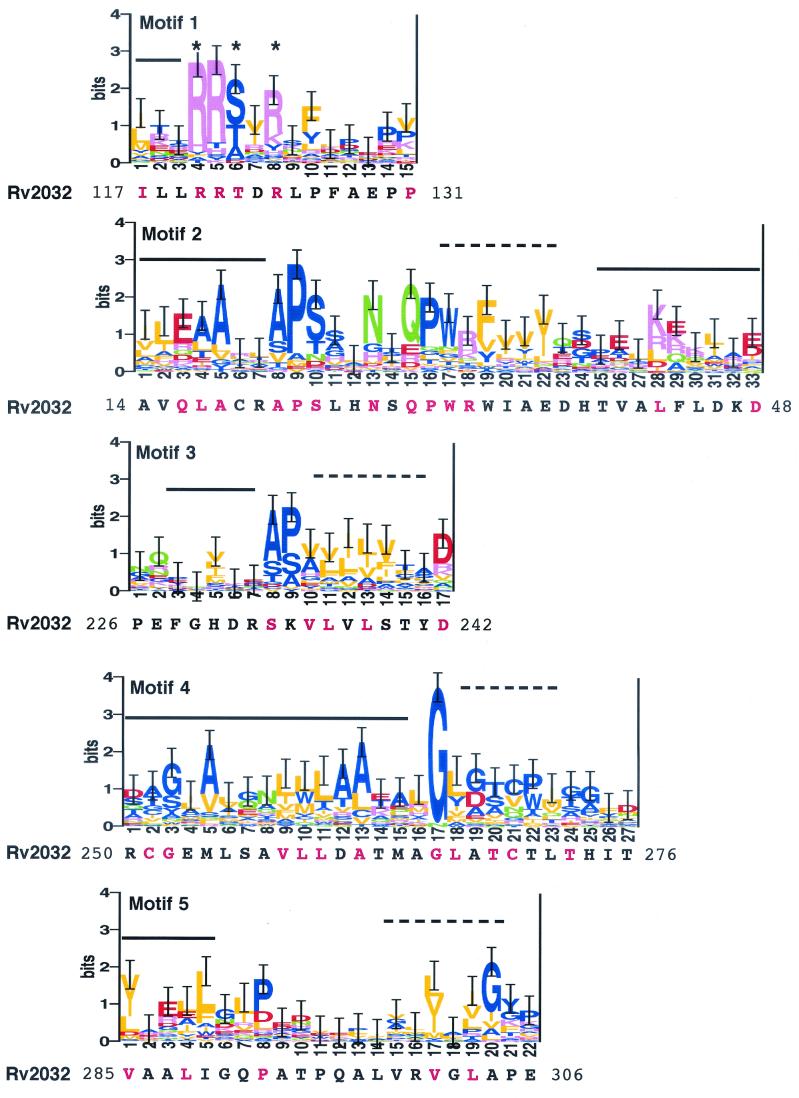
The PROBE-generated multiple sequence alignment model for this family consisted of five motifs, shown as sequence logos in Fig. 7. This model encompasses the region described as the core (or sandwich) catalytic domain of the flavoproteins Frp and Fra1 (21, 44, 45) and includes structural elements and residues known to be involved in flavin mononucleotide (FMN) cofactor binding (18, 21, 44, 45) (Fig. 7). The phosphate moiety of FMN forms hydrogen bonds with two strongly conserved arginine residues (positions 4 and 8) and the polar residue (position 6) of motif 1 (marked with asterisks in Fig. 7). Mutation studies have also implicated sequences in the region of motif 2 between positions 4 and 11 in FMN binding stability (50). Several additional strongly conserved residues in the model are of unknown function and are not in positions within known secondary structural elements (Fig. 7).
FIG.7.
The five motifs in the multiple sequence alignment model for this reductase family, represented as sequence logos (38). The logos represent the 74 family members purged at an MSP score of 150 to prevent bias from overrepresented sequences (e.g., of the three highly homologous NfnB sequences, from E. cloacae, E. coli, and S. typhimurium, only one is retained after purging). The MAP scores for the motifs are 89.53 (motif 1), 432.11 (motif 2), 11.19 (motif 3), 300.23 (motif 4), and 31.07 (motif 5). The most probable alignment of Rv2032 amino acid sequence for each motif as determined by SCAN analysis is included under each logo, with starting and ending residue positions noted at each end of the sequence. For those positions in the model with ≥1.0 information bit, the residues in Rv2032 that correspond to the conserved residues are in red. Correspondence of the motifs with known secondary structural elements is indicated above each motif, where solid lines represent alpha-helix and dashed lines denote beta-strand structures. Conserved residues known to form hydrogen bonds with the phosphate moiety of FMN in some family members are marked with asterisks at positions 4, 6, and 8 of motif 1.
An NADH binding site conforming to the GXGXXG motif that is associated with Rossmann dinucleotide binding folds (31, 39) was not detected in the primary Rv2032 sequence or the superfamily model. This is consistent with the classification of the CNR superfamily as a group distinct from the Rossmann fold-associated superfamily of dinucleotide binding proteins (http://scop.mrc-lmb.cam.ac.uk/scop/). However, the QPWHF motif (at positions 15 to 19 in motif 2) that is common to all of the CNRs (16) is similar to the SQPWPFP motif that is specific for the NADH pyrophosphatase class of NUDIX hydrolases (54), suggesting that these sequences may be associated with NAD(P)H binding in the CNR family.
The multiple sequence alignment algorithm used by PROBE requires that the motifs occur in order in the primary sequence of each protein. However, close inspection of the 74 protein sequences in this family (Rv2032 in particular) revealed that the most probable alignment for 6 of the proteins resulted in the motif order 2-1-3-4-5 in the primary sequence (N terminal to C terminal). These six proteins were from M. tuberculosis (Rv2032, Rv3127, and Rv3131) or Streptomyces coelicolor (SCJ1.11, SCJ12.27c, and RedV). All other members of this family had the motif order 1-2-3-4-5 originally identified by PROBE. This permutation of the first two motifs in Rv2032 likely resulted in Rv2032 not being recognized as a member of the Pfam CNR family (PF00881). Permutation of motifs in a protein domain has been observed previously for the glutathione S-transferase (GST) domain (22) and the α/β-hydrolase fold (A. F. Neuwald, personal communication).
DISCUSSION
We report here the results of our study on the effects of shallow standing versus shaking growth conditions on the expression of the M. tuberculosis acr and acg promoters. An important feature of this work is the identification of a previously uncharacterized M. tuberculosis gene, acg/Rv2032, that coregulates with acr and is strongly induced within macrophages. Sequence analyses of the predicted Rv2032 amino acid sequence indicated that acg belongs to a putative family of CNRs. In addition, we showed that in BCG the acr and acg promoters are induced in shallow standing cultures in a manner that resembles their behavior within macrophages and in response to low oxygen conditions. A common region of each promoter is required for its expression under all three conditions. Whether acg is also required for intracellular survival of M. tuberculosis within macrophages or under hypoxic conditions remains to be determined.
Effects of culture conditions.
These results demonstrate that culture conditions can affect mycobacterial gene expression in unexpected ways. For example, intramacrophage induction of the acr promoter in this study was similar to that observed by others (57). However, this occurred only when macrophages were infected with bacteria from shaking cultures, because both promoters were already preinduced to near-maximal levels during bacterial growth in shallow standing cultures. Two-dimensional gel electrophoresis and mass spectrometry proteomic analyses of bacteria from standing versus shaking BCG cultures have shown additional differences in the protein expression profiles of bacteria grown under these two conditions (14), suggesting that other genes may also be affected.
The increases we observed with acr and acg expression under low oxygen conditions are consistent with the findings of a recent report in which transcription of both acr and Rv2032 was strongly induced by hypoxia (40). acr expression was also previously shown to increase in response to the hypoxic conditions generated by using the Wayne model for induction of M. tuberculosis latency, in which bacteria are subjected to a microaerophilic environment as they settle to the bottom of a deep liquid culture through a self-generating oxygen gradient (9, 51, 52, 57).
The nearly identical expression patterns of the truncated promoter fragments under standing and low oxygen conditions are consistent with low oxygen tension being a key regulatory signal for expression of both acr and acg in shallow standing cultures, although other factors cannot be ruled out. This finding was unexpected, because the model used in the present study differs from the Wayne model in several key respects. Our standing cultures were very shallow (2 mm) compared to the Wayne model (41 to 70 mm) and had a substantial surface area (at least 25 cm2) exposed to the air interface of a very large headspace. The resulting headspace air/liquid ratio in our studies was 30:1, while that typically used in the Wayne model is often as low as 0.5:1.
It is likely that oxygen depletion under shallow standing conditions occurred within the immediate vicinity of the settled bacteria as a result of respiration, which may explain the length of time required for induction in the present study. The microenvironments of the bacteria under shaking versus standing conditions may also differ with respect to local pH and/or nutrient conditions, as well as the accumulation of metabolic by-products. Differences due to cell-cell interactions or quorum sensing also cannot be ruled out.
The deletion studies identified a short region for each promoter that was essential for expression under all three conditions tested. This suggests that both promoters are subject to positive regulation, consistent with a recent study that showed Rv3133c is required for acr induction under low oxygen conditions (40). The two regions we identified were nearly adjacent and may form a larger regulatory site common to both promoters. The essentiality of DNA sequences downstream of these regions was not tested. Additional studies are needed to define the precise boundaries of these regulatory sequences and the specific nucleotides that are involved. Intramacrophage expression was also affected by upstream sequences that did not influence standing or low oxygen expression, suggesting that the regulation within the macrophage may be more complex than under standing or low oxygen conditions. Additional levels of regulation for standing and low oxygen conditions are also possible.
The highest levels of GFP were obtained from the acr and acg constructs when the bacteria were transitioning from the late-log to stationary phase in standing cultures. This is consistent with the findings of others, who observed maximal acr/hspX transcription in late-log-phase bacteria subjected to microaerophilic conditions, despite continued accumulation of Acr protein (5, 9, 19, 56). In our study, these increases were due to a proportional increase in the number of bacteria with active acr and acg promoters between 5 and 7 days, rather than higher levels of expression within individual GFP-expressing bacteria (Fig. 1).
Predicted function of acg.
Advanced sequence analysis using the PROBE algorithm showed that acg/Rv2032 is part of a family of M. tuberculosis ORFs, all previously of unknown function, which belong to a CNR superfamily of enzymes. CNRs are oxygen-insensitive flavin-dependent enzymes that reduce nitroaromatic compounds, often forming toxic and/or mutagenic compounds (48-50). These data show the power of PROBE in characterizing unknown ORFs, which make up approximately 20% of the M. tuberculosis genome (7), and facilitating predictions of possible roles for these unknown gene products. Despite a lack of high overall amino acid conservation, PROBE detected critical sequence similarities between the M. tuberculosis CNR-like proteins and their homologues in unrelated organisms that were not apparent by pairwise sequence analysis (BLAST) or comparison to the multiple sequence alignments in Pfam, since these alignments are strictly linear. Comparison of the regions of similarity between Rv2032 and some of these better-characterized family members may also facilitate the further characterization of functional domains within these related proteins.
The prototypic member of the CNR family is the nitroreductase from Salmonella enterica serovar Typhimurium that plays an important role in the Ames mutagenicity test (49). Inactivation of the gene encoding this nitroreductase confers resistance to the killing effects of nitroarenes, while overexpression increases the bacterium's sensitivity to nitroaromatic compounds (50). Other well-characterized enzymes with amino acid similarity to this S. enterica serovar Typhimurium CNR include the flavin-dependent nitroreductases of Enterobacter cloacae and Escherichia coli (6, 58, 59), the flavin reductase of Vibrio fischeri (60), the NADH oxidase of Thermus thermophilus (18, 34), DrgA of Synechocystis sp. (12), and the NADPH nitroreductase RdxA of Helicobacter pylori (16).
Coregulation of acg with acr suggests that both genes' products may be needed at similar times during the M. tuberculosis infection process. The acg gene product may be involved in detoxification within macrophages and/or granulomas, where nitroaromatic compounds may be present. Members of the M. tuberculosis CNR family could also have a role in M. tuberculosis drug sensitivity. Mutations in one (rdxA) of two CNR homologues in H. pylori are the primary source of metronidazole (Mtz) resistance in this organism (16), and rdxA confers metronidazole sensitivity when expressed in E. coli (41). Similarly, inactivation of the CNR-encoding drgA gene in the cyanobacterium Synechocystis sp. results in resistance to Mtz and nitrophenolic herbicides (12). The second CNR family member in H. pylori, frxA, is also associated with Mtz resistance, although it is not a primary target (20).
Mtz kills dormant, but not actively growing, cultures of M. tuberculosis (53). A newly developed nitroimidazopyran Mtz analog, PA-824, is effective against M. tuberculosis in actively growing and static cultures, as well as in mice (43), and this drug has considerable promise as a new lead candidate for TB treatment in humans. The mode of action of the CNR family proteins that confers Mtz sensitivity in H. pylori is activation of the Mtz prodrug (11, 16). It is possible that one or more of the M. tuberculosis CNR proteins play a similar role in conferring sensitivity to this important class of nitroaromatic prodrugs, and this would seem to be an important line of future investigation.
This report describes a novel M. tuberculosis gene, acg, whose expression coregulates with acr, a gene that is thought to be important during several stages of the M. tuberculosis infection process. Identification of the regulatory loci that control the expression of these genes, as well as the specific environmental signals that affect their expression, will contribute to our understanding of M. tuberculosis gene expression within and outside of its host. In addition, future studies to confirm the predicted function of the acg gene product and determine its importance to M. tuberculosis survival within macrophages and/or granulomas may provide new insights into the constellation of factors that contributes to this pathogen's extraordinary resilience within its host.
Acknowledgments
We gratefully acknowledge the assistance of the Wadsworth Center's Computational Molecular Biology and Statistics Core; R. Dilwith of the Immunology Core; and T. Moran and M. Shutt of the Molecular Genetics Core. B. Rowland made important contributions to the construction of the pLACint vector, and plasmid DNA was generously donated by B. Gicquel and G. Hatfull. We also thank M. Florczyk, C. E. Lawrence, and K. Derbyshire for helpful discussions and critical reading of the manuscript.
This work was supported in part by National Institutes of Health grant AI4565801 (K.A.M.) and the Potts Foundation (K.A.M). L.A.M. was supported by NIH grant HG01257 (C.E.L.) and Department of Energy grant 96ER62266 (C.E.L.).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermudez, L. E., and J. Goodman. 1996. Mycobacterium tuberculosis invades and replicates within type II alveolar cells. Infect. Immun. 64:1400-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom, B. R., and C. J. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055-1064. [DOI] [PubMed] [Google Scholar]
- 5.Boon, C., R. Li, R. Qi, and T. Dick. 2001. Proteins of Mycobacterium bovis BCG induced in the Wayne dormancy model. J. Bacteriol. 183:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant, C., L. Hubbard, and W. D. McElroy. 1991. Cloning, nucleotide sequence, and expression of the nitroreductase gene from Enterobacter cloacae. J. Biol. Chem. 266:4126-4130. [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham, A. F., and C. L. Spreadbury. 1998. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J. Bacteriol. 180:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W.H.O. Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, D. I. 1979. Mechanism of antimicrobial action of metronidazole. J. Antimicrob. Chemother. 5:499-502. [DOI] [PubMed] [Google Scholar]
- 12.Elanskaya, I. V., E. A. Chesnavichene, C. Vernotte, and C. Astier. 1998. Resistance to nitrophenolic herbicides and metronidazole in the cyanobacterium Synechocystis sp. PCC 6803 as a result of the inactivation of a nitroreductase-like protein encoded by drgA gene. FEBS Lett. 428:188-192. [DOI] [PubMed] [Google Scholar]
- 13.Fenton, M. J., and M. W. Vermeulen. 1996. Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect. Immun. 64:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florczyk, M. A., L. A. McCue, R. F. Stack, C. R. Hauer, and K. A. McDonough. 2001. Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect. Immun. 69:5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbe, T. R., N. S. Hibler, and V. Deretic. 1999. Response to reactive nitrogen intermediates in Mycobacterium tuberculosis: induction of the 16-kilodalton α-crystallin homolog by exposure to nitric oxide donors. Infect. Immun. 67:460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 17.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht, H. J., H. Erdmann, H. J. Park, M. Sprinzl, and R. D. Schmid. 1995. Crystal structure of NADH oxidase from Thermus thermophilus. Nat. Struct. Biol. 2:1109-1114. [DOI] [PubMed] [Google Scholar]
- 19.Hu, Y., and A. R. M. Coates. 1999. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J. Bacteriol. 181:1380-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong, J.-Y., A. K. Mukhopadhyay, D. Dailidiene, Y. Wang, B. Velapatino, R. H. Gilman, A. J. Parkinson, G. B. Nair, B. C. Y. Wong, S. K. Lam, R. Mistry, I. Segal, Y. Yuan, H. Gao, T. Alarcon, M. L. Brea, Y. Ito, D. Kersulyte, H.-K. Lee, Y. Gong, A. Goodwin, P. S. Hoffman, and D. E. Berg. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koike, H., H. Sasaki, T. Kobori, S. Zenno, K. Saigo, M. E. Murphy, E. T. Adman, and M. Tanokura. 1998. 1.8 A crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminescent bacterium, Vibrio fischeri: overall structure, cofactor and substrate-analog binding, and comparison with related flavoproteins. J. Mol. Biol. 280:259-273. [DOI] [PubMed] [Google Scholar]
- 22.Koonin, E. V., A. R. Mushegian, R. L. Tatusov, S. F. Altschul, S. H. Bryant, P. Bork, and A. Valencia. 1994. Eukaryotic translation elongation factor 1 gamma contains a glutathione transferase domain—study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 3:2045-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, B. Y., and M. A. Horwitz. 1995. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J. Clin. Investig. 96:245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J. S., A. F. Neuwald, and C. E. Lawrence. 1999. Markovian structures in biological sequence alignments. J. Am. Statist. Assoc. 94:1-15. [Google Scholar]
- 26.Mariani, F., G. Cappelli, G. Riccardi, and V. Colizzi. 2000. Mycobacterium tuberculosis H37Rv comparative gene-expression analysis in synthetic medium and human macrophage. Gene 253:281-291. [DOI] [PubMed] [Google Scholar]
- 27.McCue, L. A., K. A. McDonough, and C. E. Lawrence. 2000. Functional classification of cNMP-binding proteins and nucleotide cyclases with implications for novel regulatory pathways in Mycobacterium tuberculosis. Genome Res. 10:204-219. [DOI] [PubMed] [Google Scholar]
- 28.McDonough, K. A., M. A. Florczyk, and Y. Kress. 2000. Intracellular passage within macrophages affects the trafficking of virulent tubercle bacilli upon reinfection of other macrophages in a serum-dependent manner. Tubercle Lung Dis. 80:259-271. [DOI] [PubMed] [Google Scholar]
- 29.McDonough, K. A., and Y. Kress. 1995. Cytotoxicity for lung epithelial cells is a virulence-associated phenotype of Mycobacterium tuberculosis. Infect. Immun. 63:4802-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonough, K. A., Y. Kress, and B. R. Bloom. 1993. The interaction of Mycobacterium tuberculosis with macrophages: a study of phagolysosome fusion. Infect. Agents Dis. 2:232-235. [PubMed] [Google Scholar]
- 31.Mittl, P. R., A. Berry, N. S. Scrutton, R. N. Perham, and G. E. Schulz. 1994. Anatomy of an engineered NAD-binding site. Protein Sci. 3:1504-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuwald, A. F., J. S. Liu, and C. E. Lawrence. 1995. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 4:1618-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuwald, A. F., J. S. Liu, D. J. Lipman, and C. E. Lawrence. 1997. Extracting protein alignment models from the sequence database. Nucleic Acids Res. 25:1665-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park, H. J., R. Kreutzer, C. O. Reiser, and M. Sprinzl. 1993. Molecular cloning and nucleotide sequence of the gene encoding a H2O2-forming NADH oxidase from the extreme thermophilic Thermus thermophilus HB8 and its expression in Escherichia coli. Eur. J. Biochem. 211:909.. [PubMed] [Google Scholar]
- 35.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 36.Raviglione, M. C., D. E. Snider, Jr., and A. Kochi. 1995. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA 273:220-226. [PubMed] [Google Scholar]
- 37.Rowland, B., A. Purkayastha, C. Monserrat, Y. Casart, H. Takiff, and K. A. McDonough. 1999. Fluorescence-based detection of lacZ reporter gene expression in intact and viable bacteria including Mycobacterium species. FEMS Microbiol. Lett. 179:317-325. [DOI] [PubMed] [Google Scholar]
- 38.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scrutton, N. S., A. Berry, M. P. Deonarain, and R. N. Perham. 1990. Active site complementation in engineered heterodimers of Escherichia coli glutathione reductase created in vivo. Proc. R. Soc. Lond. B Biol. Sci. 242:217-224. [DOI] [PubMed] [Google Scholar]
- 40.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sisson, G., J.-Y. Jeong, A. Goodwin, L. Bryden, N. Rossler, S. Lim-Morrison, A. Raudonikiene, D. E. Berg, and P. S. Hoffman. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J. Bacteriol. 182:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 43.Stover, C. K., P. Warrener, D. R. VanDevanter, D. R. Sherman, T. M. Arain, M. H. Langhorne, S. W. Anderson, J. A. Towell, Y. Yuan, D. N. McMurray, B. N. Kreiswirth, C. E. Barry, and W. R. Baker. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962-966. [DOI] [PubMed] [Google Scholar]
- 44.Tanner, J. J., B. Lei, S. C. Tu, and K. L. Krause. 1996. Flavin reductase P: structure of a dimeric enzyme that reduces flavin. Biochemistry 35:13531-13539. [DOI] [PubMed] [Google Scholar]
- 45.Tanner, J. J., S. C. Tu, L. J. Barbour, C. L. Barnes, and K. L. Krause. 1999. Unusual folded conformation of nicotinamide adenine dinucleotide bound to flavin reductase P. Protein Sci. 8:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Triccas, J. A., F. X. Berthet, V. Pelicic, and B. Gicquel. 1999. Use of fluorescence induction and sucrose counterselection to identify Mycobacterium tuberculosis genes expressed within host cells. Microbiology 145:2923-2930. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe, M., M. Ishidate, Jr., and T. Nohmi. 1990. Nucleotide sequence of Salmonella typhimurium nitroreductase gene. Nucleic Acids Res. 18:1059.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe, M., M. Ishidate, Jr., and T. Nohmi. 1989. A sensitive method for the detection of mutagenic nitroarenes: construction of nitroreductase-overproducing derivatives of Salmonella typhimurium strains TA98 and TA100. Mutat. Res. 216:211-220. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe, M., T. Nishino, K. Takio, T. Sofuni, and T. Nohmi. 1998. Purification and characterization of wild-type and mutant “classical” nitroreductases of Salmonella typhimurium. L33R mutation greatly diminishes binding of FMN to the nitroreductase of S. typhimurium. J. Biol. Chem. 273:23922-23928. [DOI] [PubMed] [Google Scholar]
- 51.Wayne, L. G. 1976. Dynamics of submerged growth of Mycobacterium tuberculosis under aerobic and microaerophilic conditions. Am. Rev. Respir. Dis. 114:807-811. [DOI] [PubMed] [Google Scholar]
- 52.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wayne, L. G., and H. A. Sramek. 1994. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, W., C. A. Dunn, and M. J. Bessman. 2000. Cloning and characterization of the NADH pyrophosphatases from Caenorhabditis elegans and Saccharomyces cerevisiae, members of a Nudix hydrolase subfamily. Biochem. Biophys. Res. Commun. 273:753-758. [DOI] [PubMed] [Google Scholar]
- 55.Yamada, M., J. J. Espinosa-Aguirre, M. Watanabe, K. Matsui, T. Sofuni, and T. Nohmi. 1997. Targeted disruption of the gene encoding the classical nitroreductase enzyme in Salmonella typhimurium Ames test strains TA1535 and TA1538. Mutat. Res. 375:9-17. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, Y., D. D. Crane, and C. E. Barry III. 1996. Stationary phase-associated protein expression in Mycobacterium tuberculosis: function of the mycobacterial α-crystallin homolog. J. Bacteriol. 178:4484-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan, Y., D. D. Crane, R. M. Simpson, Y. Q. Zhu, M. J. Hickey, D. R. Sherman, and C. E. Barry III. 1998. The 16-kDa alpha-crystallin (Acr) protein of Mycobacterium tuberculosis is required for growth in macrophages. Proc. Natl. Acad. Sci. USA 95:9578-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zenno, S., H. Koike, A. N. Kumar, R. Jayaraman, M. Tanokura, and K. Saigo. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 178:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zenno, S., H. Koike, M. Tanokura, and K. Saigo. 1996. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J. Biochem. 120:736-744. [DOI] [PubMed] [Google Scholar]
- 60.Zenno, S., K. Saigo, H. Kanoh, and S. Inouye. 1994. Identification of the gene encoding the major NAD(P)H-flavin oxidoreductase of the bioluminescent bacterium Vibrio fischeri ATCC 7744. J. Bacteriol. 176:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]