Abstract
In the present study we investigated the role of the fibronectin (FN)- and fibrinogen (FGN)-binding protein (FBPS) in the pathogenesis of Streptococcus suis serotype 2 in piglets. The complete gene encoding FBPS from S. suis serotype 2 was cloned in Escherichia coli and sequenced. The occurrence of the gene in various serotypes was analyzed by hybridization studies. The FBPS protein was expressed in E. coli and purified, and binding to human FN and FGN was demonstrated. The induction of antibodies in piglets was studied upon infection. An isogenic mutant unable to produce FBPS was constructed, and the levels of virulence of the wild-type and mutant strains were compared in a competitive infection model in young piglets. Organ cultures showed that FBPS was not required for colonization of the tonsils but that FBPS played a role in the colonization of the specific organs involved in an S. suis infection. Therefore, the FBPS mutant was considered as an attenuated mutant.
Streptococcussuis causes severe infections in piglets. The bacterial infections include meningitis, septicemia, and arthritis, and the animals often do not survive the infection (6, 28). Occasionally, S. suis causes septicemia and meningitis in humans (3). The pathogenesis of an S. suis infection is rarely understood. Sows are symptomless carriers of S. suis on their tonsils and pass the bacteria on to their piglets. The piglets cannot cope with the bacterium and subsequently develop the specific symptoms of an S. suis infection. Until now, 35 capsular serotypes of S. suis have been described (26), but serotype 2 strains are most often isolated from diseased piglets. Capsule is an important virulence factor, since piglets infected with an acapsular mutant of S. suis serotype 2 strains do not develop any clinical symptoms (22). Bacterial proteins have been suggested to play a role in the pathogenesis as well (1, 26). The expression of muramidase-released protein (MRP), extracellular factor (EF), and suilysin was shown to be strongly associated with pathogenic strains of S. suis serotype 2 (2, 29, 30). Since isogenic mutants lacking MRP and EF and isogenic mutants lacking suilysin were still pathogenic for young piglets, these proteins are not absolutely required for virulence (1, 23). Recently, a new virulence factor was identified (21) by using a complementation approach. The function of this virulence factor in the pathogenesis has to be further investigated.
Many important virulence factors are environmentally regulated and are induced at specific stages of the infection process (15). To identify these genes in S. suis, we cloned promoters and their downstream sequences that are “on” during experimental S. suis infection of piglets (20). Twenty-two in vivo-selected (ivs) genes were found. Two of the ivs genes were directly linked to virulence, since homology to genes in the database that encode for known virulence factors was found. One of these ivs genes (ivs-21) was identical to the epf gene of virulent S. suis serotype 2 strains (30). The other (ivs-31) showed homology to genes encoding fibronectin (FN)- and/or fibrinogen (FGN)-binding proteins of Streptococcus gordonii (GenBank accession no. X65164) and Streptococcus pyogenes FBP54 (8). A considerable number of FN-binding proteins of various bacterial species have been shown to be important virulence factors (12). In S. pyogenes, FBP54 was shown to be expressed in the human host and to preferentially mediate adherence to human buccal epithelial cells (7). It was recently shown that the FBP54 protein induces protective immunity against S. pyogenes challenge in mice (13).
In the present study we describe an FN- and FGN-binding protein of S. suis (FBPS). The sequence of fbps was determined. Binding studies showed that purified FBPS bound FN and FGN. A contribution of FBPS to the pathogenesis of S. suis serotype 2 was found.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. suis strains were grown in Todd-Hewitt broth (code CM 189; Oxoid, Ltd., London, United Kingdom) and plated on Columbia blood base agar plates (code CM 331; Oxoid, Ltd.), containing 6% (vol/vol) horse blood. Escherichia coli strains were grown in Luria broth (17) and plated on Luria broth containing 1.5% (wt/vol) agar. If required, the following antibiotics were added at the indicated concentrations: spectinomycin (Sigma, St. Louis, Mo.) (50 μg/ml for E. coli and 100 μg/ml for S. suis), ampicillin (Boehringer, Mannheim, Germany) (100 μg/ml for E. coli), and kanamycin (Boehringer) (25 μg/ml for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL2-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr) amy Cmr] | Stratagene |
| M15 | Nals Strs Rifs Thi− Lac− Ara+ Gal+ Mtl− F− RecA+ Uvr+ Lon+ | Qiagen |
| S. suis | ||
| 10 | Virulent serotype 2 strain | 29 |
| 10ΔFBPS | Isogenic fbps mutant of strain 10 | This work |
| Plasmids | ||
| pGEM7Zf(+) | Replication functions pUC, Ampr | Promega Corp. |
| pKUN19 | Replication functions pUC, Ampr | 14 |
| pIC19R | Replication functions pUC, Ampr | 16 |
| pDL282 | Replication functions of pBR322 and pVT736-1, Ampr Spcr | 25 |
| pIC-spc | pIC19R containing Spcr gene of pDL282 | Laboratory collection |
| pQE-30 | Replication functions pBR322, Ampr, expression vector, six-His tag | Qiagen |
| pQE-30-FBPS | pQE-30 containing the 1.8-kb fbps gene | This work |
| pREP4 | Replication functions pACYC, Kanr, lacI gene | Qiagen |
| pE194 | Emr | 11 |
| pIVS-E | Replication functions of pWV01, Spcr, promoterless erm gene of pE194 | 20 |
| pIVS-31 | pIVS-E containing 200 bp showing homology to Streptococcus gordonii flpa | 20 |
| pFBPS7-46 | pGEM7Zf(+) containing EcoRI-EcoRI fragment of fbps | This work |
| pFBPS7-47 | pFBPS7-46 in which 382-bp SalI-SalI fragment is replaced by 1.2-kb Spcr from pIC-spc | This work |
Tetr, tetracycline resistant; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant; Spcr, spectinomycin resistant; Kanr, kanamycin resistant.
DNA techniques and sequence analysis.
Routine DNA manipulations were performed as described by Sambrook et al. (19). DNA sequences were determined on a 373A DNA Sequencing System (Applied Biosystems, Warrington, Great Britain). Samples were prepared by use of an ABI Prism dye terminator cycle sequencing ready reaction kit (Applied Biosystems). Sequencing data were assembled and analyzed using the Lasergene program (DNASTAR). The BLAST software package was used to search for protein sequences homologous to the deduced amino acid sequences in the GenBank and EMBL databases.
Southern blotting and hybridization.
Chromosomal DNA was isolated as described by Sambrook et al. (19). DNA fragments were separated on 0.8% agarose gels and transferred to GeneScreen Plus hybridization transfer membrane (NEN Life Science Products, Boston, Mass.) as described by Sambrook et al. (19). DNA probes of the fbps and spc genes were labeled with [α-32P]dCTP (3,000 Ci/mmol; Amersham Life Science, Little Chalfont, Buckinghamshire, Great Britain) by use of a random primed DNA labeling kit (Boehringer). The DNA on the blots was prehybridized for at least 30 min at 65°C and subsequently hybridized for 16 h at 65°C with the appropriate DNA probes in a buffer containing 0.5 M sodium phosphate (pH 7.2), 1 mM EDTA, and 7% sodium dodecyl sulfate (SDS). After hybridization, the membranes were washed twice with a buffer containing 40 mM sodium phosphate (pH 7.2), 1 mM EDTA, and 5% SDS for 30 min at 65°C and twice with a buffer containing 40 mM sodium phosphate (pH 7.2), 1 mM EDTA, and 1% SDS for 30 min at 65°C. The signal was detected on a phosphorimager (Storm; Molecular Dynamics, Sunnyvale, Calif.).
Construction of an fbps knockout mutant.
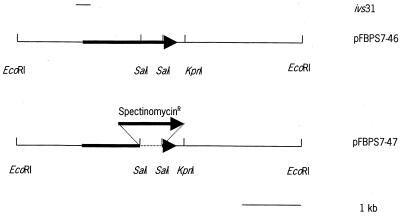
To construct the mutant strain 10Δ FBPS, the pathogenic strain 10 (27, 29) of S. suis serotype 2 was electrotransformed (24) with the plasmid pFBPS7-47. In this plasmid, the fbps gene was inactivated by the insertion of a spectinomycin resistance gene. To create pFBPS7-47 (Fig. 1) the 382-bp SalI-SalI fragment of pFBPS7-46 was replaced by the 1.2-kb EcoRV-SmaI fragment of pIC-Spc, containing the spectinomycin resistance gene, after the SalI sites of the vector were made blunt (Fig. 1). After electrotransformation of strain 10 with pFBPS7-47, spectinomycin-resistant colonies were selected on Columbia agar plates containing spectinomycin (100 μg/ml). Southern blotting and hybridization experiments were used to select for double-crossover integration events (data not shown).
FIG. 1.
Schematic presentation of the procedure used to clone the fbps gene of S. suis serotype 2 and the construction of an insertional knockout mutant in S. suis serotype 2. A 5-kb EcoRI fragment was cloned in pGEM7Zf(+), yielding pFBPS7-46. In pFBPS7-47, the 382-bp SalI-SalI fragment of pFBPS7-46 was replaced by a 1.2-kb spectinomycin resistance gene, after the vector was made blunt, to obtain an insertional knockout of fbps.
FBPS expression construct.
To construct an FBPS expression plasmid the QIA expressionist kit (Qiagen GmbH, Hilden, Germany) was used. The primers corresponded to positions 250 to 272 and positions 1911 to 1892 of the fbps gene. The sequences of these primers were 5′-GCGGATCCGATGACGATGACAAATCTTTTGACGGATTTTTTTTAC-3′ and 5′-CCCAAGCTTGGGCATGAACTAGATTTTCATGG-3′. The primers contained restriction sites for BamHI and HindIII, respectively, to amplify the fbps gene from pFBPS7-47. The amplified PCR product was digested with BamHI and HindIII, and the 1.8-kb fbps gene was cloned into pQE-30 digested with BamHI and HindIII, yielding pQE-30-FBPS. pQE-30-FBPS was transformed to M15[pREP4].
Purification of FBPS.
M15[pREP4][pQE-30-FBPS] was used to express and to purify the FBPS using the QIA expressionist (Qiagen). In short, M15[pREP4][pQE-30-FBPS] cells were grown exponentially; 1 mM IPTG was added, and the cells were allowed to grow another 4 h at 37°C. Subsequently, cells were harvested and lysed. The cleared supernatants were loaded onto Ni2+-nitrilotriacetic acid agarose columns. FBPS containing a six-His tag was bound to the Ni2+ column. The columns were washed and the protein was eluted. Different buffers were used for native and for denaturing purification. FBPS purified under denaturing conditions was renatured on an Ni2+-nitrilotriacetic acid column by using a linear urea gradient (6 M to 1 M) in 500 mM NaCl, 20% glycerol, and 20 mM Tris-HCl (pH 7.4), containing protease inhibitors (per milliliter: 25 μg of Pefabloc, 0.7 μg of pepstatin, 1 μg of aprotinin, and 0.5 μg of leupeptin). All procedures were performed according to the manufacturer's recommendations. The six-His tag was removed from the protein by incubating purified FBPS in a solution containing 20 mM Tris-HCl (pH 7.4), 50 mM NaCl, 2 mM CaCl2, and 0.5 U of light-chain enterokinase (New England Biolabs, Beverly, Mass.) for 16 h at room temperature (RT).
Immunization of rabbits with FBPS.
Purified and renatured FBPS was used to immunize two rabbits. To remove urea the protein was dialyzed against phosphate-buffered saline (136 mM NaCl, 2.68 mM KCl, 8.1 mM Na2HPO4, 2.79 mM KH2PO4 [pH 7.2]) (PBS) overnight at 4°C. Seven days before immunization, blood was collected from the rabbits to determine the natural titers of antibody against FBPS. At day 1 those rabbits with negative anti-FBPS titers were immunized intramuscularly with two injections of 0.5 ml of 100-μg/ml FBPS in a water-in-oil emulsion (Specol; ID-Lelystad, Lelystad, The Netherlands). At day 28, rabbits were immunized for the second time using the same amount of protein and the same route of immunization. Three weeks after the second immunization the rabbits were sacrificed and blood was collected. The blood was coagulated and serum was collected and used for immunodetection of FBPS.
Immunodetection of FBPS.
Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) by standard procedures (19). Proteins in the gel were visualized using SYPRO-orange (Molecular Probes, Sunnyvale, Calif.) staining according to the manufacturer's recommendations. Signals were detected on a phosphorimager (Storm; Molecular Dynamics). A known bovine serum albumin concentration range was used as a standard to calculate the amounts of protein present in the gel. The Molecular Dynamics program was used for the calculations.
Proteins were transferred to a nitrocellulose membrane by standard procedures (19). The membranes were blocked in Blotto-Tris-buffered saline (50 mM Tris-HCl [pH 7.5], 150 mM NaCl) (Blotto-TBS) containing 4% skim milk and 0.05% Tween 20, at RT for 1 h. To detect recombinant purified FBPS, membranes were incubated with a monoclonal antibody against the six-His tag (Clontech, Palo Alto, Calif.) in a 1:10,000 dilution in Blotto-TBS (1:1) at RT for 1 h, followed by an incubation with alkaline phosphatase-conjugated anti-mouse antibody in a 1:1,000 dilution in Blotto-TBS (1:1) at RT for 1 h. Reactivity of purified FBPS was tested by using a convalescent-phase serum of a pig that had survived an S. suis infection. Nitrocellulose membranes were incubated with the polyclonal pig serum in a 1:200 dilution in Blotto-TBS (1:1) at RT for 1 h, followed by an incubation at RT for 1 h with alkaline phosphatase-conjugated anti-swine antibody in a 1:2,000 dilution in Blotto-TBS (1:1). As a substrate Nitro Blue Tetrazolium (Merck, Darmstadt, Germany)-bromochloroindolyl phosphate (Sigma) was used. All washing steps were performed in Blotto-TBS (1:1).
FN and FGN binding.
Binding studies were performed by indirect Western blotting. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane as described above. The membranes were blocked in MPBS (PBS containing 4% skim milk and 0.05% Tween 20). Subsequently, the membrane was incubated with human FN (5 μg/ml; Sigma) or human FGN (5 μg/ml; Sigma) in PBS containing 5% fetal calf serum, 2% NaCl, and 0.05% Tween 80 at RT for 1 h. To detect bound FN and FGN, the membranes were incubated with horseradish peroxidase-conjugated anti-FN (DAKO) or anti-FGN (DAKO) antibodies in a 1:1,000 dilution in PBS containing 5% fetal calf serum, 2% NaCl, and 0.05% Tween 80 at RT for 1 h. The signal was visualized by using an ECL+ kit (Amersham Life Science) according to the manufacturer's recommendations. Signals were detected on a phosphorimager (Storm; Molecular Dynamics). All washing steps were performed in MPBS-PBS (1:1).
Experimental infections.
Germfree piglets, crossbreeds of Great Yorkshire and Dutch Landrace, were obtained from sows by cesarean sections. The surgery was performed in sterile flexible film isolators. Piglets were allotted to groups of 4, and were housed in sterile stainless steel incubators. Housing conditions and feeding regimens were as described before (27, 29). Six-day-old piglets were inoculated intranasally with about 107 CFU of Bordetella bronchiseptica 92932, to predispose the piglets to infection with S. suis. Two days later they were inoculated intranasally with 106 CFU of S. suis strain 10 plus 106 CFU of S. suis strain 10Δ FBPS. To determine differences in virulence between wild-type and mutant strains, 50% lethal doses should be determined. To do this, large numbers of piglets would be required. For ethical reasons this is not acceptable. To circumvent this problem we decided to perform cocolonization studies. To monitor for the presence of S. suis and B. bronchiseptica and to check for absence of contaminants, swabs taken from the nasopharynx and the feces were cultured three times a week. The swabs were plated directly onto Columbia agar containing 6% horse blood or grown for 48 h in Todd-Hewitt broth and subsequently plated onto Columbia agar containing 6% horse blood. Pigs were monitored twice a day for clinical signs and symptoms, such as fever, nervous signs, and lameness. Blood samples from each pig were collected three times a week. Leukocytes were counted with a cell counter. The piglets were killed when specific signs of an S. suis infection were observed, such as arthritis or meningitis, or when the pigs became mortally ill. The other piglets were killed 2 weeks after inoculation with S. suis and examined in the same way as the piglets that were killed based on their clinical symptoms. All piglets were examined for pathological changes. Tissue specimens from heart, lung, liver, kidney, spleen, and tonsil and from the organs specifically involved in an S. suis infection (central nervous system [CNS], serosas, and joints) were sliced with a scalpel or a tissuemizer. Tissue slices from each organ or site were resuspended in 2 to 25 ml of Todd-Hewitt broth containing 15% glycerol, depending on the size of the tissue slice. The suspension was centrifuged at 1,200 δ g for 5 min. The supernatant was collected and serial dilutions were plated on Columbia agar containing 6% horse blood, as well as on Columbia agar plates containing 6% horse blood and 100 μg of spectinomycin per ml to quantitate the number of wild-type and mutant bacteria present. The number of mutant strain 10Δ FBPS cells was determined by counting the number of CFU on the appropriate serial dilution on the selective plates; the number of wild-type strain 10 cells was determined by counting the number of CFU on the appropriate serial dilution on the Columbia agar blood plates, from which the number of CFU counted on the selective plates was subtracted. When wild-type and mutant bacteria were found in tissues, the ratio of wild-type and mutant strain was determined again, by moving about 100 individual colonies by toothpick onto both Columbia agar plates and onto Columbia agar plates containing spectinomycin (100 μg/ml).
All animal experiments were approved by the ethical committee of the Institute for Animal Science and Health, Lelystad, The Netherlands, in accordance with the Dutch law on animal experiments.
Nucleotide sequence accession number.
The nucleotide sequence data of fbps have been submitted to GenBank under accession no. AF438158.
RESULTS
Cloning of the S. suis fbps gene.
One of the in vivo-selected genes (ivs-31) (20) showed homology to the 5′ part of genes coding for FlpA and FBP54, FN binding proteins (FnBP) of S. gordonii (GenBank accession no. X65164) and S. pyogenes (8), respectively. To clone the entire fbps gene of S. suis, ivs-31 was subsequently used as a probe to identify a chromosomal DNA fragment of S. suis serotype 2 containing flanking fbps sequences. A 5-kb EcoRI fragment was identified and cloned in pGEM7Zf(+) yielding pFBPS7-46 (Fig. 1). Sequence analysis revealed that this fragment contained the entire fbps gene of S. suis serotype 2. An open reading frame of 1,659 bp coding for a polypeptide of 553 amino acids was found. The putative ATG start codon is preceded by a sequence similar to ribosome binding sites of gram-positive bacteria. Further upstream, two putative promoter sequences could be identified. Upstream of these promoter sequences of fbps an inverted repeat was found that could serve as a transcription terminator of the gene located 5′ of fbps. Downstream of fbps a gene was found that showed homology to an alpha-acetolactate decarboxylase was found. This gene is transcribed in the opposite direction of fbps. The deduced amino acid sequence was aligned with that of several previously identified FnBPs from other bacteria. As expected, FBPS was very homologous to FlpA of S. gordonii (76%) and also showed homology to FnBPs of other organisms, like Streptococcus pneumoniae (73%), S. pyogenes (69%), Lactococcus lactis (59%), and Bacillus subtilis (41%). Compared to the sequence of FBP54, FBPS has a longer N terminus with 76 additional amino acids. This longer N terminus was also seen in other organisms like S. gordonii, S. pneumoniae, and B. subtilis. In FBP54 the primary FN-/FGN-binding domain was localized to its N-terminal part, to the first 89 amino acids (8). Over this region the homology of FBPS to FBP54 is very high (80%) suggesting that FBPS can bind both FN and FGN.
Binding of FBPS to FN and FGN.
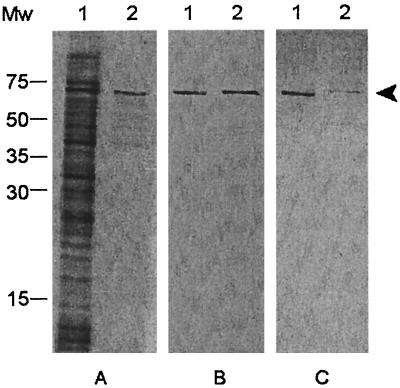
To confirm the binding of FBPS of S. suis to FN and FGN, FBPS was purified under native conditions. A protein expression construct, which expresses FBPS with a six-His tag fused to the N terminus, was used for this purification. Four hundred μg of FBPS was purified from 50 ml of exponential-phase E. coli cells after induction with IPTG. The purity of this FPBS was determined with SDS-PAGE and Western blotting (Fig. 2). The induced E. coli lysate contained a broad range of proteins, among which the 64-kDa protein FBPS was very clearly present (Fig. 2A, lane 1). After purification, highly purified FBPS with six-His tag was obtained (Fig. 2A, lane 2). When both samples were incubated with a monoclonal antibody against the six-His tag, FBPS was the only protein that was detected (Fig. 2B).
FIG. 2.
Purity and immunogenicity of FBPS purified under native conditions. SDS-PAGE analysis with SYPRO-orange, a nonspecific protein-staining dye (A), and Western blot analysis with a monoclonal antibody against the six-His tag (B) of 4 μl of E. coli M15 [pQE-30-pREP4-FBPS] lysate (lanes 1) and 165 ng of purified FBPS (lanes 2) were carried out. Convalescent-phase serum raised against S. suis strain 10 was used to test immunogenicity of FPBS present in 4 μl of E. coli M15 [pQE-30-pREP4-FBPS] lysate and 0.5 μg of purified FBPS (C) (lanes 1 and 2). Arrowhead, 64-kDa FPBS; Mw, molecular size marker (in kilodaltons).
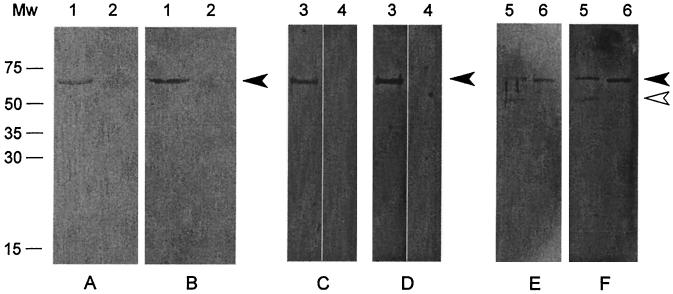
To determine whether FBPS binds FN and FGN, a Western blot containing purified FBPS was incubated with soluble human FN and human FGN (Fig. 3A and B). Specific binding of FN and FGN to FBPS was clearly detected. No binding of FN and FGN to BSA, a negative control protein, was observed. To exclude possible background signals due to immunoglobulin-binding of FBPS, the same experiment was performed without addition of FN or FGN. No binding was found (Fig. 3C and D), indicating that the binding was specific for FN and FGN. To control whether the binding of FN and FGN to FBPS, was not mediated by the six-His tag, the tag was removed by an enterokinase treatment. Figure 3E and F clearly show that FBPS without the six-His tag, still efficiently bound to FN and FGN. Therefore, it was concluded that FBPS can specifically bind to FN and FGN.
FIG. 3.
Binding studies with purified FBPS. (A and B) Gels were probed with FN (A) or FGN (B) at 5 μg/ml. Lanes 1 contain 500 ng of purified FBPS, and lanes 2 contain 500 ng of BSA. (C and D) Lanes 3 and 4 contain 500 ng of purified FBPS. Lanes 3 were probed with FN (C) or FGN (D) at 20 μg/ml, and lanes 4 were only incubated with conjugate without FN or FGN. (E and F) Gels were probed with FN (E) or FGN (F) at 20 μg/ml. Lanes 5 contain 1.8 μg of purified FBPS digested with enterokinase, and lanes 6 contain 500 ng of purified FBPS. The closed arrowhead indicates 64-kDa FBPS; the open arrowhead indicates approximately 55-kDa FBPS without the six-His tag. Mw, molecular size marker (in kilodaltons).
Immunogenicity of FBPS.
Since it was shown that FBP54 induced a protective immune response in mice against a lethal dose of S. pyogenes (13), we next determined whether purified FBPS was recognized by convalescent-phase serum of a pig that survived an S. suis infection. As shown in Fig. 2 panel C, the FBPS clearly reacted with this antiserum. When the same experiment was performed with nonimmune serum of an SPF piglet, no band of the size of FBPS was detected (data not shown). These findings indicate that FBPS is expressed in vivo and that the protein is indeed immunogenic in young pigs.
Distribution of the fbps gene among the 35 S. suis serotypes.
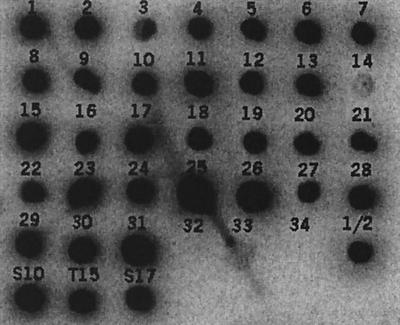
Since we were interested in a cross-protective vaccine candidate, we next analyzed the presence of the fbps gene among the various S. suis serotypes. ivs-31, the clone containing the promoter and the 5′ part of the fbps gene was radiolabeled, and chromosomal DNA of the reference strains of the 35 different S. suis serotypes was hybridized with this probe. The three different phenotypes of S. suis serotype 2—a pathogenic, a nonpathogenic, and a weakly pathogenic strain—were included in this study as well. The fbps gene was present in all S. suis serotypes and phenotypes, except for serotypes 32 and 34 (Fig. 4).
FIG. 4.
Distribution of fbps among various S. suis serotypes. Chromosomal DNA (1 μg) was spotted onto nitrocellulose membrane and hybridized with a 32P-labeled fbps probe. Serotypes were spotted as indicated. S10, S. suis serotype 2 MRP+ EF+; T15, S. suis serotype 2 MRP− EF−; S17, S. suis serotype 2 MRP+ EF* (23).
Role of FBPS in pathogenesis.
To test the role of FBPS in the pathogenesis of S. suis, an isogenic knockout mutant of FBPS was constructed in strain 10, strain 10ΔFBPS. Since upstream of fbps an inverted repeat was found that could serve as a transcription terminator and downstream of fbps a gene showing homology to an alpha-acetolactate decarboxylase was found that is transcribed in the opposite direction, polar effects on genes upstream or downstream of fbps are not expected. To verify that the mutant strain 10ΔFBPS did not produce FBPS, protoplasts of strain 10 and strain 10ΔFBPS were subjected to SDS-PAGE and Western blotting. FBPS was detected using a polyclonal antiserum raised against purified FBPS. It was shown that strain 10 41 FBPS expressed no FBPS, while strain 10 did (data not shown). Subsequently the virulence of this mutant strain was tested in an experimental infection in piglets. The mutant strain 10ΔFBPS was used in a competition challenge experiment with the wild-type strain to determine the relative attenuation of the mutant strain. Under in vitro conditions, the growth rates of the wild-type and mutant strain in Todd-Hewitt medium were found to be essentially identical (data not shown). Wild-type and mutant strains were inoculated at an actual ratio of 0.65 (1.63 × 1066 CFU of wild-type bacteria ml−1 and 3.09 × 106 CFU of mutant bacteria ml−1). During the experiment, piglets that developed specific S. suis symptoms (meningitis, arthritis, or mortal illness) were killed. Piglets that did not develop these symptoms were killed at the end of the experiment. From all piglets the ratio of wild-type strain to mutant strain in various organs was determined. As shown in Fig. 5A, similar numbers of wild-type and mutant bacteria were reisolated from tonsils. The ratio was similar to the input ratio (ratio varied from 0.33 to 0.85; average, 0.61). This clearly indicates that the efficiencies of colonization of wild-type and mutant strains on tonsils were essentially identical. Apparently, FBPS is not strictly required for colonization of the tonsils of the piglets. Three out of four piglets developed clinical signs specific for an S. suis infection. Two piglets (4664 and 4666) showed clinical signs of arthritis, and one piglet (4668) showed clear central nervous signs. The fourth piglet did not develop any clinical signs. These observations coincided with pathomorphological abnormalities of the specific organs of an S. suis infection in postmortem sections. As shown in Fig. 5B and Table 2, exclusively wild-type bacteria were reisolated from the joints of piglet 4664 and from the CNS of piglet 4668. The numbers of CFU of wild-type bacteria that were reisolated from these specific organs were very high, while absolutely no mutant bacteria were found. From the joints of pig 4666 low numbers of both wild-type and mutant bacteria were reisolated in a ratio of 0.84 (1.0 × 102 CFU of wild-type bacteria and 5.2 × 102 CFU of mutant bacteria), a ratio essentially identical to the input ratio (Fig. 5B and Table 2). Southern blot experiments, using the fbps and the spc genes as probes, confirmed that the mutant bacteria isolated from the joint of pig 4666 were indeed identical to the input mutant bacteria. Taken together, these data indicate that the FBPS mutant is capable of reaching and colonizing the specific S. suis organs (at least the joints) but that the mutant is far less efficiently recovered from organs than the wild type.
FIG. 5.
Efficiency of colonization of wild-type and mutant bacteria on various organs of infected pigs. (A) Colonization of the wild-type strain 10 (wt) and the mutant strain 10ΔFBPS of the tonsils. Symbols: ⧫, tonsil from pig 4664; ▪, tonsil from pig 4665; ▴, tonsil from pig 4666; •, tonsil from pig 4668. (B) Colonization of the specific organs. Symbols: ◊ and ⧫, pus from joints from pig 4664; ▴, pus from a joint from pig 4666; •, CNS specimen from pig 4668. Each symbol represents the numbers of wild-type or mutant bacteria isolated from one particular organ, from one piglet.
TABLE 2.
Numbers of reisolated wild-type (strain 10) and mutant (strain 10ΔFBPS) bacteria from organs of infected piglets (mean actual inoculation ratio 65% mutant strain)
| Sampled | No. of bacteria (CFU/ml) for pig no.
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4664
|
4665
|
4666
|
4667
|
||||||||||
| w.t.a | mut.b | % mut.c | w.t.a | mut.b | % mut.c | w.t.a | mut.b | % mut.c | w.t.a | mut.b | % mut.c | ||
| Tonsil | 1.77 × 105 | 3.29 × 105 | 65 | 4.35 × 105 | 2.42 × 106 | 85 | 5.34 × 104 | 8.73 × 104 | 61 | 7.94 × 105 | 3.96 × 105 | 33 | |
| Pus joint 1 | 6.75 × 104 | <10 | 0 | 1.02 × 102 | 5.2 × 102 | 84 | |||||||
| Pus joint 2 | 5.15 × 104 | <10 | 0 | ||||||||||
| CNSe | 1.88 × 105 | <10 | 0 | ||||||||||
Number of wild-type bacteria found (CFU/ml).
Number of mutant bacteria found (CFU/ml).
Percentage of mutant bacteria calculated as follows: b/(a+b) × 100%.
Only relevant organs are depicted.
CNS, central nervous system.
DISCUSSION
In this work we describe the first FBPS of S. suis. The gene encoding FBPS was cloned and sequenced, and FBPS was purified. Binding of FBPS to human FN and FGN was shown. FBPS was shown to be involved in the colonization of the organs specific for an S. suis infection in piglets, but not in the colonization of S. suis on the tonsils of piglets.
Many streptococci and staphylococci have several different FnBPs, most of which are very large, about 130 kDa (12). Until now, S. pyogenes is the only organism having a large as well as a smaller (54-kDa) FnBP (8). The existence of more than one FnBP explains why in some organisms isogenic mutants defective in only one of the FnBPs can still bind to FN and/or FGN and are not completely attenuated in vivo (10). Although, so far no FnBP other than that described here has been described for S. suis, their existence could explain the fact that the FBPS mutant is not completely attenuated in vivo.
A considerable number of FN- and FGN-binding proteins of various bacterial species have been described (12). Most of these proteins were shown to be involved in adhesion to epithelial and/or endothelial cells (5, 8, 18). Therefore, it is attractive to assume a similar role for FBPS of S. suis. Previously, Charland and coworkers used human brain microvascular endothelial cells (HBMEC) in an in vitro blood-brain barrier model to study the pathogenesis of S. suis meningitis. Since S. suis adhered to HBMEC (4) in future experiments it would be of interest to test whether FBPS is involved in binding to HBMEC and involved in crossing the blood-brain barrier.
The role of FBPS in the pathogenesis of S. suis was studied in an experimental infection model in piglets. Since we were unable to determine a 50% lethal dose for the mutant strains, it was decided to compare the virulence of the isogenic FBPS mutant to that of the wild-type S. suis strain in a competitive infection assay in piglets. This kind of cocolonization experiment has been successfully applied to determine the virulence of mutants of Actinobacillus pleuropneumoniae in piglets (9). The data clearly showed that the mutant strain was capable of colonizing the tonsil as efficiently as the wild-type strain. This strongly indicates that FBPS is not involved in the colonization of the tonsil. The data also indicated that FBPS does play a role in the colonization of specific organs, since in the competition assay joints and the CNS were more efficiently colonized by wild-type than by mutant bacteria. In addition, higher numbers of wild-type bacteria were reisolated from the specific organs compared to the numbers of mutant bacteria, indicating that the mutant strain is attenuated in vivo. Although the number of pigs used for this experiment was low, these data indicate that the FBPS mutant is less virulent than the wild-type strain. Loss of virulence of S. suis was also described by Allen et al. (1). They constructed an isogenic knockout mutant of suilysin in a pathogenic serotype 2 strain of S. suis, and tested this mutant in an experimental animal model in pigs. From their findings, it was concluded that suilysin might play a role in reaching higher levels of colonization of various organs after S. suis has gained entrance into the bloodstream.
We were able to demonstrate that FBPS reacted with a convalescent-phase serum of a pig that survived an S. suis infection. Therefore FBPS is immunogenic in pigs, and this finding clearly demonstrates that FBPS of S. suis is expressed under in vivo conditions. Recently, it was reported that FBP54 of S. pyogenes is expressed in the human host (7). The in vivo expression of FBPS confirms the selection of the fbps gene from a gene library under in vivo conditions, as described by Smith and coworkers (20).
We showed that the fbps gene was present in all known serotypes of S. suis (except for two), as well as in all three phenotypes of serotype 2. This suggests that the fbps gene is present among most serotypes. However, the expression of FBPS in all serotypes and phenotypes was not studied. Therefore, it is possible that although all strains, except for serotypes 32 and 34, possess the fbps gene, not all strains express FBPS. Based on the facts that FBPS is immunogenic in pigs and that the fbps gene is present in all prevailing S. suis serotypes except for serotypes 32 and 34, FBPS is a very attractive candidate for a cross-protective vaccine against all serotypes. Since the mutant strain 10 Δ FBPS is not completely attenuated, this vaccine should be based on purified protein with a suitable adjuvant. This idea is supported by recent data from Kawabata et al. (13) which showed that vaccination with purified FBP54 can protect mice against a S. pyogenes infection.
Acknowledgments
We thank the people from the Animal and Laboratory Services and the people from Pathobiology of the ID-Lelystad for their assistance during the animal experiments.
REFERENCES
- 1.Allen, A. G., S. Bolitho, H. Lindsay, S. Khan, C. Bryant, P. Norton, P. Ward, J. Leigh, J. Morgan, H. Riches, S. Eastty, and D. Maskell. 2001. Generation and characterization of a defined mutant of Streptococcus suis lacking suilysin. Infect. Immun. 69:2732-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allgaier, A., R. G., H. J. Wisselink, H. E. Smith, and P. Valentin-Weigand. 2001. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arends, J. P., and H. C. Zanen. 1988. Meningitis caused by Streptococcus suis in humans. Rev. Infect. Dis. 10:131-137. [DOI] [PubMed] [Google Scholar]
- 4.Charland, N., V. Nizet, C. Rubens, K. Kim, S. Lacouture, and M. Gottschalk. 2000. Streptococcus suis interactions with human brain microvascular endothelial cells. Infect. Immun. 68:637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chia, J. S., C. Y. Yeh, and J. Y. Chen. 2000. Identification of a fibronectin binding protein from Streptococcus mutans. Infect. Immun. 68:1864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clifton-Hadley, F. A. 1983. Streptococcus suis type 2 infections. Br. Vet. J. 139:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Courtney, H. S., J. B. Dale, and D. L. Hasty. 1996. Differential effects of the streptococcal fibronectin-binding protein, FBP54, on adhesion of group A streptococci to human buccal cells and Hep-2 tissue culture cells. Infect. Immun. 64:2415-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courtney, H. S., Y. Li, J. B. Dale, and D. L. Hasty. 1994. Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62:3937-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller, T. E., S. Martin, J. F. Teel, G. R. Alaniz, M. J. Kennedy, and D. E. Lowery. 2000. Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microb. Pathog. 29:39-51. [DOI] [PubMed] [Google Scholar]
- 10.Greene, C., D. McDevitt, P. François, P. Vaudaux, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1065-1076. [DOI] [PubMed] [Google Scholar]
- 11.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide and streptogramin type B antibiotics. J. Bacteriol. 150:804-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joh, D., E. R. W. Wann, B. Kreikemeyer, P. Speziale, and M. Höök. 1999. Role of fibronectin-binding MSCRAMMS in bacterial adherence and entry into mammalian cells. Matrix Biol. 18:211-223. [DOI] [PubMed] [Google Scholar]
- 13.Kawabata, S., E. Kunitomo, Y. Terao, I. Nagakawa, K. Kiruchi, K.-I. Totsuka, and S. Hamada. 2001. Systemic and mucosal immunizations with fibronectin-binding protein FBP54 induce protective immune responses against Streptococcus pyogenes challenge in mice. Infect. Immun. 69:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konings, R. N. H., E. J. M. Verhoeven, and B. P. H. Peeters. 1987. pKUN vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 153:12-34. [DOI] [PubMed] [Google Scholar]
- 15.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 16.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Peacock, S. J., T. J. Foster, B. J. Cameron, and A. R. Berendt. 1999. Bacterial fibronectin-binding proteins and endothelial cell surface fibronectin mediate adherence of Staphylococcus aureus to resting human endothelial cells. Microbiology 145:3477-3486. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Smith, H. E., H. Buijs, R. de Vries, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 2001. Environmentally regulated genes of Streptococcus suis: identification by the use of iron-restricted conditions in vitro and by experimental infection of piglets. Microbiology 147:271-280. [DOI] [PubMed] [Google Scholar]
- 21.Smith, H. E., H. Buijs, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 2001. Selection of virulence-associated determinants of Streptococcus suis serotype 2 by in vivo complementation. Infect. Immun. 69:1961-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, H. E., M. Damman, J. van der Velde, F. Wagenaar, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 60:1750-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, H. E., U. Vecht, H. J. Wisselink, N. Stockhofe-Zurwieden, Y. Biermann, and M. A. Smits. 1996. Mutants of Streptococcus suis types 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect. Immun. 64:4409-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, H. E., H. J. Wisselink, U. Vecht, A. L. J. Gielkens, and M. A. Smits. 1995. High-efficiency transformation and gene inactivation in Streptococcus suis type 2. Microbiology 141:181-188. [DOI] [PubMed] [Google Scholar]
- 25.Sreenivasan, P. K., D. L. LeBlanc, L. N. Lee, and P. Fives-Taylor. 1991. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect. Immun. 59:4621-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Cheganppa. 1995. Streptococcus suis: past and present. Vet. Res. Comm. 21:381-407. [DOI] [PubMed] [Google Scholar]
- 27.Vecht, U., J. P. Arends, E. J. van der Molen, and L. A. M. G. van Leengoed. 1989. Differences in virulence between two strains of Streptococcus suis type 2 after experimentally induced infection of newborn germfree pigs. Am. J. Vet. Res. 50:1037-1043. [PubMed] [Google Scholar]
- 28.Vecht, U., L. A. M. G. van Leengoed, and E. R. M. Verheyen. 1985. Streptococcus suis infections in pigs in The Netherlands (part I). Vet. Q. 7:315-321. [DOI] [PubMed] [Google Scholar]
- 29.Vecht, U., H. J. Wisselink, J. E. van Dijk, and H. E. Smith. 1992. Virulence of Streptococcus suis type 2 strains in newborn germfree pigs depend on phenotype. Infect. Immun. 60:550-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vecht, U., H. J. Wisselink, M. L. Jellema, and H. E. Smith. 1991. Identification of two proteins associated with virulence of Streptococcus suis type 2. Infect. Immun. 59:3156-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]