Abstract
To identify the major outer surface proteins of Streptococcus agalactiae (group B streptococcus), a proteomic analysis was undertaken. An extract of the outer surface proteins was separated by two-dimensional electrophoresis. The visualized spots were identified through a combination of peptide sequencing and reverse genetic methodologies. Of the 30 major spots identified as S. agalactiae specific, 27 have been identified. Six of these proteins, previously unidentified in S. agalactiae, were sequenced and cloned. These were ornithine carbamoyltransferase, phosphoglycerate kinase, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase, purine nucleoside phosphorylase, enolase, and glucose-6-phosphate isomerase. Using a gram-positive expression system, we have overexpressed two of these proteins in an in vitro system. These recombinant, purified proteins were used to raise antisera. The identification of these proteins as residing on the outer surface was confirmed by the ability of the antisera to react against whole, live bacteria. Further, in a neonatal-animal model system, we demonstrate that some of these sera are protective against lethal doses of bacteria. These studies demonstrate the successful application of proteomics as a technique for identifying vaccine candidates.
Streptococcus agalactiae (group B streptococcus [GBS]) is the causal agent of a broad range of human diseases. Of these, the most prominent are septicemia, pneumonia, and meningitis of neonates. The major component of the cell surface is a polysaccharide coat, which is serotype specific. Embedded throughout this coat is a complement of proteins. To date, only a few such proteins have been identified. Predominant of these are the tandem repeat proteins, including Cα protein (40), R proteins (21), Rib protein (39), the Cβ protein (35), and X proteins (33). Other identified outer surface proteins include glutamine synthetase (37), α-enolase (32), and Hsp70 (13). Sip, an antigen capable of raising a protective immune response, has also been localized to the cell surface (5). The mapping of proteins to the bacterial surface may well suggest alternate functions for proteins with other, well-established roles—for example, the characterization of the glycolytic enzyme α-enolase as a plasmin binding protein on the outside of the bacterial cell (32). The identification of proteins located on the outer surface of S. agalactiae may be of use in suggesting potential vaccine candidates.
There is a history of over 2 decades of research into vaccines for the prevention of neonatal GBS infections. The first human trials, including one performed in pregnant women, were performed in the 1980s using unconjugated capsular polysaccharides (2, 3). These unconjugated polysaccharides were shown to be insufficiently immunogenic, so subsequent studies have been performed with protein-conjugated vaccines in order to boost antibody responses (C. M. Mink, H.-K. Guttormsen, K. R. Lottenbach, J. C. Cannon, L. C. Paoletti, P. McInnes, and D. L. Kasper, Abstr. 35th Intersci. Conf. Antimicrob. Agents Chemother, abstr. G6, 1995). There are a number of technical difficulties to be overcome with capsule-containing conjugate vaccines: multiple serotypes are needed, an appropriate protein conjugate needs to be identified and validated, and regulatory issues such as potential cross-reaction with human tissues needs to be addressed (19). As an alternative to capsule-based vaccines, a number of workers have examined the possibility of using protein vaccines. Antigens that have been examined include some of the surface-localized proteins mentioned above, including a family of laddering proteins (Cα and R) that are restricted to particular serotypes, as well as protein present on most or all GBS strains (5, 7, 22). The research described in this paper was undertaken to identify surface proteins of GBS that could be used as novel vaccine candidates.
Proteomic techniques were employed, and several proteins were identified, including six whose sequence was previously unpublished. We confirmed the identity of several of these proteins as outer surface proteins. Antisera raised against two of the identified proteins were capable of providing a degree of protection against a lethal challenge of S. agalactiae in a neonatal-mouse model, demonstrating both the presence of the protein on the cell surface and the utility of this proteomic strategy in identifying possible vaccine candidates.
MATERIALS AND METHODS
Strain and culture conditions.
S. agalactiae serotypes and strains used were type III (strain M732 [8]), type Ia/c (RF76, an isolated pathogenic strain which, due to ethical considerations, we are unable to release), type Ib/c (ATCC strain H36b), type II (NCTC strain 18RS21), type IV (ATCC strain R80405), and type V (ATCC Prague strain). All strains used were maintained on Columbia horse blood agar plates (Oxoid, Basingstoke, United Kingdom). Liquid cultures for sample preparation were grown overnight in Todd-Hewitt broth (Oxoid) at 37°C. Prior to harvesting, cells were checked for group specificity by latex agglutination test (Oxoid).
Outer surface protein preparation.
Outer surface proteins were prepared using a slightly modified protocol (6, 17). Following cell harvest and washing in phosphate-buffered saline, cells from a 400-ml culture were resuspended in 50 ml of osmotic digestion buffer (20% sucrose in 20 mM Tris-HCl, pH 7.0, 10 mM MgCl2, and protease inhibitor cocktail [comprised of {final concentrations} 1 mM phenylmethylsulfonyl fluoride, 10 μM iodoacetic acid, 1 μM pepstatin A, and 10 mM 1,10-phenanthroline], and 4,000 U of mutanolysin [Sigma, St. Louis, Mo.]). Enzymatic digestion was allowed to proceed for 2 h at 37°C. The majority of intact protoplasts were removed by centrifugation at 7,000 × g for 15 min. The supernatant was then subjected to ultracentrifugation at 28,000 rpm (Beckman Ti50.2) to remove cell debris and remaining protoplasts. The supernatant was concentrated by ultrafiltration to a final volume of approximately 1.5 ml.
2-D gel electrophoresis and in-gel digestion.
A Genomic Solutions Investigator two-dimensional (2-D) electrophoresis system (Genomic Solutions, Inc., Ann Arbor, Mich.) running Tris/Tricine chemistry was used to separate the outer surface proteins. The proteins were focused through the first dimension over a pH range of 3 to 10, prior to application on a second-dimension sodium dodecyl sulfate (SDS)-10% polyacrylamide gel. Gels were run using loadings between 300 μg and 1 mg and were stained using GelCode blue (Perbio Science UK Ltd., Cheshire, United Kingdom). Separated gel spots were subjected to in-gel tryptic digestion (porcine trypsin; Sigma) and were digested for 16 h at 37°C in ammonium bicarbonate buffer (BDH Merck). The resulting peptides were extracted from the gel and purified by reversed-phase high-performance liquid chromatography on a C18 microtrap cartridge (Michrom BioResource, Auburn, Calif.). The peptides were eluted with 30% acetonitrile (Rathburn Chemicals, Peebleshire, United Kingdom)-1% aqueous trifluoroacetic acid (Sigma).
MS.
Aliquots of purified digest mixtures were analyzed by delayed-extraction matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) using a PE Biosystems Voyager-Elite operating in reflectron mode with a matrix of α-cyano-4-hydroxycinnamic acid (Sigma). Digest mixtures were then screened in the MS mode using electrospray ionization on a Q-TOF mass spectrometer (Micromass, Manchester, United Kingdom) by examining 1 or 2 μl of the 30% acetonitrile-0.1% aqueous trifluoroacetic acid solution in a nanospray needle as described previously (26, 27, 38). Selected peptides, usually those suggested by the MALDI-MS data, were fragmented by collisionally activated decomposition, and the spectra were interpreted according to the known fragmentation pathways of peptides under soft-ionization, low-energy decomposition (25). This information was used to search the nonredundant sequence database OWL (4), enabling identification of many of the proteins.
Reverse genetics.
Protein spots whose identity remained unknown following a database search were subjected to reverse genetic techniques. At least two peptide sequences were obtained and were used to design degenerate oligonucleotide PCR primers. PCR was carried out using Taq polymerase (MBI, Sunderland, United Kingdom) and a series of optimization buffers (MBI). In all cases, the conditions for PCR were as follows: 92°C for 2 min, 25 cycles of 92°C for 30 s, 55°C for 30 s, 72°C for 1 min, and then a final step of 72°C for 5 min prior to being held at 4°C. Amplified fragments were cloned into pCR2.1 (InVitrogen Inc., Groningen, The Netherlands) and were sequenced using an ABI 310 automated sequencer (ABI, Warrington, United Kingdom) running ABI big-dye terminator mix chemistry (ABI). The sequence data obtained were used to search the EMBL database and enabled the encoded proteins to be identified. Primers were designed from the revealed sequence and were used to sequence genomic S. agalactiae DNA in an “outward” direction (i.e., towards the 5′ and 3′ ends of the gene). The full-length sequence was determined at the 5′ end by the presence of a ribosome binding site (36) positioned appropriately in relation to a potential “start” codon, at the 3′end by repeated sequencing of putative termination codons, and by comparison with published sequences. Once the sequence was established, a further set of primers was designed to amplify the full-length gene by PCR. These primers included restriction endonuclease recognition sites to enable subsequent cloning into a modified lactococcal expression vector. Following PCR amplification, the full-length products were cloned into the vector pCR2.1 (InVitrogen Inc.).
Expression and purification of recombinant proteins and raising of polyclonal antisera.
Full-length genes were excised from pCR 2.1 using NcoI and EcoO109I and were cloned into a nisin-inducible expression vector, pNZ80482HT. pNZ80482HT is a modified form of pNZ8048 (9), harboring two 10× His tags which allow for fusion to either the C or N terminus, separated by a multicloning site. Digestion of this plasmid with NcoI and EcoO109I excises one of the 10× His tags, allowing insertion of a gene in its place. Further details of the construction of this plasmid are available on request from the authors. Fusion of the plasmid backbone to the digested S. agalactiae DNA resulted in a gene with an appropriate lactococcal translation start recognition signal upstream of the ATG codon included in the NcoI recognition site, fused to a C-terminal 10× His tag, followed by a translation termination codon. Recombinant plasmids were transformed into Lactococcus lactis NZ9000. Overexpressed protein was obtained by induction with nisin (produced from the NZ9800 strain of L. lactis) and was released from the bacteria by bead beating. Cleared bacterial lysate was passed over a metal affinity Talon column (Clontech, Basingstoke, United Kingdom) to trap His-tagged proteins, which were eluted with imidazole according to the manufacturer's instructions.
Fractions containing recombinant protein were used to immunize New Zealand White rabbits (AbCam Ltd., Cambridge, United Kingdom). Antisera were tested for specificity by probing Western blots of S. agalactiae outer surface proteins. Proteins were analyzed on a Criterion SDS-12% polyacrylamide gel (Bio-Rad, Hertfordshire, United Kingdom) prior to blotting onto a 0.2-μm-pore-size nitrocellulose membrane (Bio-Rad). Blots were blocked with 3% bovine serum albumin prior to probing with 1:5,000 dilutions of the test or preimmune sera. Blots were decorated with goat anti-rabbit alkaline phosphatase-conjugated secondary antibody, and the blot was developed with Western blue-stabilized substrate (Promega, Southampton, United Kingdom).
Whole-cell ELISA.
The sera directed against the recombinant proteins were used to challenge S. agalactiae in whole-cell enzyme-linked immunosorbent assays (ELISAs). Six strains of S. agalactiae representing the major clinical serotypes were used. The cells were grown to mid-log phase, harvested, washed with phosphate-buffered saline, and used to coat ELISA plates (Nunc Immunosorb; Life Technologies Ltd., Paisley, United Kingdom). The supernatant from the wells was carefully aspirated following overnight coating and was assayed for the presence of the intracellular marker aldolase (17). After blocking with 3% bovine serum albumin, the plates were probed with serial dilutions of pre- and postimmune sera. The antibody was decorated with alkaline-phosphatase conjugated anti-rabbit immunoglobulin G (Sigma), which was in turn detected with p-nitrophenyl phosphate. Plate contents were incubated at 37°C for 30 min and were then read at 405 nm.
Animal protection experiments.
Sera were tested for their ability to protect against a lethal infection of S. agalactiae by using a neonatal-mouse model (34). Prior to use, complement was inactivated by treating the sera at 56°C for 30 min. S. agalactiae M732 was grown to mid-log phase and was counted by microscopy. Bacteria (n = 106) were mixed with 50 μl of antisera (pre- or postimmune) and were then injected by the intraperitoneal route into newborn (0 to 24 h old) BALB/c mice. Mice were monitored and scored for survival over a 48-h period.
As a positive control for the experiment, sera raised against the challenge strain (anti-whole-cell) by using the method of Lancefield (20) were used. As with the test sera, they were mixed with the bacteria prior to injection.
The animals were housed and maintained in accordance with the Code of Practice for the Housing and Care of Animals used in Scientific Procedures issued by the United Kingdom Home Office. The experiments were carried out under the authority of a Project Licence granted under the Animals (Scientific Procedures) Act 1986.
Nucleotide sequence accession number.
The nucleotide sequences of the novel genes described in this publication have been submitted to GenBank. They have been assigned the following numbers: nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase, AF439646; ornithine carbamolytransferase, AF439647; phosphoglycerate kinase, AF439648; enolase, AF439649; purine nucleoside phosphatase, AF439650; and glucose-6-phosphate isomerase, AF439651.
RESULTS
2-D analysis of extracellular proteome: assignment by peptide sequence.
Outer surface proteins were stripped from S. agalactiae, using previously published methods. To check for the possibility of intracellular contamination, the protein preparations were examined for the presence of DNA. No DNA was detected (data not shown), giving confidence that the proteins predominantly represented those present on the outer surface of S. agalactiae. Proteins were separated by 2-D gel electrophoresis. Figure 1 shows a representation of Coomassie blue-stained spots, identified with differing concentrations of protein being loaded on the first dimension gel. The proteins from all the spots shown were subjected to in-gel tryptic digestion prior to MALDI-MS analysis of the digest mixtures produced. In general, some MALDI-MS masses are known to be either trypsin autodigest products or keratin-derived contamination. Probable sample derived peptides were then chosen for sequence analysis by nanospray-MS/MS on a quadrupole orthogonal acceleration time-of-flight mass spectrometer. Fourteen of the proteins could be identified following a search of the OWL database, and these are shown in Table 1.
FIG. 1.
Representation of the outer surface proteins of S. agalactiae M732 separated by 2-D gel electrophoresis. Proteins were isolated from the outer surface of S. agalactiae by enzymatic digestion. Following concentration and dialysis, samples of protein (300 μg to 1 g) were analyzed through the first dimension over a pH range of 3 to 10 prior to application on a second-dimension SDS-10% polyacrylamide gel. The gel was stained using GelCode blue (Pierce). The representation is a compilation of gels which were loaded with differing amounts of protein.
TABLE 1.
Peptide sequences and protein identification/assignation from proteins derived from 2-D gel separation of S. agalactial outer surface proteinsa
| Spot no. | Peptide sequence(s)b | Identification |
|---|---|---|
| 1 | AYIDLYDVK | GBS Rib |
| AEDSIGNLPDLPK | GBS Rib | |
| 2 | AEDSIGNLPDLPK | GBS Rib |
| IDPLQLITLNSPDLK | GBS Rib | |
| 3 | AEDSIGNLPDLPK | GBS Rib |
| DQQVNVGETPK | GBS Rib | |
| 4 | TD(I/L)DNNPK | GBS Rib |
| AEVISGSAVTLNTNMTK | GBS Rib | |
| 5 | QIPEDIR | GBS superoxide dismutase |
| VNELYQAAK | GBS superoxide dismutase | |
| STANQDTPIM*EGK | GBS superoxide dismutase | |
| LADVSQIPEDIR | GBS superoxide dismutase | |
| 6 | AFFEIINWNK | GBS superoxide dismutase |
| STANQDTPIM*EGK | GBS superoxide dismutase | |
| 7 | FQLTDIPAAPR | GBS dnaK-Hsp70 |
| 8 | FQLTDIPAAPR | GBS dnaK-Hsp70 |
| EQHIVIQSNSGLTDEEIDK | GBS dnaK-Hsp70 | |
| 9 | EIFPA | Exonuclease ABC, subunit B |
| VGNDGVITIEESR | Hsp60 | |
| 10 | GSPLITNDGVTIAK | Hsp60 |
| NVTAGANPIGIR | Hsp60 | |
| F(I/L)DEYL | Acid glycoprotein | |
| 11 | Y(I/L)FTHH | Acid glycoprotein |
| 12 | (I/L)A(I/L)AM(I/L)ED | Cysteine synthase |
| 13 | DAF(I/L)AGVGT | Cysteine synthase |
| 14 | SGTTTEPAIAFR | Glucose-6-phosphate isomerase |
Assignation of proteins was possible by searching the OWL and Swiss-Prot database with the MS-derived peptide sequences.
M*, methionine sulfoxide.
Identification of proteins whose identity could not be assigned by peptide sequence.
Peptide sequences which produced no hit from the protein databases were used to design degenerate primers for PCR. A codon preference table generated using the GCG program codon preference was used to design primers for PCR. Primers were synthesized in both orientations, and assay PCRs were carried out using all possible primer combinations (including single primer controls) and in a range of buffer conditions provided by a PCR Optimisation kit (MBI Fermentas, Helena BioSciences, Sunderland, United Kingdom). Positive PCR amplification products were cloned into TOPO vectors and sequenced. Searches of the databases with the generated sequence identified the proteins. These are shown, together with the peptide sequences in Table 2.
TABLE 2.
Peptide sequences and protein identification/assignation from proteins derived from two-dimensional gel separation of S. agaldctiae outer surface proteinsa
| Spot no. | Peptide sequence | Identification |
|---|---|---|
| 15 | AIGLVIPELNGK | Glyceraldehyde-3-phosphate dehydrogenase |
| IQNVEGVEVTR | ||
| 16 | AIGLVIPELNGK | Glyceraldehyde-3-phosphate dehydrogenase |
| IQNVEGVEVTR | ||
| 17 | IQNVEGVEVTR | Glyceraldehyde-3-phosphate dehydrogenase |
| 18 | PAFHD | Ornithine carbamoyltransferase |
| HFQDA | ||
| 19 | FPAEE(I/L)VK | Ornithine carbamoyltransferase |
| EVTD | ||
| 20 | YNQ(I/L)(I/L)R | Enolase |
| FEGTEDRE | ||
| ETAVG | ||
| 21 | VVEEVADE(L/I)AEK | Nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase |
| GPDNFPF(L/I)G(L/I)K | ||
| 22 | (L/I)GQDVVF | Phosphoglycerate kinase |
| (L/I)GGGDSAAA | Phosphoglycerate kinase | |
| 23 | (I/L)A(I/L)AM(I/L)ED | Purine nucleoside phosphatase |
| DAF(I/L)AGVGT | Purine nucleoside phosphatase |
Assignation of proteins was possible only after reverse genetic analysis. The peptide sequences shown were used to synthesize oligonucleotides, which were in turn used for PCR amplification of the gene from S. agalactiae genomic DNA. The DNA sequence was used to search the database and assign the protein identification.
Full-length sequence of six new genes of S. agalactiae.
Using the genomic sequencing methods described, we successfully derived the nucleic acid sequence of six genes previously unidentified in S. agalactiae. In silico translation of these sequences enabled a search of the trEMBL database. At publication, the highest homologies shown to these proteins are with those from Streptococcus pneumoniae (15) and are given as percentages in parentheses. The proteins were identified as being ornithine carbamoyltransferase (85%), phosphoglycerate kinase (94%), nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (71%), purine nucleoside phosphatase (76%), enolase (93%), and glucose-6-phosphate isomerase (87%). The sequences of these genes and derived proteins are available through GenBank (Materials and Methods).
Sera directed against ornithine carbamoyltransferase and phosphoglycerate kinase react against whole S. agalactiae.
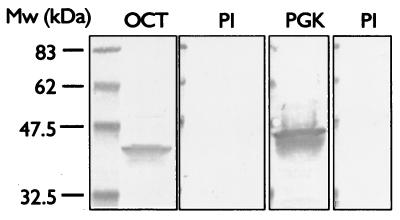
Overexpressed His-tagged ornithine carbamoyltransferase and phosphoglycerate kinase were purified from bacterial lysate using metal affinity chromatography. Individual proteins were used to immunize New Zealand White rabbits using a schedule of an initial immunization followed by three boosts over a 60-day period. Figure 2 shows Western blots using the sera, reacted against blots of a preparation of outer surface proteins, prepared as described. As can be seen, the sera react in an appropriate fashion, decorating bands migrating close to the calculated mass of 38 kDa in the case of ornithine carbamoyltransferase and of 42 kDa for phosphoglycerate kinase.
FIG. 2.
Western blot analysis of S. agalactiae outer surface proteins probed with preimmune (PI), anti-phosphoglycerate kinase (PGK), and anti-ornithine carbamoyltransferase (OCT) antisera. Outer surface proteins were analyzed by SDS-polyacrylamide gel electrophoresis (Criterion 22%; Bio-Rad), blotted to nitrocellulose, and probed with a 1:5,000 dilution of preimmune-phase sera or antisera. Blots were decorated with anti-rabbit alkaline phosphatase conjugate secondary antibody and were developed with Western blue substrate (Promega). Mw, molecular weight.
Using the same sera, whole S. agalactiae cells were screened in an ELISA both to give additional confirmation that the antigens were indeed situated on the outside of the cell and also that they were present in representative strains from the major S. agalactiae serotypes. To confirm that the cells had maintained their integrity on the ELISA plate, the supernatant (following coating) was carefully aspirated and assayed for the presence of the intracellular marker aldolase (17). Comparing the result obtained to that for a lysed cell extract treated in the same way indicated that less than 3% of the cells lysed during this period and that the cells were essentially whole.
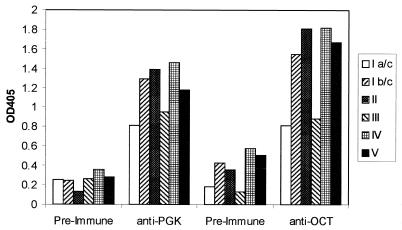
Results of the ELISA are shown in Fig. 3. Higher antiserum titers against the S. agalactiae cells were seen when screened with postimmune sera from both proteins than when they were screened with preimmune sera, demonstrating the presence of the antigens on the cell surface. Further, strong reactivity was observed with strains from all six serotypes tested. No reactivity was seen for the test sera against wells which did not contain cells (blocked only).
FIG. 3.
Sera directed against S. agalactiae phosphoglycerate kinase (PGK) and ornithine carbamoyltransferase (OCT) recognize S. agalactiae in a whole-cell ELISA. Cells of six S. agalactiae serotypes, type Ia/c (RF76a), type Ib/c (H36b), type II (18RS21, type III (M732), type IV (R80405), and type V (Prague strain), were used to coat ELISA plates which were subsequently challenged with rabbit preimmune sera or antisera raised against PGK or OCT. The reacting antibody was decorated with secondary (goat anti-rabbit, alkaline phosphatase-conjugated) antibody, and the reaction was developed with p-nitrophenyl phosphate and read at 405 nm. OD405, optical density at 405 nm.
Sera directed against ornithine carbamoyltransferase and phosphoglycerate kinase protect neonatal animals from S. agalactiae infection.
As the antigens were confirmed as being situated on the outside of the cell, it was hypothesized that they may make targets for vaccination. To test this, a passive immunization experiment was performed, using the antisera which reacted to the whole cells.
Neonatal mice were challenged by the intraperitoneal route with a mixture of S. agalactiae and sera. The sera were either preimmune or that raised against ornithine carbamoyltransferase or phosphoglycerate kinase. A positive control consisting of sera directed against whole killed S. agalactiae M732 was also included in the experiment. Table 3 shows the results of the neonatal protection experiments. Antisera directed against both ornithine carbamoyltransferase and phosphoglycerate kinase offer a degree of protection in this neonatal-animal model compared to the preimmune sera, though not to the level offered by antibodies raised against type III carbohydrate.
TABLE 3.
Protection against S. agalactiae infection offered by anti-ornithine carbamoyltransferase (anti-OCT) and anti-phosphoglycerate kinase (anti-PGK) sera in a neonatal protection model compared to anti-whole-cell and preimmune seraa
| Serum type | No. of sera in experiment | % Dead | % Alive |
|---|---|---|---|
| Preimmune | 41 | 98 | 2 |
| Whole-cell | 74 | 36 | 64 |
| Anti-OCT | 48 | 54 | 46 |
| Anti-PGK | 54 | 65 | 35 |
Complement-inactivated rabbit sera were mixed with 106 cells of S. agalactiae M732 and were injected by the intraperitoneal route into neonatal mice. They were monitored for 48 h, and the number of deaths was recorded. Results shown are the totals for three separate experiments.
DISCUSSION
The outer surface of bacteria is of great importance to the understanding of bacterial pathogenesis. Not only are there bacterial defense mechanisms, but elements of the surface perform virulence-related functions, such as adhesion, invasion, direct injury, and induction of septic shock.
The surface of S. agalactiae comprises two distinct sets of carbohydrate antigens, the group carbohydrate and type-specific capsular polysaccharides. In addition there are protein antigens present on the surface. Those identified to date include the well-characterized type-specific tandem repeat, containing proteins such as Cα, Rib, and Cβ. Other, previously identified outer surface-located proteins include glutamine synthetase (37) and α-enolase (32).
In this study we have identified major proteins on the surface of S. agalactiae. These include several proteins which have been shown to be present on the surface of other organisms, as well as others not previously localized to the outer surface of bacterial cells. Whole-cell ELISAs, using antisera directed against the identified proteins, confirmed the presence of these proteins on the surface of the cell. Of further interest is the protective activity that some of these antisera showed against streptococcal infection in a mouse neonatal model. While the degree of protection seen is not as high as that observed for the anti-whole-cell antisera, it is nonetheless encouraging, and unlike the serotype-specific proteins, such as Cα, Cβ, and Rib, these proteins are present on at least six serotypes and may be present across a range of S. agalactiae serotypes. Thus, it is possible that some of these proteins may represent vaccine candidates with the potential to protect against infection caused by a range of serotypes.
The finding that a number of enzymes that one would expect to see in the cytoplasm appear instead on the surface is at first somewhat surprising. This is especially true when one takes into consideration the fact that these proteins have no N-terminal signal peptide (by PSORT computer sequence analysis) and do not possess the canonical gram-positive anchor motif LPXTG. In this, they join a growing list of such proteins (11), though it is unclear what the mechanism of attachment of these proteins to the cell wall may be. N-terminal attachment and charge/hydrophobic interactions have been suggested as possibilities (11). However, in addition to the immunochemical and protective data presented here, there are other data locating these proteins at the cell surface, albeit in other organisms. α-Enolase has been reported to be present on several streptococcal types other than GBS (31, 32). Phosphoglycerate kinase has been identified as a protective antigen in Schistosoma mansoni (23). Also in Schistosoma, a specific epitope of the enzyme glyceraldehyde-3-phosphate dehydrogenase has been identified as a vaccine candidate (1). Interestingly, the same protein has been identified as a plasmin(ogen) binding protein on Streptococcus equisimilis (12), the same function assigned to surface-located enolase in Streptococcus pyogenes (23, 30). Demonstrating the multiple functions that the same protein may possess, glyceraldehyde-3-phosphate dehydrogenase has also been identified as a transferrin binding protein (24). Ornithine carbamoyltransferase has been isolated as a putative adhesin from a surface molecule preparation of Staphylococcus epidermidis (16). Superoxide dismutase has been identified as a virulence factor located in the periplasm of Salmonella enterica serovar Typhimurium (10) and Listeria monocytogenes (14). It has also provided protection in mice against infection with Brucella abortus (29). Finally, heat shock proteins have been suggested to be vaccine candidates. Hsp70 has been shown to protect against streptococcal infection (13), though it has been suggested that this antigen is not surface located in S. pneumoniae (18). Hsp60 has been suggested as a vaccine candidate against Yersinia enterocolitica (28), though in the case of both Hsp70 and Hsp60, the potential use of these proteins as human vaccines is questionable due to the close similarity between the bacterial and human proteins, raising issues of autoimmunity.
The ability of antisera directed against the single proteins ornithine carbamoyltransferase and phosphoglycerate kinase to protect against infection in a neonatal murine model is encouraging. This data not only confirms the presence of the proteins on the surface of the cell, validating this approach, but also suggests that there may be proteins on the surface of S. agalactiae which may be candidates as vaccines against infection.
This work describes the presence of these and other proteins on the surface of S. agalactiae. Despite the assigned function of the protein (by their names), they may well have some other role to play in the bacteria. This is clearly demonstrated in cases such as enolase, when the protein was identified as an outer surface protein after a function had been ascribed to it. The identification in this work of unexpected proteins with an outer surface location may indicate secondary functions for these proteins. In addition, the protection data may also indicate that such proteins may make good vaccine candidates against infection by this bacteria. The proteomic techniques used here have thus proved to be a valuable method to identify proteins present in particular subcellular locations. Indeed, given the lack of N-terminal signal sequence or C-terminal anchor sequence on the proteins identified in this study, the use of a direct biochemical method is demonstrably advantageous to in silico searches of genome sequences.
Acknowledgments
The plasmid pNZ8048 and the Lactococcus sp. strain NZ9000 were obtained under licence from NIZO, P.O. Box 20 6710 BA Ede, The Netherlands.
REFERENCES
- 1.Argiro, L. L., S. S. Kohlstadt, S. S. Henri, H. H. Dessein, V. V. Matabiau, P. P. Paris, A. A. Bourgois, and A. J. Dessein. 2000. Identification of a candidate vaccine peptide on the 37 kDa Schistosoma mansoni GAPDH. Vaccine 18:2039-2048. [DOI] [PubMed] [Google Scholar]
- 2.Baker, C. J., and D. L. Kasper. 1985. Vaccination as a measure for prevention of neonatal GBS infection. Antibiot. Chemother. (Basel) 35:281-290. [DOI] [PubMed]
- 3.Baker, C. J. M. A. Rench, M. S. Edwards, R. J. Carpenter, B. M. Hays, and D. L. Kasper. 1988. Immunization of pregnant women with a polysaccharide vaccine of group B streptococcus. N. Engl. J. Med. 319:1180-1185. [DOI] [PubMed] [Google Scholar]
- 4.Bleasby, A. J., and J. C. Wootton. 1990. Construction of validated, non-redundant composite protein sequence databases. Protein Eng. 3:153-159. [DOI] [PubMed] [Google Scholar]
- 5.Brodeur, B. R., M. Boyer, I. Charlebois, J. Hamel, F. Couture, C. R. Rioux, and D. Martin. 2000. Identification of group B streptococcal Sip protein, which elicits cross-protective immunity. Infect. Immun. 68:5610-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calandra, G. B., and R. M. Cole. 1980. Lysis and protoplast formation of group B streptococci by mutanolysin. Infect. Immun. 28:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, Q., B. Carlson, S. Pillai, R. Eby, L. Edwards, S. B. Olmsted, and P. Cleary. 2001. Antibody against surface-bound C5a peptidase is opsonic and initiates macrophage killing of group B streptococci. Infect. Immun. 69:2302-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Cueninck, B. J., T. K. Eisenstein, T. S. McIntosh, G. D. Shockman, and R. M. Swenson. 1982. Type-specific protection of neonatal rats from lethal group B streptococcal infection by immune sera obtained from human volunteers vaccinated with type III-specific polysaccharide. Infect. Immun. 37:961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J.-C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulence Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti, V. A. 2000. Surface proteins on gram-positive bacteria, p. 11-24. In V. A. Fischetti, R. P. Novic, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 12.Gase, K., A. Gase, H. Schirmer, and H. Malke. 1996. Cloning, sequencing and functional overexpression of the Streptococcus equisimilis H46A gapC gene encoding a glyceraldehyde-3-phosphate dehydrogenase that also functions as a plasmin(ogen)-binding protein. Purification and biochemical characterization of the protein. Eur. J. Biochem. 239:42-51. [DOI] [PubMed] [Google Scholar]
- 13.Hamel, J., B. Brodeur, D. Martin, and C. Rioux. December1996. Streptococcal heat shock proteins members of the Hsp70 family. U.S patent WO9640928.
- 14.Hess, J., G. Dietrich, I. Gentschev, D. Miko, W. Goebel, and S. H. Kaufmann. 1997. Protection against murine listeriosis by an attenuated recombinant Salmonella typhimurium vaccine strain that secretes the naturally somatic antigen superoxide dismutase. Infect. Immun. 65:1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 83:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain, M., G. Peters, G. S. Chhatwal, and M. Herrmann. 1999. A lithium chloride-extracted, broad-spectrum-adhesive 42-kilodalton protein of Staphylococcus epidermidis is ornithine carbamoyltransferase. Infect. Immun. 67:6688-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kling, D. E., L. C. Madoff, and J. L. Michel. 1999. Subcellular fractionation of group B streptococcus. BioTechniques 27:24-26. [DOI] [PubMed] [Google Scholar]
- 18.Kolberg, J., E. A. Hoiby, A. Aase, K. Sletten, G. Rodal, T. E. Michaelsen, and A. Bucher. 2000. Streptococcus pneumoniae heat shock protein 70 does not induce human antibody responses during infection. FEMS Immunol. Med. Microbiol. 29:289-294. [DOI] [PubMed] [Google Scholar]
- 19.Korzeniowska-Kowal, A., D. Witkowska, and A. Gamian. 2001. Molecular mimicry of bacterial polysaccharides and their role in etiology of infectious and autoimmune diseases. Postêpy Hig. Med. Dosw. 55:211-232. [PubMed] [Google Scholar]
- 20.Lancefield, R. C. 1932. A serological differentiation of human and other groups of hemolytic streptococci. J. Exp. Med. 57:571-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancefield, R. C., and G. E. Perlmann. 1952. Preparation and properties of a protein (R antigen) occurring in streptococci of group A, type 28 and in certain streptococci of other serological groups. J. Exp. Med. 96:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson, C., M. Stalhammar-Carlemalm, and G. Lindahl. 1999. Protection against experimental infection with group B streptococcus by immunization with a bivalent protein vaccine. Vaccine 17:454-458. [DOI] [PubMed] [Google Scholar]
- 23.Lee, K. W., A. Thakur, A. M. Karim, and P. T. LoVerde. 1995. Immune response to Schistosoma mansoni phosphoglycerate kinase during natural and experimental infection: identification of a schistosome-specific B-cell epitope. Infect. Immun. 63:4307-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris, H. R., M. Panico, M. Barber, R. S. Bordoli, R. D. Sedgewick, and A. N. Tyler. 1981. Fast atom bombardment: a new mass spectrometric method for peptide sequence analysis. Biochem. Biophys. Res. Commun. 101:623-631. [DOI] [PubMed] [Google Scholar]
- 26.Morris, H. R., T. Paxton, A. Dell, J. Langhorn, M. Berg, R. S. Bordoli, J. Hoyes, and R. H. Bateman. 1996. High sensitivity collisionally-activated decomposition tandem mass spectroscopy on a novel quadrupole/orthogonal-accelerated time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 10:889-896. [DOI] [PubMed] [Google Scholar]
- 27.Morris, H. R., T. Paxton, M. Panico, R. McDowell, and A. Dell. 1998. A novel geometry mass spectrometer, the quadrupole orthogonal acceleration time of flight instrument, for the femtomole/attomole range bipolymer sequencing, p. 53-80. In B. Larsen and C. McEwan (ed.), Mass spectrometry of biological materials, vol. 2. Marcel Dekker, Inc., New York, N.Y. [Google Scholar]
- 28.Noll, A., and I. B. Autenrieth. 1996. Immunity against Yersinia enterocolitica by vaccination with Yersinia HSP60 immunostimulating complexes or Yersinia HSP60 plus interleukin-12. Infect. Immun. 64:2955-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onate, A. A., R. Vemulapalli, E. Andrews, G. G. Schurig, S. Boyle, and H. Folche. 1990. Vaccination with live Escherichia coli expressing Brucella abortus Cu/Zn superoxide dismutase protects mice against virulent B. abortus. Infect. Immun. 67:986-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancholi, V., and V. A. Fischetti. 1997. A novel plasminogen/plasmin binding protein on the surface of group A streptococci. Adv. Exp. Med. Biol. 418:597-599. [DOI] [PubMed] [Google Scholar]
- 32.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 33.Rainard, P., Y. Lautrou, P. Sarradin, and B. Poutrel. 1991. Protein X of Streptococcus agalactiae induces opsonic antibodies in cows. J. Clin. Microbiol. 29:1842-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodewald, A. K., A. B. Onderdonk, H. B. Warren, and D. L. Kasper. 1992. Neonatal mouse model of group B streptococcal infection. J. Infect. Dis. 166:635-639. [DOI] [PubMed] [Google Scholar]
- 35.Russell-Jones, G. J., E. C. Gotschlich, and M. S. Blake. 1984. A surface receptor specific for human IgA on group B streptococci possessing the Ibc protein antigen. J. Exp. Med. 160:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suvorov, A. N., A. E. Flores, and P. Ferrieri. 1997. Cloning of the glutamine synthetase gene from group B streptococci. Infect. Immun. 65:191-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng-Umnuay, P., H. R. Morris, A. Dell, M. Panico, T. Paxton, and C. M. West. 1998. The cytoplasmic F-box binding protein SKP1 contains novel pentasaccharide linked to hydroxyproline in Dictyostelium. J. Biol. Chem. 273:18242-18249. [DOI] [PubMed] [Google Scholar]
- 39.Wastfelt, M., M. Stalhammar-Carlemalm, A. M. Delisse, T. Cabezon, and G. Lindahl. 1996. Identification of a family of streptococcal surface proteins with extremely repetitive structure. J. Biol. Chem. 271:18892-18897. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson, H. W., and R. G. Eagon. 1971. Type-specific antigens of group B type Ic streptococci. Infect. Immun. 4:596-604. [DOI] [PMC free article] [PubMed] [Google Scholar]