Abstract
Differential fluorescence induction (DFI) technology was used to identify promoters of Streptococcus pneumoniae induced under various in vitro and in vivo conditions. A promoter-trap library using green fluorescent protein as the reporter was constructed in S. pneumoniae, and the entire library was screened for clones exhibiting increased gfp expression under the chosen conditions. The in vitro conditions used were chosen to mimic aspects of the in vivo environment encountered by the pathogen once it enters a host: changes in temperature, osmolarity, oxygen, and iron concentration, as well as blood. In addition, the library was used to infect animals in three different models, and clones induced in these environments were identified. Several promoters were identified in multiple screens, and genes whose promoters were induced twofold or greater under the inducing condition were mutated to assess their roles in virulence. A total of 25 genes were mutated, and the effects of the mutations were assessed in at least two different infection models. Over 50% of these mutants were attenuated in at least one infection model. We show that DFI is a useful tool for identifying bacterial virulence factors as well as a means of elucidating the microenvironment encountered by pathogens upon infection.
The gram-positive pathogen Streptococcus pneumoniae is a major cause of community-acquired infections, including those of the upper and lower respiratory tract, otitis media, bacteremia, and meningitis (1, 4). The ability of this organism to disseminate from localized sites of infection to cause more serious invasive disease renders infections particularly difficult, yet imperative, to treat (24). Particularly vulnerable to pneumococcal infection are small children and the elderly, and the organism is usually a major cause of pneumonia and meningitis in these populations (7).
Only a few pneumococcal virulence determinants have been associated with disease, including pneumolysin, autolysin, capsule, adhesins, and other surface molecules (1, 5, 6, 15, 21, 24). The expression of these factors, for the most part, is unknown in vivo, and the temporal requirements for these factors during infection have not been determined. It has been hypothesized that surface factors play a role early in infection by preventing phagocytosis and allowing the bacteria to grow; later, continued growth in host tissues leads to the production of autolysin, and the subsequent release of pneumolysin results in inflammation. This progression of events during infection, culminating in high levels of inflammation, is probably the reason for the high morbidity and mortality for S. pneumoniae infections, even with antibiotic therapy (4). Indeed, in animals treated with pneumococcal cell wall components or with purified pneumolysin, similar levels of inflammation have been observed (17).
Fortunately, relevant animal models are available which can mimic most diseases caused by pneumococci, including pneumonia, otitis media, meningitis, and bacteremia. These models allow the assessment of the virulence of specifically engineered strains in ways not possible with tissue culture cells. Because animal infection models are the only direct means of measuring S. pneumoniae virulence, researchers have developed several clever strategies to screen large bacterial populations for inducible genes important for virulence or to negatively select for avirulent mutants. Such methods as in vivo expression technology (11) and signature-tagged mutagenesis (12, 18) have been applied previously to different streptococcal infection models; here, we report the use of differential fluorescence induction (DFI) to identify S. pneumoniae promoters induced under a variety of in vitro and in vivo conditions. A number of gram-negative and gram-positive bacterial pathogens have been studied using DFI, including Escherichia coli (2), Salmonella enterica serovar Typhimurium (22, 23), Listeria monocytogenes (25), and Staphylococcus aureus (W. P. Schneider, submitted for publication).
DFI relies on the use of a reporter gene, gfp, to monitor promoter activity by subjecting a library of DNA fragments cloned upstream of a promoterless gfp gene to a specific environment. The cells are then sorted by flow cytometry (fluorescence-activated cell sorting [FACS]) based on their fluorescence levels, which are indicative of gfp expression. In a previous study, a plasmid-based promoter-probe library containing S. pneumoniae DNA fragments was constructed and screened for gfp activity in a proof-of-principle study aimed at identifying promoters induced under conditions for competence (3). Here, to follow up those experiments, we report that this library was subjected to in vitro conditions of high osmolarity, temperature shift, change in CO2 concentration, change to blood agar medium, and iron limitation, as well as in vivo conditions of otitis media, respiratory tract infection (RTI), and intraperitoneal chamber implant. Our intent was to identify promoters of genes that are induced under the above-mentioned conditions, chosen so as to mimic in vivo conditions, with the expectation that such genes would play an important role in pneumococcal pathogenesis. Indeed, it was found that when deletions were constructed in these genes, over 50% of them were attenuated in at least one animal infection model.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The S. pneumoniae strains used in these studies are derivatives of D39, an encapsulated serotype 2 strain (20). Growth conditions for S. pneumoniae were either brain heart infusion broth (BHIB; Difco Laboratories) supplemented with 5% yeast extract or tryptic soy agar (TSA) plates containing 5% defibrinated sheep blood (BBL) at 37°C in an atmosphere with 7.5% CO2. For S. pneumoniae, antibiotics were used at the following concentrations: spectinomycin, 500 μg/ml; erythromycin, 0.3 μg/ml. E. coli was routinely grown in Luria broth medium containing 100 μg of apramycin/ml or 40 μg of apramycin and 40 μg of spectinomycin/ml, as appropriate.
Construction of the S. pneumoniae promoter-probe library.
The S. pneumoniae promoter-trap library in vector pNE1 gfp has been described previously (3). Briefly 200- to 500-bp DNase I fragments of S. pneumoniae D39 DNA were cloned upstream of the promoterless gfp gene in pNE1 gfp. The ligation was then electroporated into E. coli RR1. Plasmid DNA was prepared and used to transform S. pneumoniae D39 as described previously. The quality and randomness of the library is described in detail in reference 3. The copy number of vector pNE1 gfp is between 15 and 25 per cell (13).
DFI conditions.
In all cases the number of bacteria analyzed was approximately 106, which has been calculated to represent full coverage of the library (3). Induced and uninduced cells were prepared as described below, and at least 40,000 fluorescent events were collected using a FACStar machine (Becton Dickinson Immuno Cytometry Systems, San Jose, Calif.) equipped with an argon laser emitting at 488 nm. The gate was adjusted to sort only bacteria fluorescing above background, as determined by uninduced cells and/or cells containing only the pNE1 gfp vector. The sorted cells were propagated in BHIB-spectinomycin and grown on TSA-5% sheep blood agar plates containing spectinomycin. From 800 to 1,000 individual colonies were then screened for differential gfp expression in duplicate wells under noninducing and inducing conditions in a 96-well format. The clones were grown overnight in BHIB-spectinomycin, washed three times in phosphate-buffered saline (PBS), and resuspended, and duplicate cultures were diluted 1:10. The duplicate cultures were incubated for 4 h under inducing and noninducing conditions and then washed and resuspended in PBS-bovine serum albumin (BSA). Samples were analyzed on a FACS Caliber machine (Becton Dickinson Immuno Cytometry Systems) equipped with an argon laser emitting at 488 nm. Fluorescence, side-scatter, and forward-scatter data were collected using logarithmic amplifiers. Clones showing induction (mean channel fluorescence [MCF] of induced versus uninduced of twofold or greater) were rescreened in quadruplicate. Inserts of clones showing induction on repeat analysis were amplified by PCR using primers DB9 (5′ CCAAGCTTGCATGCCTGCAG) and WPS205 (5′ GTGAAAAGTTCTTCTCCTTTACTC), which correspond to sequences on either side of the multiple cloning site in pNE1 gfp, and the products were purified using the QIAQuick PCR purification system (Qiagen) in preparation for sequencing.
(i) Low iron.
Iron-limiting medium was prepared by stirring BHIB-spectinomycin medium with 2% Chelex-100 (Sigma, St. Louis, Mo.) for 16 h (9). The medium was then passed through a 0.2-μm-pore-size filter, and CaCl2 and MgSO4 (stocks prepared in Chelex-100-treated H2O) were added to final concentrations of 100 and 1.0 μM, respectively. The media were tested for iron depletion using the iron and total iron-binding capacity UIBC kit from Sigma-Aldrich (catalog no. 565U50P). The S. pneumoniae D39 promoter-trap library was grown to early logarithmic phase (A600 = 0.05) and washed three times in iron-limiting medium. The culture was then diluted to an A600 of 0.02 in 3 ml of iron-limiting medium and incubated for 4 h at 37°C and 7.5% CO2 in air without aeration. The induced culture was washed once in PBS containing 0.5% BSA (Sigma) and resuspended in PBS-BSA in preparation for FACS. For screening individual clones, the uninduced condition was growth to the same A600 in BHIB.
(ii) High osmolarity.
The condition used to screen for clones induced at high osmolarity was growth in BHIB containing 0.2 M NaCl. An aliquot of the library was thawed and incubated for 2 h in BHIB containing 500 μg of spectinomycin/ml. This culture was then serially diluted in both BHIB-spectinomycin and BHIB-0.2 M NaCl-500 μg of spectinomycin/ml, and dilutions were incubated overnight at 37°C with 7.5% CO2 in air. A separate set of cultures was prepared with the vector control strain. The A600s of the diluted cultures were determined, and cultures in logarithmic phase were pelleted and resuspended in PBS-BSA for sorting. Following sorting, the collected cells were plated onto TSA-5% sheep blood-spectinomycin plates, and after overnight growth, colonies were picked into wells of duplicate microtiter plates containing either BHIB-spectinomycin or BHIB-0.2 M NaCl-spectinomycin. The plates were incubated until the cultures reached logarithmic phase and were analyzed by flow cytometry.
(iii) Blood agar.
The S. pneumoniae D39 promoter-trap library was inoculated onto TSA-5% defibrinated sheep blood plates containing spectinomycin, and as a control for sorting, the vector control strain D39/pNE1 gfpm2 was spread onto another set of plates. Both sets of plates were incubated for 16 h at 37°C in an atmosphere of 7.5% CO2, and the cell growth was resuspended in PBS-BSA in preparation for sorting. For screening individual clones, the sorted population was inoculated onto TSA-spectinomycin plates, and after overnight growth, colonies were picked and patched onto two sets of plates: TSA-5% sheep blood-spectinomycin (induced condition) and TSA-spectinomycin (uninduced condition).
(iv) Carbon dioxide.
For the CO2 screen, a representative aliquot of the S. pneumoniae D39 promoter-trap library was thawed and used to inoculate 10 TSA plates containing 5% defibrinated sheep blood and 500 μg of spectinomycin/ml. One plate was incubated overnight at 37°C in a normal atmosphere without CO2, and the other nine were incubated overnight at 37°C in an atmosphere containing 7.5% CO2. As a control, the vector control strain was plated and grown overnight. Bacterial growth was resuspended in PBS-BSA and sorted; the negative control was the vector control strain, and the uninduced control was the library plated and grown in the absence of CO2. For screening individual clones, colonies were picked into wells of duplicate microtiter plates, which were then incubated overnight at 37°C with and without CO2.
(v) Static temperature shift.
The S. pneumoniae D39 promoter-trap library was used to inoculate TSA-5% defibrinated sheep blood plates containing spectinomycin and incubated at 37°C in 7.5% CO2 in air for 16 h. As controls to set the gate for sorting, an aliquot of the library and also the vector control strain were plated and incubated at 30°C in 7.5% CO2 for 16 h. The bacterial growth on each set of plates was resuspended separately in PBS-BSA, and FACS was performed as described above. For screening individual clones, the uninduced condition was growth at 30°C.
RTI.
The S. pneumoniae D39 promoter-trap library was used to infect groups of three mice (female; CD-1; age, 6 weeks) via intranasal instillation (see below); as a negative control for sorting, D39 carrying the promoterless gfp fusion plasmid was used to infect another set of mice. Both sets of mice were sacrificed 24 h postinfection, and their lungs were subjected to lavage with 1 ml of sterile PBS. The titer of an aliquot of each recovered sample was determined to enumerate bacteria inside the lungs; the remainder was pooled and used for sorting. The sample containing the negative control was also analyzed to adjust the gate for background fluorescence. The sorted cells were collected in BHIB containing spectinomycin, and this suspension was spread onto TSA plates containing 5% sheep blood and spectinomycin. After overnight growth, the cells were swabbed into BHIB, and aliquots were frozen at −80°C. Two more rounds of infection and sorting, after which the resulting population was plated for single colonies, enriched the sorted pool, and 96 individual clones were picked for PCR and sequencing analysis of inserts. Individual clones were screened for in vivo induction in the intraperitoneal chamber implant system (see below). All procedures were performed in accordance with the Institutional Animal Care and Use Committee Guidelines of Protein Design Labs, Inc.
Otitis media infection.
To screen for promoters induced during otitis media infection, groups of male Mongolian gerbils (weighing 25 to 30 g each) were infected with the S. pneumoniae D39 promoter-trap library or with D39/pNE1 gfpm2 via intrabulla injection (see below). The gerbils were sacrificed after 24 h and injected with 100 μl of PBS through the tympanic membrane to collect middle ear exudates. Titers of aliquots of the exudates were determined individually to assess bacterial growth, and the remainder was pooled and sorted. The negative control sample was used to set the gate. The sorted cells were collected in BHIB-spectinomycin, and this suspension was spread onto TSA-5% sheep blood-spectinomycin plates for overnight growth. Colonies were then swabbed into BHIB, and aliquots were frozen at −80°C. This infection and sorting was repeated twice more to enrich the fluorescent population, after which 96 individual clones were grown for PCR and sequencing to analyze inserts. Individual clones were screened for in vivo induction in the intraperitoneal chamber implant system (see below). All procedures were performed in accordance with the Institutional Animal Care and Use Committee Guidelines of Protein Design Labs, Inc.
Intraperitoneal chamber implant model.
The intraperitoneal chamber implant model was used both to identify in vivo-induced promoters and to screen clones identified under other conditions for in vivo induction. In the first application, the S. pneumoniae promoter-trap library was grown overnight in BHIB-5% yeast extract-spectinomycin to logarithmic phase. The cells were washed in PBS and used to load chambers, which were prepared as follows. Chambers were constructed from 1-cm-long sections of a 1-ml syringe. Millipore MF 0.22-μm-pore-size filters were cut to the diameter of the syringe barrels and attached to the ends of the barrels by melting them on a hot plate and pressing the molten plastic onto the filter. The entire sealed chamber was then sterilized by autoclaving. Bacteria were injected into the chamber with a needle, and the injection site was sealed with a heated glass rod. Mice (female CD-1; age, 7 to 8 weeks) were anesthetized with 3 to 4% isoflurane, and their abdomens were shaved and swabbed with betadine. A small longitudinal incision (2 to 3 cm) was made in the abdomen, and one to four chambers were placed in the peritoneal cavity. The abdominal wall was then sutured closed using 3/0-gauge silk sutures, and the skin was stapled closed. The animals were allowed to recover from anesthesia and given food and water ad libitum. An aliquot of the bacterial inoculum, representing the uninduced control, was analyzed for fluorescence by flow cytometry on a FACS Caliber machine with an argon laser emitting at 488 nm. As a negative control, the D39 strain carrying the promoterless gfp fusion plasmid was also loaded into chambers and implanted. After 22 h, the chambers were removed and bacteria were collected. The recovered bacteria were sorted as for the other conditions and were used to load fresh chambers and implanted. Three rounds of implantation followed by sorting were performed in order to enrich for in vivo-induced clones. After each round of sorting, 96 random clones were chosen for PCR amplification of inserts and sequencing. Clones that were isolated multiple times were reimplanted individually and analyzed for promoter induction in vivo by flow cytometry as described above, and their fluorescence was compared to that prior to implantation. Clones induced twofold or greater were analyzed by bioinformatics, and downstream genes were mutated as described below.
In the second application of the chamber model, individual clones were used to load the chambers, which were then implanted into mice and harvested after 22 h. The recovered bacteria were analyzed by flow cytometry as described above. All procedures were performed in accordance with the Institutional Animal Care and Use Committee Guidelines of Protein Design Labs, Inc.
Construction of replacement mutants.
Plasmid pCZA342 (10) was used to generate replacement mutations in DFI-identified genes for virulence assessment in various animal infection models. Plasmid pCZA342 encodes apramycin (for selection in E. coli) and erythromycin (for selection in S. pneumoniae) resistance markers. The strategy for constructing null mutants requires generation of a spectinomycin resistance marker (14) flanked by PCR-generated fragments upstream and downstream of the gene to be mutated cloned into pCZA342. Plasmid pCZA342 can replicate in E. coli but not in S. pneumoniae, so E. coli transformants are screened for an erythromycin-sensitive, spectinomycin-resistant phenotype on Luria-Bertani agar plates containing 40 μg of spectinomycin/ml and 40 μg of apramycin/ml. The primers used for amplification of the cloning fragments were based on the S. pneumoniae genome sequence generated by the Institute for Genomic Research (http://www.tigr.org/).
S. pneumoniae D39 bacterial cells were transformed with the plasmid DNA of the recombinant pCZA342 derivatives that had been purified from E. coli DH12S electrocompetent cells (Gibco BRL catalog no. 18312-017). The transformation of S. pneumoniae required the bacterial cells to be in early log phase (A600 = 0.05 to 0.1) in BHIB containing synthetic CSP-1 at a final concentration of 0.1 μg/ml, 10 mM glucose, and 10% horse serum (Sigma). The bacteria were cultured on TSA plates with 5% sheep blood and 0.5g of spectinomycin/liter (the plates were prepared to specification by Becton Dickinson [catalog no. 292755]) in an atmosphere of 7.5% CO2 at 37°C. Two independent transformations were performed, and an isolate from each was characterized for each mutant generated.
In vivo infection models.
Each mutant was assessed for virulence in at least two infection models, and mutants showing attenuation in either model were examined in two other models. The primary screen was a murine systemic infection, and each mutant was used to infect mice at two inocula: the D39 50% lethal dose (LD50) and 1,000 times the D39 LD50. Mutants that showed attenuation were used to infect larger groups of mice at multiple doses in order to calculate an LD50. In addition, all mutants were tested in a murine RTI model. Mutants showing a significant reduction in virulence in either model were tested in a gerbil model of otitis media as well as in an intraperitoneal chamber implant model. Finally, secondary isolates of all mutants were screened in the primary systemic infection to confirm that our results were due to the constructed alteration and not to an unrelated mutation due to the mutagenesis process.
(i) Murine RTI.
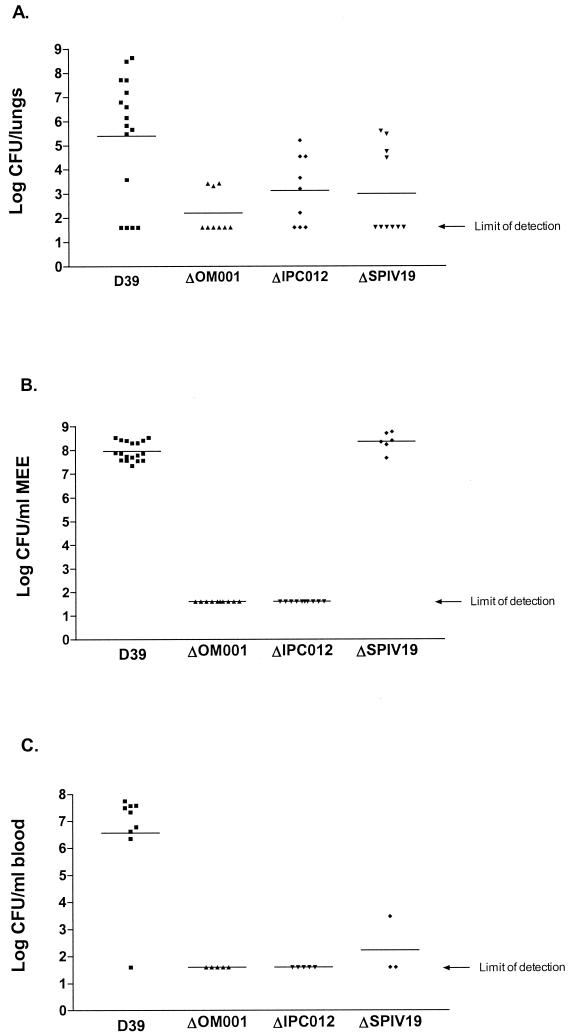
Bacterial cultures for infection were prepared by inoculating TSA-5% sheep blood plates containing appropriate antibiotics with bacteria from frozen stocks. The plates were incubated overnight, and the growth was swabbed into PBS. Bacterial suspensions were adjusted so that the A600 of a 1:10 dilution was approximately 0.3. Female CD-1 mice (age, 6 weeks) were anesthetized with isoflurane (4% in O2) and infected with 50 μl of bacterial suspension via intranasal instillation. Five to 10 animals were routinely infected per group. The animals were allowed to recover from anesthesia and were given food and water ad libitum, and after 24 or 48 h, they were euthanatized by CO2 overdose. Blood was collected via cardiac puncture, and titers were determined for viable bacteria; the lungs were aseptically removed and homogenized in 1 ml of PBS in a stomacher (Labconco), titers were determined, and the homogenate was plated for viable counts. Each experiment was performed a minimum of two times, and the data in the accompanying figure (see Fig. 3A) are a composite of all experiments.
FIG. 3.
In vivo characterization of mutants generated after identification in DFI screens. (A) Results of murine respiratory tract infection. The points represent the log number of bacteria harvested from mouse lungs 48 h postinfection. (B) Results of otitis media infection in gerbils. The points represent the log number of bacteria recovered from gerbil middle ear exudates (MEE) 4 days postinfection. (C) Results of otitis media infection in gerbils. The points represent the log number of bacteria found in the blood 4 days following infection. The limit of detection for all experiments is 40 organisms, and the graphs shown are composites of results from at least two individual experiments.
(ii) Murine systemic infection.
Bacterial cultures for infection were prepared by inoculating TSA-5% sheep blood plates containing appropriate antibiotics with bacteria from frozen stocks. The plates were incubated overnight, and the growth was swabbed into PBS. The bacterial suspensions were adjusted so that the A600 of a 1:10 dilution was approximately 0.3 and were serially diluted 10-fold in PBS to the appropriate concentrations such that the mice would receive between 10 and 106 organisms. Female CD-1 mice aged 6 weeks were given 200 μl of diluted bacterial suspension via intraperitoneal injection using a 28-gauge tuberculin syringe. Six to 10 mice were infected per group, the mice were monitored twice daily, and deaths were recorded. LD50 calculations were based on the number of survivors in each group at 24 h postinfection. For each mutant, a second independent isolate was used to infect groups of mice in order to ensure that the observed results were due to the engineered mutation and not to a second event. The secondary transformants were injected into mice at the D39 LD50 and at 1,000 times the LD50, and in all cases the numbers of survivors corroborated the results seen with the primary transformants (data not shown).
(iii) Otitis media infections.
Bacterial cultures for infection were prepared by inoculating TSA-5% sheep blood plates containing appropriate antibiotics with bacteria from frozen stocks. The plates were incubated overnight, and the growth was swabbed into PBS. The bacterial suspensions were adjusted after the A600 of a 1:10 dilution was measured and were diluted to approximately 105 bacteria per ml. Male Mongolian gerbils weighing 25 to 30 g were anesthetized with isoflurane (5% in O2), and 30 μl of a bacterial suspension (usually 103 to 104 organisms) was injected through the bones of both the right and left bullae using a 25-gauge needle attached to a 0.5-ml syringe. The animals were returned to their cages and allowed food and water ad libitum and were sacrificed 96 h postinfection by carbon dioxide overdose. Upon sacrifice, the middle ear aspirates were extracted following injection of 100 μl of PBS into each middle ear cavity through the tympanic membrane and withdrawal of fluid. The bacteria contained in this aspirate were enumerated following serial dilution and plating for CFU on TSA-5% sheep blood plates containing appropriate antibiotics. In addition, blood was collected via cardiac puncture and titers of viable bacteria were determined.
RESULTS
Construction of promoter-trap library for S. pneumoniae.
The S. pneumoniae promoter-trap library used in these studies and its analysis were previously described (3). Based on our calculations, we estimated the library to have fivefold coverage of the S. pneumoniae genome, both upon its construction in E. coli and after transfer to S. pneumoniae D39.
Inducing conditions and identification of differentially expressed clones.
Five in vitro inducing conditions were chosen for DFI analysis: low iron, high osmolarity, carbon dioxide, blood agar, and temperature shift. These were chosen in an effort to simulate individual aspects of the host environment that S. pneumoniae might encounter upon infection while being amenable to both DFI analysis and S. pneumoniae viability. The overall strategy used to identify differentially induced clones under in vitro conditions is shown in Fig. 1. Following sorting after incubation under a particular in vitro condition, up to 1,000 sorted clones were individually screened for induction. A total of 7,751 clones sorted from all five in vitro inducing conditions combined were individually screened for differential induction under the condition from which they were originally identified. Figure 2 illustrates the strategy used to identify in vivo-induced clones. The number of clones found to be induced twofold or greater under the in vivo or in vitro conditions was 86. When the inserts contained in these clones were sequenced, 78 unique sequences were identified. Descriptions of these 78 clones can be found in Table 1. Sixteen of these were identified in more than one screen, and 15 others had no match in the database or no downstream open reading frame (ORF). In cases where a clone was identified in more than one screen, they were usually overlapping clones rather than siblings. Out of the 78 unique clones identified, 23 were screened for differential expression in vivo using the intraperitoneal chambers (Table 1). Twenty-three clones were found to be induced twofold or greater when reexamined individually under the in vitro conditions under which they were identified; the most highly induced of these were STS002 (48-fold), STS011 (10.8-fold), CO2001 (4.8-fold), and ISIP454 (3.7-fold). In addition, a number of these clones were found to be highly induced in vivo when the MCF of cells recovered from the chambers was compared to that of broth-grown cells: IPC012 (6.8-fold), IPC018 (8.3-fold), IPC020 (5.8-fold), and IPC022 (2.6-fold). Out of the 23 clones examined for in vivo expression, 12 (52%) were induced twofold or greater (Table 1).
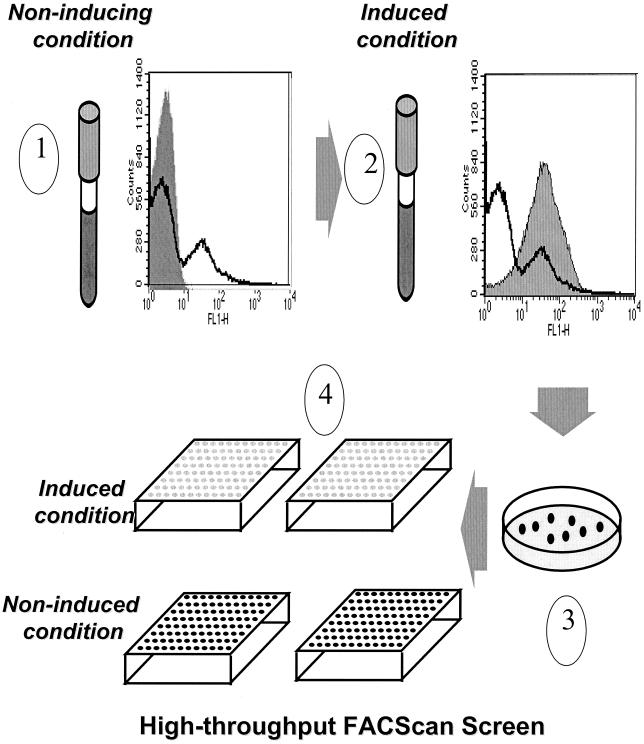
FIG. 1.
High-throughput method of screening individual clones following sorting of the promoter-probe library in S. pneumoniae under a variety of in vitro conditions. When the library is grown under noninducing conditions and analyzed by flow cytometry, the histogram shows peaks of high and low levels of fluorescence (1, heavy line). The shaded histogram represents the vector control strain. When the library is grown under a given inducing condition (see Materials and Methods) and analyzed by flow cytometry, the histogram is shifted, indicating increased fluorescence levels (2, shaded area; the heavy line indicating the uninduced library is shown for comparison). The sorted library is plated for single colonies (3), and individual colonies are picked into wells of microtiter plates for screening under inducing and noninducing conditions (4).
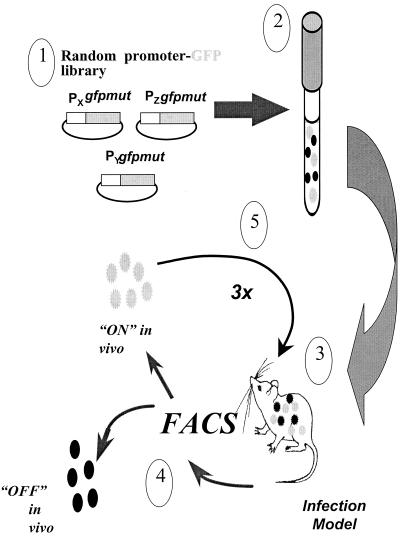
FIG. 2.
Strategy for isolating in vivo-induced promoters. The promoter-probe library (1) in S. pneumoniae is grown to logarithmic phase (2) and used to infect animals in either the respiratory tract, otitis media, or intraperitoneal chamber implant infection model (see Materials and Methods) (3). Bacteria are harvested from the animals and sorted by flow cytometry (4); clones with increased fluorescence are used to reinfect animals for enrichment (5). The sequence of infection and sorting is performed three times, and individual clones harvested from the final round of infection are analyzed as described in Materials and Methods.
TABLE 1.
Descriptions of S. pneumoniae promoters identified by DFI
| Promoter designationa | Induction ratiob | No. of ORFs in operon | ORF descriptionc | Mutant constructedd |
|---|---|---|---|---|
| BA001(NC_003028) | 1.48 | 0 | No ORF downstream | N |
| STS006 | 1.98 | |||
| BA002 (AE007482) | 1.5 | 1 | (AAK75982); hypothetical protein | N |
| BA003 (AE007410) | 2.32 | 3 | ORF1; PRPP synthetase | Ye |
| OM005 | 1.9 (CI) | ORF2; aminotransferase (AAK75206) | Yg | |
| CO2003 | 4.8 | ORF3; hypothetical protein (AAK75205) | ||
| BA004 (AE007451) | 1.8 | 1 | Pyridine nucleotide-disulfide oxidoreductase family protein (AAK75650) | N |
| IPC001 (AE007373) | 2.6 | 1 | Serine protease (AAK74791) | Y |
| ISIP456 | 1.5 | |||
| IPC003 (AE007435) | 11 | ORF1; conserved hypothetical protein (AAK75476) | Ye | |
| ORF2; 3-dehydroquinate dehydratase (AAK75475) | Yo | |||
| ORF3; shikimate dehydrogenase (AAK75474) | ||||
| ORF4; 3-dehydroquinate synthase (AAK75473) | ||||
| ORF5; chorismate synthase (AAK75472) | ||||
| ORF6; prephenate dehydrogenase (AAK75471) | ||||
| ORF7; hypothetical protein (AAK75470) | ||||
| ORF8; 3-phosphoshikimate, 1-carboxyvinyl transferase (AAK75469) | ||||
| ORF9; shikimate kinase (AAK75468) | ||||
| ORF10; prephenate dehydratase (AAK75467) | ||||
| ORF11; psr protein (AAK75466) | ||||
| IPC002 (AE007451) | 2 | ORF1; hypothetical protein (AAK75645) | Ye | |
| ORF2; deg V (AAK75644) | Yo | |||
| IPC004 (AE007391) | 1.3 | 1 | ORF1; DNA topoisomerase IV (AAK74981) | N |
| RTI004 | ||||
| IPC005 (AE007448) | 1 | 6 | ORF1; Cof family protein/peptidyl-prolyl cis-trans isomerase (AAK75626) | Ye |
| ORF2; general stress protein (AAK75625) | Yo | |||
| ORF3; conserved hypothetical protein (AAK75624) | ||||
| ORF4; conserved hypothetical protein (AAK75623) | ||||
| ORF5; Mn-dependent pyrophosphatase (AAK75622) | ||||
| ORF6; hypothetical ORF (AAK75621) | ||||
| IPC006 (AE007466) | 0.3 | 4 | ORF1; conserved hypothetical protein (AAK75807) | Ye |
| ORF2; hypothetical protein (AAK75806) | Yo | |||
| ORF3; IS1381 transposase OrfB (AAK75996) | ||||
| ORF4; hypothetical protein (AAK75805) | ||||
| IPC022 (NC_003028) | 2.6 | 6 | Multiple very small ORFs (also known as transcriptional repressor) | Ye |
| CO2002 | 3.21 | |||
| SPIV016 | ||||
| SPIV005 | ||||
| IPC007 (AE007368) | 1.9 | 3 | ORF1; phenylalanyl tRNA synthetase, alpha chain (AAK74733) | N |
| ORF2; acetyltransferase, GNAT family (AAK74734) | ||||
| ORF3; phenylalanyl-tRNA synthetase, beta chain (AAK74735) | ||||
| IPC008 (AE007321) | 32 | 1 | Phosphoribosylaminoimidazole-succinocarboxamide synthase (AAK74233) | Y |
| SPIV014 | ||||
| IPC009 (AAK7423) | 1.6 | 1 | Integral membrane protein (AAK76084) | Y |
| SPIV027 | ||||
| IPC010 (AE007507) | 0.5 | 2 | ORF1; chaperonin; heat shock protein (AAK76239) | Ye |
| ORF2; NifR3/Smm1 in Bacillus subtilis | Yo | |||
| IPC011 (AE007349) | 0.25 | 2 | ORF1; recombination protein U (AAK74537) | Ye |
| ORF2; penicillin binding pbpla gene in S. pneumoniae (AAK74536) | Yo | |||
| IPC012 (AE007405) | 6.8 | 11 | ORF1; thymidine kinase (AAK75133) | Yg |
| ORF2; spermidine acetyl transferase (AAK75134) | ||||
| ORF3; peptide chain release factor (AAK75135) | ||||
| ORF4; protoporphyrinogen oxidase/HEMK protein homolog (AAK75136) | ||||
| ORF5; Sua5/YciO/YrdC family protein (AAK7513) | ||||
| ORF6; acetyltransferase, GNAT family (AAK75138) | ||||
| ORF7; serine hydroxymethyl transferase (AAK75139) | ||||
| ORF8; hypothetical protein (AAK75140) | ||||
| ORF9; conserved hypothetical protein (AAK75141) | ||||
| ORF10; conserved hypothetical protein (AAK75142) | ||||
| ORF11; RNA methyl transferase (AAK75144) | ||||
| IPC013 (AE007487) | 1.4 | 1 | Putative membrane-spanning protein (AAK76039) | Ye |
| OM002 | 2.1 (CI) | |||
| IPC014 (AE007354) | 0.2 | 2 | ORF1; conserved hypothetical protein (AAK74596) | N |
| OM003 | 2.1 (CI) | ORF2; translation elongation factor EF-P (AAK74597) | ||
| ORF3; transcription termination protein (AAK99194) | ||||
| IPC015 (AE007318) | 1.1 | 3 | ORF1; GTP binding protein (AAK74197) | Ye |
| OM006 | ORF2; peptidyl tRNA hydrolase (PTH gene) (AAK74198) | Yo | ||
| ORF3; transcriptional-repair coupling factor (AAK99194) | ||||
| IPC016 (AE007386) | 2.2 | 1 | Conserved hypothetical protein (AAK74938) | Ye |
| IPC017 (AE007458) | 2.3 | 1 | Endopeptidase O (AAK75727) | Y |
| SPIV001 | (Zn metalloprotease) | |||
| SPIV012 | ||||
| IPC018 (AE007356) | 8.3 | 2 | ORF1; amino acid ABC transporter; amino acid-binding protein/permease protein (AAK74614) | Ye |
| ORF2; amino acid ABC transporter; ATP-binding protein (AAK74613) | Yo | |||
| IPC020 (AE007442) | 5.8 | 3 | ORF1; spoU rRNA methylase family protein (AAK75551) | Ye |
| ORF2; tRNA guanosine methyl transferase (AAL00115) | Yo | |||
| ORF3; no match in database | ||||
| IPC021 (AE007341) | 2.1 | 1 | Cof family protein (AAK74464) | N |
| SPIV002 | ||||
| SPIV026 | ||||
| CO2001 (AE007386) | 4.8 | 3 | ORF1; aminopeptidase N (AAK74934) | Yo |
| ORF2; regulator ciaR (AAK74935) | ||||
| ORF3; histidine kinase ciaH (AAK74936) | ||||
| CO2004 (NC_003028) | 4.3 | No ORF downstream | N | |
| CO2006 (NC_003028) | 4.5 | No ORF downstream | N | |
| HOI021 (AE007337) | 0.2 | 3 | ORF1; lactose-specific IIA component of PTS system (AAK74427) | Yg |
| ISIP069 | 2 | ORF2; cellobiose phosphotransferase system enzyme (AAK74428) | ||
| ORF3; PTS system; cellobiose-specific IIC component (AAK74429) | ||||
| HOI042 (AE007509) | 0.75 | 10 | ORF1; conserved hypothetical protein (AAK76273) | Ye |
| ORF2; peptidase, M16 family (AAK76272) | Yg | |||
| ORF3; conserved hypothetical protein (AAK76271) | ||||
| ORF4; phosphotidylglycerophosphate synthase (AAK76270) | ||||
| ORF5; ATP-binding protein (AAK76269) | ||||
| ORF6; ATP-binding protein (AAK76268) | ||||
| ORF7; conserved hypothetical protein (AAK76267) | ||||
| ORF8; cell shape-determining protein (AAK76266) | ||||
| ORF9; rod shape-determining protein MreD; putative (AAK76265) | ||||
| ORF10; putative secreted protein (AAK76264) | ||||
| HOI101 (AE008513) | 0.2 | 1 | Adenine phosphoribosyl transferase (AAL00239) | Y |
| ISIP454 (AE007321) | 3.7 | 2 | ORF1; competence factor transporting ATP-binding/permease protein ComA (AAK74231) | Yg |
| ORF2; competence factor transport protein ComB (AAK74232) | ||||
| ISIP741 (AE007364) | 1.7 | 4 | ORF1; ABC transporter (CAC18585) | Yg |
| ORF2; ABC transporter (CAC18584) | ||||
| ORF3; ABC transporter (CAC18583) | ||||
| ORF4; ABC transporter (AAK74687) | ||||
| ISIP409 (AE007325) | 1.5 | 1 | Uridine phosphorylase (AAK74264) | Y |
| ISIP485 (NC_003028) | 1.5 | 3 | ORF1; no match in database | Yg |
| ORF2; no match in database | ||||
| ORF3; no match in database | ||||
| ISIP497 (AE007320) | 1.6 | 4 | ORF1; hypothetical protein (AAK74215) | Yg |
| SPIV010 | ORF2; no match in database | |||
| SPIV011 | ORF3; hypothetical protein (AAK74217) | |||
| ORF4; PRPP synthetase (AAK74218) | ||||
| ISIP747 (AE007449) | 1.7 | 4 | ORF1; polypeptide deformylase (AAK75636) | N |
| SPIV003 | ORF2; hypothetical protein (AAK75635) | |||
| SPIV028 | ORF3; hypothetical protein (AAL00210) | |||
| ISIP870 (AE007337) | 2.1 | 2 | ORF1; transcriptional regulator, DeoR family (AAK74425) | Yg |
| ORF2; transcriptional regulator (AAK74426) | ||||
| OM001 (AE007492) | 2.1 (CI) | 3 | ORF1; preprotein translocase, YajC subunit (AAK76094) | Yo |
| SPIV017 | ORF2; protein tyrosine phosphatase (AAK76093) | |||
| ORF3; conserved hypothetical protein (AAK76092) | ||||
| OM004 (AE007443) | 2.3 (CI) | 2 | ORF1; peptidoglycan GLcNAc deacetylase (AAK75573) | Ye |
| ORF2; oxidoreductase; aldo/keto reductase family (AAK75572) | ||||
| RTI001 (AE007401) | 2 | ORF1; oligopeptidase F (AAK75100) | N | |
| ORF2; methyltransferase (AAK75101) | ||||
| RTI002 (NC_003028) | 1 | No match in database | N | |
| RTI005 (AE007434) | 5 | ORF1; homoserine kinase homolog (AAK75458) | N | |
| ORF2; hypothetical protein | ||||
| ORF3; peptide methionine sulfoxide reductase (AAK75457) | ||||
| ORF4; ATP-binding protein (AAK75456) | ||||
| ORF5; putative ABC transporter (AAK75455) | ||||
| RTI006 (AE007477) | 2 | ORF1; choline transporter (AAK75933) | Yg | |
| ORF2; choline transporter (AAK75932) | ||||
| RTI007 (NC_003028) | 1 | No match in database | N | |
| RTI008 (AE007440) | 3 | ORF1; conserved hypothetical protein (AAK75533) | N | |
| ORF2; cation ABC transporter (AAK75534) | ||||
| ORF3; Spn1 putative transposase (NP_345810) | ||||
| RTI009 (AE007511) | 1 | SPSpoJ (AAK76287) | Y | |
| RTI010 (NC_003028) | 1 | No match in database | N | |
| RTI011 (NC_003028) | 1 | No match in database | N | |
| RTI012 (NC_003028) | 0 | No match in database | N | |
| RTI013 (NC_003028) | 0 | No match in database | N | |
| SPIV004 (AE007376) | 2 | ORF1; hypothetical protein (AAK74808) | Yg | |
| SPIV009 | ORF2; putative zinc metalloproteinase (AAK74809) | |||
| SPIV015 | ||||
| SPIV007 (AE007330) | 4 | ORF1; unknown protein (AAK74324) | N | |
| SPIV023 | ORF2; conserved domain protein (AAK74325) | |||
| ORF3; hypothetical protein (AAK74326) | ||||
| ORF4; conserved hypothetical protein (AAK74327) | ||||
| SPIV008 (AE007367) | 3 | ORF1; licT (AAK74730) | N | |
| SPIV025 | ORF2; beta-glucoside-specific IIABC PTS system component (AAK74731) | |||
| ORF3; beta-glucosidase (AAK74732) | ||||
| SPIV013 (AE007496) | 1 | ABC transporter; ATP-binding protein (AAK76137) | Y | |
| SPIV018 (AE007497) | 3 | ORF1; response regulator pnpR (AAK76142) | Ye | |
| ORF2; histidine kinase pnpS (AAK76143) | Yg | |||
| ORF3; phosphate-binding protein pstS (AAL00697) | ||||
| SPIV019 (AE007352) | 2 | ORF1; RNase HII (AAK74566) | Yo | |
| SPIV029 | ORF2, signal peptidase 1; lep (AAK74565) | |||
| SPIV020 (NC_003028) | 0 | No ORF downstream | N | |
| SPIV021 (AE007429) | 1 | Flavodoxin (AAK75401) | N | |
| SPIV022 (AE007405) | 1 | 4-Oxalocrotonate tautomerase (AAK75132) | N | |
| SPIV024 (NC_003028) | 0 | No match in database | N | |
| SPIV030 (AE007402) | 1 | Hypothetical protein (AAK75106) | N | |
| STS001 (AE008461) | 7.96 | 3 | ORF1; dihydroorotate dehydrogenase electron transfer subunit (AAK99669) | Yg |
| ORF2; dihydroorotate dehydrogenase B, catalytic subunit (AAK99670) | ||||
| ORF3; endo-beta-N-acetylglucosaminidase (AAK19156) | ||||
| STS002 (AE007325) | 44.77 | 1 | PII-type cell wall-associated proteinase precursor (AAK74270) | Ye |
| STS003 (AE007481) | 2.73 | 1 | RNA methyl transferase homolog (AAK75972) | N |
| STS004 (AE007356) | 2.07 | 1 | Hypothetical protein (AAK74615) | Y |
| STS005 (AE007359) | 24.1 | 1 | Lactose permease IIBC component, lactose-specific PTS system (AAK74637) | Ye |
| STS007 (AE008443) | 2.03 | 1 | Unknown conserved protein (AAK99455) | N |
| STS008 (AE007485) | 2.51 | 1 | Nucleoside diphosphate kinase (AAK76026) | Y |
| STS009 (AE007433) | 1.92 | 2 | ORF1; no match in database | N |
| ORF2; no match in database | ||||
| STS010 (NC_003028) | 2.9 | 0 | No ORF downstream | N |
| STS011 (NC_003028) | 10.8 | 1 | No match in database | N |
| STS012 (AE007379) | 2.49 | 2 | ORF1; orotidine 5′- phosphate decarboxylase (pyrF) (AAK74844) | Yo |
| ORF2; orotate phosphoribosyl transferase (pyrE) (AAK74845) | ||||
| STS013 (AE007355) | 2.81 | 3 | ORF1; glutamyl tRNA amidotransferase subunit A (AAK74599) | Ye |
| ORF2; glutamyl tRNA amidotransferase subunit B (AAK74598) | Yo | |||
| ORF3; translation elongation factor P (AAK74597) | ||||
| STS014 (AE007350) | 2.47 | 4 | ORF1; FMN-dependent dehydrogenase family protein (AAK74551) | Ye |
| ORF2; hypothetical protein (AAK74552) | Yo | |||
| ORF3; sensor histidine kinase; putative (AAK74553) | ||||
| ORF4; response regulator (AAK74554) | ||||
| STS016 (AE007482) | 2.88 | 2 | ORF1; hypothetical protein (AAK75983) | N |
| ORF2; hypothetical protein (AAK75982) | ||||
| STS017 (AE007438) | 1.66 | 4 | ORF1; Hpr (Ser) kinase/phosphatase (AAK75511) | N |
| ORF2; prolipoprotein diacylglyceryl transferase (AAK75510) | ||||
| ORF3; conserved hypothetical protein (AAK75509) | ||||
| ORF4; conserved hypothetical protein (AAK75508) | ||||
| STS018 (AE007333) | 2.76 | 4 | ORF1; riboflavin biosynthetic protein ribD (AAK74359) | Yo |
| ORF2; riboflavin synthase alpha chain (AAK74358) | ||||
| ORF3; 3,4-dihydroxy-2-butanone 4-phosphate synthase/GTP cyclohydrolase II (AAK74357) | ||||
| ORF4; riboflavin synthase beta chain (AAK74356) |
Prefix indicates identifying DFI screen (CO2, carbon dioxide shift; HOI, high osmolarity; IPC, intraperitoneal chamber; ISIP, iron starvation; OM, otitis media infection; RTI and SPIV, respiratory tract infection; STS, static temperature shift); accession number (in parentheses) refers to chromosomal location.
Ratio of induced MCF to uninduced MCF under conditions under which clone was originally identified, except for clones labeled “CI,” which were assayed in vivo in the intraperitoneal chamber implant.
Accession number (in parentheses) refers to protein.
Result of replacement mutagenesis. Y, replacement mutant constructed; Yg, mutant constructed by replacing first gene in operon; Yo, mutant constructed by replacing entire operon; Ye, replacement mutant construction was attempted but was unsuccessful, indicating that downstream gene is essential; N, no replacement mutant was constructed because downstream gene(s) was essential by literature search, there was no ORF downstream of clone homology, or there was no sequence homology to anything in the S. pneumoniae database downstream of clone homology.
Three in vivo inducing conditions were chosen: RTI, otitis media infection, and intraperitoneal chamber implant. Unlike the first two, the last model is an incubation of the organism within the mouse, without the requirement for dissemination or avoidance of host defense. A strategy similar to that for isolation of in vitro-induced clones was used for the in vivo conditions; however, following three enrichment rounds of infection followed by sorting each time, the cells were plated for single colonies, and random colonies were picked for analysis of inserts by PCR. In some cases, when a promoter was identified numerous times, unique clones were chosen for individual in vivo screening in the murine intraperitoneal chamber implant model (Table 1).
From our bioinformatics analysis of the sequences downstream of the regions identified in our clones, 23 were found to have a single ORF downstream and 42 clones had multiple ORFs downstream.
Generation of replacement mutants.
We wanted to determine whether the genes induced under our chosen in vitro and in vivo conditions contributed to virulence. Genes downstream of identified promoters were deleted via replacement mutagenesis. For 33 of the 78 identified sequences, no attempt was made to construct a mutant, either because there was no match for the sequence in the S. pneumoniae database, no downstream ORF could be identified, or a review of the literature indicated the downstream gene(s) would be essential. For 20 of the 78 sequences, our attempts at generating a mutant were unsuccessful, indicating that the genes are likely to be essential. Twenty-five replacement mutants were generated, and all were characterized for the ability to cause infection.
In vivo characterization of mutants.
All viable mutants were assessed for virulence in two infection models: RTI and systemic infection. In cases where we saw a significant decrease in virulence in both models, we also ascertained virulence in the otitis media model. The results of these studies are shown in Table 2, and Fig. 3 shows the levels of attenuation for mutants OM001, IPC012, and SPIV019. Twenty-five mutants were examined in at least two infection models. The LD50s for 14 of these (56%) were determined to be at least 10-fold higher than that of D39, with mutants HOI101, IPC012, and OM001 showing the highest levels of attenuation. In the RTI model, 11 mutants (44%) were found to colonize the lungs at least 10-fold less than D39. Mutants IPC012, OM001, and STS012 are severely attenuated in their abilities to cause lung infection in this model. Finally, 10 mutants were examined for virulence in the otitis media infection model; two of these, IPC012 and OM001, were completely attenuated, and two, HOI101 and SPIV019, were found to be capable of wild-type levels of growth in the middle ear cavity but could not disseminate to the blood (Table 2). Some of these results are represented graphically in Fig. 3.
TABLE 2.
In vivo characterization of replacement mutants
| Mutanta | LD50b | Attenuation levelc
|
|
|---|---|---|---|
| Respiratory tract infection | Otitis media infection | ||
| ΔCO2001 | 3.28 | WT | |
| ΔHOI101 | >5.6 | 1-log unit decrease | WT; 3-log unit decrease in blood |
| ΔIPC009 | 4.08 | WT | |
| ΔIPC012 | >6.26 | 3.5-log unit decrease | 6 log units; undetectable in blood |
| ΔISIP409 | 4.53 | ∼1-log unit decrease | WT |
| ΔISIP454 | 4.65 | ∼WT | |
| ΔISIP456 | 4.84 | WT | |
| ΔISIP485 | ND | ∼WT | |
| ΔISIP497 | 3.69 | ∼WT | |
| ΔISIP069 | ND | ∼WT | |
| ΔISIP741 | ND | ∼WT | |
| ΔISIP870 | ND | ∼WT | |
| ΔOM001 | >5.97 | 4-log unit decrease | 6 log units; undetectable in blood |
| ΔRTI006 | 2.52 | 1-log unit decrease | WT |
| ΔRTI009 | 3.65 | 1-log unit decrease | WT |
| ΔSPIV012 | 2.85 | WT | |
| ΔSPIV013 | ND | WT | |
| ΔSPIV014 | 5.79 | 1-log unit decrease | WT; 1-log unit decrease in blood |
| ΔSPIV015 | 2.9 | WT | |
| ΔSPIV019 | 5.54 | 1-log unit decrease | WT; 4.5-log unit decrease in blood |
| ΔSTS001 | ND | WT | |
| ΔSTS004 | ND | WT | |
| ΔSTS008 | ND | 1.5 log units | |
| ΔSTS012 | 4.40 | 2.5-log unit decrease | WT |
| ΔSTS018 | 3.65 | 2 log units | WT |
| No./totals (%) | 14/25 (56)d | 11/25 (44)e | 2/25 (8)f |
Prefix indicates DFI screen in which clone was identified: CO2, carbon dioxide shift; HOI, high osmolarity; IPC, intraperitoneal chamber; ISIP, iron starvation; OM, otitis media infection; RTI and SPIV, respiratory tract infection; STS, static temperature shift.
Represents the log value of the LD50. The LD50 of wild-type D39 is approximately 100 organisms. ND, not done.
Denotes attenuation level compared to D39 (WT).
Number of mutants with LD50 ≥10-fold higher than that of D39.
Number of mutants colonizing lungs ≥10-fold less than D39.
Number of mutants completely attenuated.
DISCUSSION
DFI has proven to be an effective method for identifying bacterial promoters induced under specific conditions. It was our aim to identify promoters of virulence genes, using the rationale that such genes would be expressed in vivo or under in vitro conditions designed to mimic individual aspects of the in vivo environment. Once such genes were identified, their roles in virulence were ascertained by constructing deletion mutants and testing them in a number of in vivo models. In previous work (3), we showed that DFI could be used successfully to identify novel S. pneumoniae genes involved in competence. In that study, known competence genes, as well as novel ones, were identified; a number of these were determined to have a role in pathogenesis as well, providing further evidence for the connection between virulence and competence. Our present results with S. pneumoniae, using both in vivo and in vitro conditions for DFI, confirm these earlier studies and further help to validate this approach as a method for identifying pathogenesis factors.
The in vitro conditions were chosen to simulate the in vivo environment encountered by S. pneumoniae upon infection. Our results show that several promoters were identified in more than one screen; for example, a promoter found in the screen for CO2 induction was also found in both the chamber implant screen and the RTI screen (CO2002, IPC022, SPIV005, and SPIV016 [Table 1]). Another promoter was identified in the blood agar, carbon dioxide, and otitis media screens (BA003, CO2003 and OM005 [Table 1]). This validates our two central hypotheses: that in vitro conditions could mimic those in vivo and that genes induced under these conditions might be important for virulence. In cases where a gene was identified in both in vitro and in vivo screens, the use of defined in vitro conditions could help to uncover the function of an unknown gene(s) and, in addition, establish the element(s) of the in vivo environment sensed by S. pneumoniae that can induce expression of that gene. The low-iron in vitro condition yielded three clones that were also identified in in vivo screens for both the chamber implant and RTI. It is interesting to speculate that, based on these results, the lungs and peritoneal cavity are low-iron environments. Mutants generated from these clones either were attenuated (ISIP456 and ISIP497) or the gene was found to be essential (ISIP747). Presumably, the more clones analyzed in these screens, the more such correlations can be made. The screens were most likely not saturating, in that we chose to individually screen only 800 to 1,000 clones for induction after sorting.
Two known two-component signal transduction systems were identified in our screens: ciaR-ciaH (CO2001) (8) and pnpR-pnpS (SPIV018) (16). In addition, genes downstream of the clone homology in STS014 encode an unknown histidine kinase and response regulator pair. The ciaR-ciaH pair was identified previously in a genome-wide search for two-component signal transduction systems in S. pneumoniae (19), and a ciaR-ciaH double mutant was found to be highly attenuated in a murine RTI model (19). Our corresponding mutant, CO2001, showed attenuation in our systemic-infection model but none in our RTI model, perhaps reflecting differences in pneumococcal strains, methods of generating mutants, or even mouse strains used.
In addition, the same report (19) showed that a pnpR-pnpS double mutant was strongly attenuated in the RTI model; we were unable to construct this mutant (SPIV018) and assume it is essential in D39. Again, our results most likely indicate strain differences between D39 and 0100993, the strain studied by Throup et al.
Our screens did not identify such known pneumococcal virulence determinants as capsule biosynthetic genes and the genes for pneumolysin and autolysin, perhaps because these genes are necessary for S. pneumoniae growth and therefore would not be differentially expressed or, as in the case of autolysin, are presumed to be expressed in a growth-stage-specific manner. However, a number of clones identified genes encoding binding proteins and proteases, which have been implicated in virulence. Penicillin binding protein (IPC011) was identified in the intraperitoneal chamber implant screen, and as we were unable to delete the two genes in this operon, we believe that at least one of the genes is essential. Two clones were identified as promoters of protease-encoding genes (IPC017/SPIV001/SPIV012, and SPIV004/SPIV009/SPIV015); when mutated, these were found to moderately affect virulence (Table 2).
We chose two primary infection models, which offer two different challenges to the invading pathogen, in which to test our mutants. The systemic-infection model is an intraperitoneal injection with rapid spread to more distal sites, resulting in bacteremia and organ failure, usually within 48 h. The RTI model is a more localized infection, requiring the bacteria to avoid alveolar macrophages and grow in the lungs over a 48-h period. This model will also lead to blood dissemination, and mice infected in this way will rarely survive past 72 h. Many of the mutants tested (7 out of 28 [29%]) were attenuated in only one infection model, which perhaps underscores the different requirements for each. It is possible that the mutants attenuated in only one model involve genes required for pathogenesis in only that model or host environment.
The fact that so many promoters were identified in more than one screen could be due to several possible factors. First, it is possible that these promoters are constitutively highly expressed and so were positively sorted under all conditions. This is unlikely, since no clone was identified in all screens. Second, it could be that induction of these promoters is due to an environmental feature common to those particular conditions. Finally, perhaps these promoters drive expression of genes involved in the S. pneumoniae stress response and so were isolated from a number of screens. Again, more saturating screens resulting in the analysis of larger numbers of clones would help to address this point.
Our in vivo DFI screens—chamber implant, RTI, and otitis media—yielded many clones induced under two of the three conditions. Ten such clones were identified, indicating that even though the screens represent three different infection sites (peritoneal cavity, lungs, and middle ear, respectively), there are common elements to which the bacteria are responding. Alternatively, these 10 identified genes could represent factors involved in the S. pneumoniae general stress response.
From the results shown in Table 2, it appears that the most attenuated mutants in either model (OM001 or IPC012) are also severely attenuated in the other two models examined. It is possible that both sets of genes represented by these mutants are required for some aspect of bacterial growth or survival in these three models. Our strategy for constructing mutants does not allow us to definitively determine which gene(s) in these operons is required for pathogenesis; further work is needed in the form of precise deletion mutants to address this issue. However, since kinase genes are predicted to be in both operons, it is interesting to speculate that the mutants lack an important signaling mechanism to respond to the host environment or defense response.
We set out to identify S. pneumoniae genes important for virulence in a number of infection models by using DFI. We identified a number of essential genes (20 out of 78 [26%]). Several unknown genes were also identified. Our results indicate that DFI can be a valuable tool for virulence gene identification. In addition, by using a combination of in vivo and in vitro screening conditions, it is possible to identify bacterial genes required for pathogenesis and to correlate this with a particular in vitro condition, perhaps offering some elucidation of the microenvironment encountered by a pathogen upon host infection.
Acknowledgments
We thank Stanley Falkow and Rafael Valdivia for helpful discussions and reagents, Don LeBlanc and Patti Treadway for reagents and guidance, and Patricia Lekas, Dixie Polakoff, Jon Yang, Christine Johnson, and Roman Moniz for technical assistance.
Editor:E. I. Tuomanen
REFERENCES
- 1.Alonso DeVelasco, E., A. F. M. Verheul, J. Verhoef, and H. Snippe. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol. Rev. 59:591-603. [DOI] [PMC free article] [PubMed]
- 2.Badger, J., C. Wass, and K. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. [DOI] [PubMed]
- 3.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromockyj. 2000. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence peptide. Mol. Microbiol. 39:126-135. [DOI] [PubMed]
- 4.Boulnois, G. J. 1992. Pneumococcal proteins and the pathogenesis of disease caused by Streptococcus pneumoniae. J. Gen. Microbiol. 138:249-259. [DOI] [PubMed]
- 5.Canvin, J. R., A. P. Marvin, M. Sivakumaran, J. C. Paton, G. J. Boulnois, P. W. Andrew, and T. J. Mitchell. 1995. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J. Infect. Dis. 172:119-123. [DOI] [PubMed]
- 6.Friedland, I. R., M. M. Paris, S. Hickey, S. Shelton, K. Olsen, J. C. Paton, and G. H. McCracken. 1995. The limited role of pneumolysin in the pathogenesis of pneumococcal meningitis. J. Infect. Dis. 172:805-809. [DOI] [PubMed]
- 7.Gray, B. M., G. M. Converse, and H. C. Dillon. 1979. Serotypes of Streptococcus pneumoniae causing disease. J. Infect. Dis. 140:979-984. [DOI] [PubMed]
- 8.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. A. Hakenbeck. 1994. Two-component signal transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed]
- 9.Hasan, A., J. Holland, A. Smith, and P. Williams. 1997. Elemental iron does repress transferrin, haemopexin and haemoglobin receptor expression in Haemophilus influenzae. FEMS Microbiol. Lett. 150:19-26. [DOI] [PubMed]
- 10.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed]
- 11.Kiliç, A., M. Herzberg, M. Meyer, Z. Zhao, and L. Tao. 1999. Streptococcal reporter gene-fusion vector for identification of in vivo expressed genes. Plasmid 42:67-72. [DOI] [PubMed]
- 12.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed]
- 13.LeBlanc, D., L. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed]
- 14.LeBlanc, D., L. Lee, and J. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase ADD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed]
- 15.Mitchell, T. J., and P. W. Andrew. 1997. Biological properties of pneumolysin. Microb. Drug Resist. 3:19-26. [DOI] [PubMed]
- 16.Novak, R., A. Cauwels, E. Charpentier, and E. Tuomanen. 1999. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J. Bacteriol. 181:1126-1133. [DOI] [PMC free article] [PubMed]
- 17.Paton, J. C. 1996. The contribution of pneumolysin to the pathogenicity of Streptococcus pneumoniae. Trends Microbiol. 4:103-106. [DOI] [PubMed]
- 18.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferreri, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed]
- 19.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed]
- 20.Tiraby, G., M. S. Fox, and H. Bernheimer. 1975. Marker discrimination in deoxyribonucleic acid-mediated transformation of various pneumococcus strains. J. Bacteriol. 121:608-618. [DOI] [PMC free article] [PubMed]
- 21.Tuomanen, E., R. Austrian, and H. R. Masure. 1995. Pathogenesis of pneumococcal infection. N. Engl. J. Med. 332:1280-1284. [DOI] [PubMed]
- 22.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed]
- 23.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed]
- 24.Watson, D. A., D. M. Musher, and J. Verhoef. 1995. Pneumococcal virulence factors and host immune responses to them. Eur. J. Clin. Microbiol. Infect. Dis. 14:479-490. [DOI] [PubMed]
- 25.Wilson, R. L., A. R. Tvinnereim, B. D. Jones, and J. T. Harty. 2001. Identification of Listeria monocytogenes in vivo-induced genes by fluorescence-activated cell sorting. Infect. Immun. 69:5016-5024. [DOI] [PMC free article] [PubMed]