Abstract
Our understanding of the virulence of Burkholderia cepacia complex lung infections in cystic fibrosis patients is incomplete. There is a great deal of variability in the clinical course, from simple colonization to severe and often fatal necrotizing pneumonia, termed cepacia syndrome. Multiple subspecies (called genomovars) have been identified, and these genomovars may hold the key to understanding the variable pathogenicity. Thirty-one B. cepacia complex isolates belonging to five of the seven genomovars were examined by using a gentamicin protection assay of invasion with A549 cells. The level of epithelial cell invasion by B. cepacia in the A549 model was relatively low compared with the data obtained for other pathogens and was often variable from assay to assay. Thus, a statistical approach was used to determine invasiveness. When this model was used, one of four genomovar I strains (25%), three of eight genomovar II strains (37.5%), seven of nine genomovar III strains (77.8%), one of four genomovar IV strains (25%), and none of the four genomovar V strains examined were defined as invasive. All other strains were categorized as either noninvasive or indeterminate. Invasive, noninvasive, and indeterminate isolates belonging to genomovars II and III were subsequently tested for splenic invasion with the mouse agar bead model. Correlation between the models for six strains was demonstrated. Our results indicate that a statistical model used to determine invasiveness in an in vitro invasion assay can be used to predict in vivo invasiveness.
The Burkholderia cepacia complex, originally identified as a phytopathogen, is now recognized as an important pulmonary pathogen in cystic fibrosis (CF) patients. This ubiquitous gram-negative rod-shaped bacterium infects up to 5% of people with CF, 20% of whom develop cepacia syndrome, which is frequently fatal (3, 6, 8). In addition to the overwhelming acute infection, chronic infection with some B. cepacia complex strains has been linked to increased morbidity, as reflected by decreased lung function and increased antibiotic usage (2). The wide variation in clinical presentation and disease severity, from transient colonization to necrotizing pneumonia and sepsis, reflects the complex nature of the bacterium-host relationship in CF in general and with B. cepacia specifically. It is therefore critically important to develop models for identification of strains that are associated with severe disease.
Identification of multiple genomic species of B. cepacia has resulted in a broad classification scheme that groups B. cepacia complex strains on the basis of genetic relatedness. There are currently seven genomic species, called genomovars, that have been identified by pulsed-field gel electrophoresis, whole-cell protein composition analysis, DNA polymorphism analysis, and DNA-DNA hybridization analysis (25). Four genomovars have the following species designations: genomovar II, Burkholderia multivorans; genomovar IV, Burkholderia stabilis; genomovar V, Burkholderia vietnamiensis; and genomovar VI, Burkholderia ambifaria. When large collections of B. cepacia complex clinical isolates have been analyzed to determine genetic relatedness, the majority of CF isolates have been found to be members of two genomovars (genomovar II [B. multivorans] and genomovar III) (J. LiPuma, personal communication). However, serial respiratory tract cultures from CF patients resulted in association of transient colonization with B. multivorans strains, while genomovar III strains were associated with severe and chronic infection (13). The overrepresentation of genomovars II and III among CF clinical isolates suggests that there may be genomovar-specific genetic factors that influence both acute and chronic infections.
Studies of the pathogenesis of B. cepacia have not identified specific markers that causally link specific strains or genomovars with disease severity in CF patients. Identification of such markers would aid in our understanding of this opportunistic and often invasive organism. Epidemiological investigation has determined that genomovar III strains can be highly transmissible (9, 12), and there is a clear association between interpatient spread and the presence of a genetic marker (B. cepacia epidemic strain marker [BCESM]) that may play a role in transcriptional regulation (11). Transmissibility has also been associated with expression of the cable pilus (21), which is encoded by cblA. Pilin-associated adhesins may be responsible for B. cepacia binding to respiratory mucins or epithelial cell surfaces (18, 19, 20). Both of these markers may be associated with CF lung infections, but a link between these markers and pathogenesis has yet to be established.
Other research has examined the interaction of B. cepacia with macrophages and has documented that inhibition of phagocytic function occurs in the presence of B. cepacia lipase activity (24), while the expression of ATP-dependent enzymes by B. cepacia clinical isolates has been shown to promote macrophage cell death, which allows the organisms to evade host defenses (5, 15). These characteristics have also been proposed to be potential virulence factors.
The virulence of B. cepacia has been investigated by using in vitro and in vivo models to evaluate a strain's potential to invade respiratory epithelial cells and to cause systemic infection, respectively. In vitro studies have documented that B. cepacia invades and replicates within respiratory epithelial cells (1, 14), that this organism persists in murine macrophages for up to 5 days (17), and that it invades and replicates within the free-living amoeba Acanthamoeba polyphaga (7). In vivo studies with C57/Black 6 mice have documented that persistent infection of the reticuloendothelial system occurs (23).
We utilized the A549 cell model (1) and a modified murine agar bead model to investigate the correlations among genomovar, genetic markers, and in vitro and in vivo invasion. Because of the wide variation in disease presentation in CF patients infected with different strains of B. cepacia, we were interested in addressing the following questions. (i) Which strains show evidence of being invasive? (ii) Which genomovars show evidence of being more invasive, as defined by the proportion of strains in a genomovar that show evidence of invasion in an in vitro model? (iii) Is in vitro invasiveness correlated with the ability to invade in an animal model? A number of well-characterized B. cepacia strains were assayed in vitro, and each strain was evaluated to determine its relative invasiveness compared with that of control strains. The association between invasiveness in the in vitro model and the presence of BCESM and cblA was also analyzed. Selected strains were evaluated with the in vivo model to investigate the correlation between in vitro invasiveness and splenic infection in the mouse agar bead model.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. cepacia isolates used in this study included organisms from the B. cepacia experimental panel (kindly provided by Eshwar Mahenthiralingam, University of Wales, Cardiff) (10). This panel consists of 30 strains that belong to genomovars I to V and were obtained from both environmental sources and CF and non-CF clinical sources (Table 1). The control strains were a genomovar III CF clinical isolate, K61-3 (kindly provided by Pam Sokol, University of Calgary, Calgary, Alberta, Canada), and 69NH, a spontaneous nonhemolytic mutant of a genomovar III clinical isolate, strain 69 (kindly provided by Mike Vasil, University of Colorado, Denver) (27). These control strains were selected previously based on an approximately 2-log difference in invasiveness in the in vitro model and differential splenic invasion with a mixed infection in the mouse agar bead model (data not shown).
TABLE 1.
Strains belonging to the B. cepacia complex experimental panel used
| Strain | No. of replicates | No. of days on which assays were performed | % Invasiona | Source | cblA | BCESM |
|---|---|---|---|---|---|---|
| Genomovar I strains | ||||||
| CEP509 | 18 | 6 | 1.243 (0.622) | CF | − | − |
| ATCC 25416 | 12 | 4 | 0.040 (0.690) | Onion | − | − |
| ATCC 17759 | 18 | 6 | 0.01 (0.010) | Soil | − | − |
| LMG17997 | 15 | 5 | 0.299 (0.622) | UTIb | − | − |
| Genomovar II (B. multivorans) strains | ||||||
| C5393 | 18 | 6 | 0.279 (0.341) | CF | − | − |
| C1576 | 18 | 6 | 3.208 (3.489) | CF | − | − |
| JTC | 21 | 7 | 0.052 (0.079) | CGDc | − | − |
| C1962 | 15 | 5 | 0.368 (0.553) | Abscess | − | − |
| 249-2 | 9 | 3 | 0.135 (0.196) | Laboratory | − | − |
| ATCC 17616 | 9 | 3 | 0.427 (0.354) | Soil | − | − |
| LMG13010 | 18 | 6 | 0.097 (0.115) | CF | − | − |
| CP-A1-1 | 12 | 4 | 0.38 (0.577) | CF | − | − |
| Genomovar III strains | ||||||
| J2315 | 18 | 6 | 0.271 (0.160) | CF | + | + |
| BC7 | 12 | 4 | 0.381 (0.582) | CF | + | + |
| K56-2 | 18 | 6 | 0.328 (0.304) | CF | + | + |
| C5424 | 12 | 4 | 0.903 (0.812) | CF | + | + |
| C1394 | 18 | 6 | 0.447 (0.590) | CF | − | + |
| PC184 | 9 | 3 | 1.722 (2.213) | CF | − | + |
| CEP511 | 18 | 6 | 3.613 (3.008) | CF | − | + |
| J415 | 18 | 6 | 1.861 (1.637) | CF | − | + |
| ATCC 17765 | 9 | 3 | 0.006 (0.010) | UTI | − | + |
| Genomovar IV (B. stabilis) strains | ||||||
| FC367 | 21 | 7 | 0.992 (1.326) | CF | − | − |
| C7322 | 15 | 5 | 0.098 (0.106) | CF | − | − |
| FC779 | 15 | 5 | 0.418 (0.700) | Infection | − | − |
| FC472 | 21 | 7 | 0.777 (1.561) | Ventilator | − | − |
| Genomovar V (B. vietnamiensis) strains | ||||||
| CEP40 | 18 | 6 | 1.928 (3.728) | CF | − | − |
| FC466 | 18 | 6 | 0.214 (0.278) | CF | − | − |
| FC441 | 15 | 5 | 0.195 (0.378) | CGD | − | − |
| FC369 | 18 | 6 | 0.003 (0.005) | Rice | − | − |
| Control strains | ||||||
| K61-3 | 42 | 14 | 1.081 (0.987) | |||
| 69NH | 78 | 25 | 0.025 (0.039) |
Mean (standard deviation) based on the results of three A549 cell invasion assays.
UTI, urinary tract infection.
CGD, chronic granulomatous disease.
All strains were maintained in 10% glycerol at −70°C. Prior to assays, bacteria were streaked from frozen stock preparations onto Luria-Bertani (LB) agar plates and incubated at 37°C for 24 to 48 h. Individual colonies were inoculated into LB media and grown with shaking to the mid-logarithmic phase (optical density at 600 nm, 0.5).
Epithelial cell invasion assay.
The A549 cells used for in vitro assays were grown in RPMI 1640 containing 1% glutamine and 10% fetal bovine serum at 37°C in the presence of 5% CO2 to obtain confluent monolayers in 24-well tissue culture plates. Cells were grown for 5 to 10 days in culture and used in modified gentamicin protection invasion assays performed in triplicate as previously described (1). The presence of a higher concentration of gentamicin (1 mg/ml) and addition of ceftazidime (500 μg/ml) were previously shown to result in significant killing of the control strains (1). This was confirmed with the strains tested in this study. Individual strains and genomovars were assayed on multiple days (Table 1). Each monolayer was infected with bacteria that had been grown to the mid-log phase and diluted in tissue culture medium by using inocula ranging from 5 × 104 to 5 × 106 bacteria per monolayer.
Statistical analysis methods.
Descriptive statistics were used to summarize cell invasion assay results in terms of the percent invasion for each strain, including the positive and negative control strains. The percent invasion value was calculated for each assay replicate as previously described (1).
To compare the degrees of invasiveness of strains, analysis of covariance models were used. The first analysis examined whether there was significant heterogeneity in the degree of invasion among strains belonging to a particular genomovar. The number of bacteria recovered (log10 CFU per milliliter) was modeled by using an analysis of covariance model to see if the number of bacteria recovered was significantly different for strains belonging to the same genomovar after we adjusted for the number of bacteria inoculated and day effects.
The next analysis examined which strains showed evidence of being invasive compared to the positive and negative controls (K61-3 and 69NH, respectively). The analysis was performed separately for each genomovar. The data for all strains belonging to a particular genomovar and the positive and negative control strains were included in the following model:
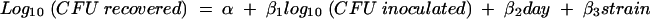 |
where day and strain were modeled as categorical variables. The model was then used to estimate the mean number of bacteria recovered for each strain, adjusted for the number of bacteria inoculated and day effects. Using these adjusted means, we made pairwise comparisons between each of the strains belonging to the genomovar of interest and the positive control strain and between each of the strains belonging to the genomovar of interest and the negative control strain. Comparisons were made by using a two-sample t test for the adjusted means derived from the model described above. The significance level of the tests was adjusted for multiple comparisons by using the Bonferroni correction method. If a strain's adjusted mean was significantly greater than that of the negative control and not significantly different from that of the positive control, the strain was defined as invasive. A strain whose adjusted mean was not significantly different from that of the negative control but was significantly less than that of the positive control was defined as noninvasive. An indeterminate strain was a strain whose adjusted mean was significantly greater than that of the negative control and significantly less than that of the positive control. The proportion of strains in each genomovar that showed evidence of being invasive according to the statistical model was then calculated.
Mouse agar bead model.
C57/Black 6 mice were infected with B. cepacia strains by using the agar bead model of lung infection (26) and were assayed for the presence of lung and splenic infections. Bacteria were incorporated into agar beads, and 105 CFU was inoculated into each mouse intratracheally. The animals were sacrificed 72 h postinfection, and the lungs and spleens were harvested, homogenized, and cultured on sheep blood agar and LB agar.
RESULTS
Invasiveness of strains in the in vitro model.
Experimental panel strains were evaluated by using A549 cell invasion assays (Table 1). Overall, there was great variability, with the mean percent invasion ranging from 0.003 to 3.613%. Using the statistical model described above, we analyzed the results of in vitro invasiveness experiments by comparing the panel strains and strains K61-3 (positive control) and 69NH (negative control) (Table 2). In this analysis, only one strain (FC369, a rice paddy isolate) was found to be significantly less invasive than the negative control (P < 0.0006), while no strain was found to be statistically more invasive than the positive control. Eight strains, including members of all five genomovars, were defined as noninvasive. Twelve strains belonging to genomovars I, II, III, and IV showed evidence of being invasive, and the remaining nine strains belonging to these genomovars were indeterminate, as their degrees of invasiveness were not statistically significantly different from the value obtained for either the negative control strain or the positive control strain.
TABLE 2.
Statistical model results for individual strains
| Strain | Genomovar |
P value
|
|
|---|---|---|---|
| Less invasive than positive control | More invasive than negative controla | ||
| Invasive strains | |||
| CEP509 | I | NSb | <0.00625 |
| C1962 | II | NS | <0.003125 |
| C1576 | II | NS | <0.003125 |
| ATCC 17616 | II | NS | <0.003125 |
| K56-2 | III | NS | <0.0025 |
| J2315 | III | NS | <0.0025 |
| C5424 | III | NS | <0.0025 |
| J415 | III | NS | <0.0025 |
| C1394 | III | NS | <0.0025 |
| PC184 | III | NS | <0.0025 |
| CEP511 | III | NS | <0.0025 |
| FC367 | IV | NS | <0.00625 |
| Noninvasive strains | |||
| ATCC 25416 | I | <0.00625 | NS |
| ATCC 17759 | I | <0.00625 | NS |
| 249-2 | II | <0.003125 | NS |
| CP-A1-1 | II | <0.003125 | NS |
| ATCC 17765 | III | <0.0025 | NS |
| C7322 | IV | <0.00625 | NS |
| FC369c | V | <0.00625 | NS |
| FC441 | V | <0.00625 | NS |
| Indeterminate strains | |||
| LMG17997 | I | <0.00625 | <0.00625 |
| JTC | II | <0.003125 | <0.003125 |
| C5393 | II | <0.003125 | <0.003125 |
| LMG13010 | II | <0.003125 | <0.003125 |
| BC7 | III | <0.0025 | <0.0025 |
| FC779 | IV | <0.00625 | <0.00625 |
| FC472 | IV | <0.00625 | <0.00625 |
| FC466 | V | <0.00625 | <0.00625 |
| CEP40 | V | <0.00625 | <0.00625 |
Except for strain FC369, all strains that were significantly different from the negative control were significantly more invasive than the negative control.
NS, Not significantly different from the control strain.
FC369 was the only strain that was significantly less invasive than the negative control.
Variability of invasiveness among genomovars in the in vitro model.
There was significant variability of invasiveness among strains belonging to a genomovar (P = 0.0001) after we adjusted for the number of bacteria inoculated and day effects, which were also significant (both P = 0.0001). Table 3 shows the percentage of invasive strains in each genomovar. Genomovar III strains appeared to be the most invasive, as we obtained evidence that seven of nine strains (78%) were invasive. In contrast, no invasive genomovar V strains were identified. Genomovar I, II, and IV strains, on the other hand, did not have uniformly invasive or noninvasive phenotypes.
TABLE 3.
Percentage of invasive strains by genomovar
| Genomovar | No. of invasive strains/no. of strains tested (%) | Clinical source(s) of invasive isolates |
|---|---|---|
| I | 1/4 (25) | CF |
| II | 3/8 (37.5) | Abscess, CF, soil |
| III | 7/9 (77.8) | CF |
| IV | 1/4 (25) | CF |
| V | 0/4 (0) | NAa |
NA, not applicable.
Correlation of invasiveness with expression of cblA and BCESM
Only genomovar III isolates from the experimental panel had cblA and BCESM (Table 1). All nine of the genomovar III strains had BCESM, and four of these strains also had cblA. Three of the four strains that had both potential virulence markers were defined as invasive in the in vitro model. Four of the remaining five isolates that had only BCESM were characterized as invasive.
Correlation between in vitro and in vivo models.
Animal experiments were performed by using one strain each of the genomovar II and III isolates that were characterized as invasive, noninvasive, and indeterminate in the A549 model. For these genomovars, strains that were invasive in the A549 model produced splenic infections in the mice, and strains that were not invasive generally did not (Table 4). Of the two strains that were defined as indeterminate in the in vitro model, the genomovar II strain appeared to be invasive in the mouse model and the genomovar III strain did not. This finding supports the use of an indeterminate category for the in vitro assay and indicates that further investigation of these strains is needed.
TABLE 4.
Recovery of B. cepacia from the spleens of C57/Black 6 mice infected in the agar bead lung infection model
| Genomovar | Results of A549 assay | Strain | No. of animals with splenic infection/total no. of animals |
|---|---|---|---|
| II | Invasive | C1576 | 4/4 |
| II | Indeterminate | C5393 | 4/5 |
| II | Noninvasive | 249-2 | 0/4 |
| III | Invasive | CEP511 | 4/4 |
| III | Indeterminate | BC7 | 0/4 |
| III | Noninvasive | FC663 | 1/3 |
DISCUSSION
In this study, in which in vitro and in vivo models of B. cepacia CF lung infection were compared, we examined a gentamicin protection assay and a murine agar bead model of CF lung infection. The in vitro results obtained with A549 cells showed that the percentages of invasion obtained for B. cepacia isolates were relatively low compared with the values obtained for other pathogens and that there was great variation in the abilities of the isolates to invade epithelial cells. This variation was observed both within a single genomovar and for multiple genomovars. Invasion by individual bacterial strains was also different on different assay dates, possibly because of variables that included differences between lots of growth media, subtle differences in the growth phases and numbers of the bacteria inoculated, and differences in epithelial monolayer maturity. Biological assay variation is difficult to quantify and has not been well described. When an organism has intrinsically low invasiveness, variability can dramatically affect the ability to demonstrate differences among strains. That is why a statistical model was used. The methods developed for this study account for day-to-day assay variability when the invasiveness of strains is examined with the in vitro model.
In the mouse model, invasion can be characterized in a qualitative, yes-or-no fashion on the basis of the ability of a strain to invade the spleen. However, in the A549 tissue culture model, invasion is not as clear-cut. In this model, invasiveness is characterized with a quantitative scale by determining the number of bacteria recovered from the cells in each well, which is dependent on the number of bacteria in the inoculum. Invasiveness is typically described by the percent invasion, which is calculated by determining the ratio of the number of bacteria inoculated to the number of bacteria recovered. In most previously described assays of bacterial invasion, experiments were standardized by using control strains selected for specific assays under defined conditions to help guide the interpretation of data sets. We propose that the best method to determine whether strains exhibit invasiveness in the in vitro model is an analysis which determines whether the degree of invasiveness is consistent with that of strains that are known to be qualitatively invasive or noninvasive in the mouse model (i.e., positive and negative control strains).
It has been shown that there can be significant variability in invasiveness among strains in the in vitro model; most of the variability is attributable to experimental conditions that vary from assay to assay or day to day. Because B. cepacia has the additional problem of low-level invasiveness (1), which is as much as 1 log lower than the invasiveness of Salmonella, Yersinia, and other well-characterized invasive pathogens (22), this variability may be quite problematic. The intrinsic variability needs to be accounted for when the consistency of the degree of invasiveness of a particular strain with that of a positive control is determined. One way to account for this variability and to objectively compare strains to positive and negative control strains is to use an analysis of variance model which models the number of bacteria recovered and adjusts for the number of bacteria inoculated and day effects. The adjusted means for strains can then be examined to see which strains produce data that are consistent with data for the positive control strain, the negative control strain, or neither control strain.
We used a systematic statistical method to analyze our biological assay data, which allowed us to draw some conclusions about the invasive phenotypes of clinical and environmental B. cepacia complex isolates belonging to multiple genomovars. This analysis yielded three categories for the strains tested on multiple assay dates: invasive, noninvasive, and indeterminate. Genomovar III was the most consistently invasive genomovar in vitro (seven of nine isolates [77.8%] were invasive), and one genomovar III strain was in the indeterminate group. The noninvasive genomovar III isolate, FC633, was a non-CF urinary tract isolate. Clinically, genomovar III strains cause between 50 and 80% of B. cepacia infections in CF patients (16). Genomovar II strains were the next most invasive strains, and the percentage of indeterminate genomovar II strains (37.5% [three of eight strains]) was higher than the percentages of indeterminate strains for the other genomovars. Genomovar II strains are found in between 15 and 40% of CF lung infections, and some of these strains have been associated with severe disease in outbreaks (4). Of the 12 strains belonging to the other three genomovars examined, 2 were in the invasive group (both were CF isolates) and 6 were in the indeterminate group. In summary, genomovar II and III strains accounted for 80% of the invasive strains, which correlates well with clinical reports. Within genomovar III, there was no correlation between relative invasiveness and either cblA or BCESM.
To begin to verify the potential for invasiveness for strains identified in the in vitro assay, splenic invasion was assessed with the murine agar bead model. Because the primary pathogens in CF lung infections are genomovar II and III strains, we focused on isolates belonging to these genomovars. The correlation was good for the six strains examined. The isolates that were defined as invasive based on the in vitro assay resulted in splenic infections, and the isolates that were characterized as noninvasive generally did not. The indeterminate strains sometimes invaded the spleen and sometimes did not. These results may also have been consistent with the sources of the isolates. C1576 was a genomovar II isolate from an outbreak in Great Britain that was associated with the cepacia syndrome (25, 28). Thus, despite the fact that this strain is a member of a genomovar that is thought to be less virulent, the finding that it is invasive in vitro and in vivo is not surprising. The noninvasive genomovar II isolate was a laboratory strain. Thus, it is unlikely that this strain would cause invasive clinical disease. Of the genomovar III isolates examined, the invasive isolate was a CF epidemic strain from Canada, as was the indeterminate strain. Interestingly, the indeterminate genomovar III strain, BC7, appeared to be noninvasive in the mouse model and had previously been reported to be noninvasive in the A549 cell assay (U. Sajjan, personal communication). The noninvasive genomovar III isolate was a urinary tract isolate and might be expected to be noninvasive in a lung model of infection.
There are some limitations of the methods used in this study. Many of the limitations of the in vitro assay were addressed by using the statistical model. The statistical model accounts for the variability and relatively low-level invasiveness shown by B. cepacia in the in vitro model. However, the statistical model is limited by the selection of control strains, since the definitions of invasive, noninvasive, and indeterminate strains are dependent on the control strains used. The controls used for our statistical analysis were selected specifically because of the difference in invasiveness in the murine model, as well as the consistent 100-fold difference in the A549 assay. If other controls were used, additional experiments would be needed to determine which categories strains fall into.
Using a statistical analysis of in vitro invasion assay data, we demonstrated that there are distinct differences in relative invasiveness between individual isolates and between different genomovars. These differences correlated with the results obtained with the mouse agar bead model and, to some extent, with the clinical sources of the isolates. The statistical model which we developed is a useful tool that allows more objective identification of invasion in vitro. The present study is an important contribution to the study of the pathogenesis of B. cepacia, an organism that exhibits low-level invasiveness. Because specific virulence factors of B. cepacia have not been clearly linked to CF lung disease, the ability to utilize an in vitro model rather than an animal model is invaluable. The in vitro model should permit screening of libraries and examination of mutants for invasiveness without the sacrifice of large numbers of animals, since the statistical model permits comparisons of strains that exhibit relatively low-level and variable invasiveness in vitro. Once screening assays are performed, the mouse agar bead model with quantitation of splenic invasion may be useful for confirming the in vitro results and for testing clones in the indeterminate category. Ongoing studies are examining the use of this model for characterization of the effect of type III secretion on the virulence of B. cepacia.
Acknowledgments
This work was supported by Public Health Service grant 5T32 HD07233 from the National Institutes of Health (to M.V.C.) and by an award from the Cystic Fibrosis Foundation (to J.L.B.).
REFERENCES
- 1.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frangolias, D. D., E. Mahenthiralingam, S. Rae, J. M. Raboud, A. G. Davidson, R. Wittmann, and P. G. Wilcox. 2000. Burkholderia cepacia in cystic fibrosis. Variable disease course. Am. J. Respir. Crit. Care Med. 160:1572-1577. [DOI] [PubMed] [Google Scholar]
- 3.Glass, S., and J. R. Govan. 1986. Pseudomonas cepacia—fatal pulmonary infection in a patient with cystic fibrosis. J. Infect. 13:157-158. [DOI] [PubMed] [Google Scholar]
- 4.Govan, J. R., J. E. Hughes and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-397. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson, M. L., I. R. Poxton, and J. R. W. Govan. 1998. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect. Immun. 66:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 7.Landers, P., K. G. Kerr, T. J. Rowbotham, J. L. Tipper, P. M. Keig, E. Ingham, and M. Denton. 2000. Survival and growth of Burkholderia cepacia within the free-living amoeba Acanthamoeba polyphaga. Eur. J. Clin. Microbiol. Infect. Dis. 19:121-123. [DOI] [PubMed] [Google Scholar]
- 8.LiPuma, J. J. 1998. Burkholderia cepacia. Management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 9.LiPuma, J. J., S. E. Dasen, D. W. Nielson, R. C. Stern, and T. L. Stull. 1990. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet 336:1094-1096. [DOI] [PubMed] [Google Scholar]
- 10.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahenthiralingam, E., P. Vandamme, M. E. Campbell, D. A. Henry, A. M. Gravelle, L. T. Wong, A. G. Davidson, P. G. Wilcox, B. Nakielna, and D. P. Speert. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469-1475. [DOI] [PubMed] [Google Scholar]
- 14.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnikov, A., O. Zaborina, N. Dhiman, B. S. Prabhakar, A. M. Chakrabarty, and W. Hendrickson. 2000. Clinical and environmental isolates of Burkholderia cepacia exhibit differential cytotoxicity towards macrophages and mast cells. Mol. Microbiol. 36:1481-1493. [DOI] [PubMed] [Google Scholar]
- 16.Pitt, T. L., M. E. Kaufmann, P. S. Patel, L. C. Benge, S. Gaskin, and D. M. Livermore. 1996. Type characterization and antibiotic susceptibility of Burkholderia cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J. Med. Microbiol. 44:103-110. [DOI] [PubMed] [Google Scholar]
- 17.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 18.Sajjan, U. S., and J. F. Forstner. 1993. Role of a 22-kilodalton pilin protein in binding of Pseudomonas cepacia to buccal epithelial cells. Infect. Immun. 61:3157-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sajjan, U. S., and J. F. Forstner. 1992. Identification of the mucin-binding adhesin of Pseudomonas cepacia isolated from patients with cystic fibrosis. Infect. Immun. 60:1434-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sajjan, U. S., M. Corey, M. A. Karmali, and J. F. Forstner. 1992. Binding of Pseudomonas cepacia to normal human intestinal mucin and respiratory mucin from patients with cystic fibrosis. J. Clin. Investig. 89:648-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sajjan, U. S., F. A. Sylvester, and J. F. Forstner. 2000. Cable-piliated Burkholderia cepacia binds to cytokeratin 13 of epithelial cells. Infect. Immun. 68:1787-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Small, P. L. C., R. R. Isberg, and S. Falkow. 1987. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within Hep-2 cells. Infect. Immun. 55:1674-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speert, D. P., B. Steen, K. Halsey, and E. Kwan. 1999. A murine model for infection with Burkholderia cepacia with sustained persistence in the spleen. Infect. Immun. 67:4027-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straus, D. C., M. K. Lonon, and J. C. Hutson. 1992. Inhibition of rat alveolar macrophage phagocytic function by a Pseudomonas cepacia lipase. J. Med. Microbiol. 37:335-340. [DOI] [PubMed] [Google Scholar]
- 25.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 26.van Heeckeren, A., R. Walenga, M. W. Konstan, T. Bonfield, P. B. Davis, and T. Ferkol. 1997. Excessive inflammatory response of cystic fibrosis mice to bronchopulmonary infection with Pseudomonas aeruginosa. J. Clin. Investig. 100:2810-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasil, M. L., D. P. Krieg, J. S. Kuhns, J. W. Ogle, V. D. Shortridge, R. M. Ostroff, and A. I. Vasil. 1990. Molecular analysis of hemolytic and phospholipase C activities of Pseudomonas cepacia. Infect. Immun. 58:4020-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whiteford, M. L., J. D. Wilkinson, J. H. McColl, F. M. Conton, J. R. Michie, T. J. Evans, and J. Y. Paton. 1995. Outcome of Burkholderia (Pseudomonas) cepacia colonization in children with cystic fibrosis following a hospital outbreak. Thorax 50:1194-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]


