Abstract
A comparative study was performed to determine the effects of pH, osmolarity, and human urine on the transcription of several fim genes, as well as the overall expression of type 1 pili. Several fim-lacZYA fusions were constructed on single-copy plasmids to test a range of pHs and a range of osmolarities. Growth in acidic medium slightly reduced expression from all of the fim promoters (fimA, fimB, and fimE). Increased osmolarity in neutral-pH medium repressed fimA and fimB transcription by approximately 50% when 400 mM NaCl was used and nearly threefold when 800 mM NaCl was used, whereas fimE transcription rose slightly as the osmolarity increased. This effect was more pronounced in high-osmolarity acidic media; fimB and fimA expression decreased fivefold in growth media containing 800 mM NaCl compared to expression in growth media without added NaCl. Moreover, fimE expression doubled under the same high-osmolarity conditions compared to expression in a low-osmolarity acidic environment. When a fimB-lacZ or fimE-lacZ fusion was inserted into the chromosome of strain AAEC189, fimE expression changed slightly as the osmolarity increased, but fimB expression decreased by 50% in a low-pH high-osmolarity environment. When strain AAEC189 with either a plasmid-borne fimB-lacZ fusion or a plasmid-borne fimE-lacZ fusion was grown in human urine, similar changes in the levels of fimB and fimE expression were observed. Limiting-dilution reverse transcription-PCR confirmed that these changes in fim expression occurred in clinical isolates of uropathogenic Escherichia coli grown in media with different pHs and different osmolarities. Furthermore, the invertible switch region in uropathogenic strain NU149 shifted from favoring the phase-on position in a neutral-pH low-osmolarity environment to favoring the phase-off position in a low-pH high-osmolarity environment. Results obtained with an ompR mutant strain demonstrated that fimB expression was derepressed and that OmpR may neutralize repression by an acid response regulator of fimE expression in a low-pH environment. In addition, H-NS was verified to be important in regulation of fimB, but it had only a slight effect on fimE under the specific pH and osmotic growth conditions tested. Enzyme immunoassays with anti-type 1 pilus antibody and hemagglutination assays showed that fewer type 1 pili were detected with cells in a low-pH high-osmolarity environment. Together, these observations demonstrate that a combination of low pH and high osmolarity regulates the transcription of fim genes, which favors a shift in the invertible element to the phase-off orientation and a loss of type 1 pilus expression. Taken together, our data suggest that the environmental cues that we tested may regulate expression of type 1 pili in specific in vivo niches, such as murine kidneys and possibly human kidneys.
Uropathogenic Escherichia coli (UPEC) strains are the most frequent cause of urinary tract infections in humans. These bacteria are able to sense a variety of environmental cues that can include differences in temperature, carbon and amino acid sources, pH, osmolarity, and amount of oxygen, as well as other stimuli. The signals that these bacteria collect in various environments help them regulate their gene expression. An important niche for UPEC strains is the human urinary tract awash with urine. Human urine can promote the growth of UPEC bacteria, but it is a poor source of iron, it can have extreme fluctuations in pH, the osmolarity may vary considerably, and it contains high levels of organic acids (39). The ability of these bacteria to grow in the human urinary tract or the most common animal model, the murine urinary tract, has clinical significance. The osmolalities in human urine can range from 0.038 to 1.4 mol/kg (58), although the osmolalities in murine urine can reach 3 mol/kg (43, 61). Moreover, the osmolality of urine in the kidneys exceeds the osmolality of urine held in the bladder (58). Strains of UPEC can grow in medium that contains up to 0.7 M NaCl (>1.4 mol/kg) (37), but the concentration tolerated increases to 0.9 M when glycine betaine is present (9) and to 1.2 M when Trypticase soy broth or urine (39) is the growth medium. In addition to osmotic variations, the pH of human urine can vary between 5.0 and 8.0, depending on physiological constraints, the diet of the individual, and what type of microorganism has colonized the urinary tract (39, 58). Kidney urine typically has a lower pH than bladder urine because of the dilution effect in the bladder (58).
Bacterial colonization and growth in a urinary tract bathed with urine are important in the disease process. Adherence to uroepithelial cells is a critical step in the ability of bacteria to cause infections. A variety of fimbriae that allow the bacteria to mediate this adherence are expressed by UPEC strains (34). One prevalent variety of fimbriae produced by UPEC strains is the type 1 fimbriae that recognize mannose receptors. More than 85% of E. coli strains possess the genetic information for type 1 pilus expression (8, 51), and more than 70% of E. coli strains express type 1 pili on their surfaces (37).
Several fim genes are involved in the production of type 1 pili. Expression of type 1 pili is mediated by a process called phase variation, where the bacteria can switch between piliated and nonpiliated states (18). Phase variation of type 1 pili is controlled by a site-specific recombination system that determines the orientation of a 314-bp invertible DNA element (1). This invertible element contains the promoter for the fimA structural gene and is flanked by inverted repeats. Two recombinases are responsible for inversion of the invertible element; these enzymes, FimB and FimE (38, 45, 46), belong to the Int class of recombinases (13, 20). The FimB recombinase facilitates switching from phase-off to phase-on, as well as switching from phase-on to phase-off, whereas previously FimE was thought to cause only switching from phase-on to phase-off. Recent studies have suggested that FimE may naturally initiate limited switching from phase-off to phase-on (67) or that this may be possible following amino acid substitution (65). Other cofactors, including integration host factor (13, 20), leucine-responsive protein (6, 23, 57), and the histone-like protein H-NS (11, 35, 53), are also involved in the switching mechanism.
Previous studies on the regulation of type 1 pilus expression have included testing the modulating effect of temperature (7, 14, 23, 54), static broth growth compared to agar growth (17, 23, 31, 52, 63, 64), stationary phase growth (16), glucose effects (19), and the addition of aliphatic amino acids (24, 57) but not pH; the effect of osmolarity has been studied only to a limited degree (41). Phenotypic studies have also been performed with urea (49) and urine (40, 49). There has been no previous examination of fim gene regulation in growth media in which pH fluctuations have been combined with osmotic differences.
In this study, we attempted to answer the following three questions. Do pH, osmolarity, and human urine affect the transcription of key fim genes? If transcriptional regulation does occur, does this regulation affect the position of the invertible element? And do H-NS and OmpR contribute to this regulation under these specific growth conditions? We obtained evidence that pH and osmotic changes in growth media have minor effects on fim gene expression when they are examined individually. However, a low pH combined with a high osmolarity appears to have a synergistic effect manifested by significant repression of fimA and fimB transcription, biasing the invertible element in the phase-off position and resulting in a loss of type 1 pili on the surface of the bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table 1. E. coli strain J96 (28) was provided by Richard Hull, Baylor University. Strains NU149 (30), NU14, NU2-25, NU2-38, and NU2-39 (31) were provided by James Duncan, Northwestern University. The Δfim operon Δlac operon strain AAEC189 (5) was supplied by Ian Blomfield, Wake Forest University. Transformations were initially done in E. coli strain DH5α (Gibco/BRL, Gaithersburg, Md.), and subsequent β-galactosidase assays were performed with this strain, as well as with strain AAEC189. The unmarked ompR mutant strain MH1160 (26) and wild-type strain MC4100 were gifts from Linda Kenney, Oregon Health Sciences University. The hns mutant strain RR1 bglY and parental strain RR1 were provided by Staffan Normark, Karolinska Institute. Plasmids pPR274, pPP2-6, pBB2-1, pP5-48, pAON-1, and pUTE1 were supplied by James Duncan. The pUJ8 and pUT-Tc plasmids (10) were gifts from Kenneth Timmis. All of the wild-type E. coli strains were grown in Luria-Bertani (LB) broth at 37°C or were passaged on LB agar plates incubated at 37°C. For recombinant E. coli strains, antibiotics (all obtained from Sigma Chemical Co., St. Louis, Mo.) were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 12.5 μg/ml; and tetracycline, 10 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| J96 | Clinical isolate | 28 |
| NU149 | Clinical isolate | 30 |
| NU14 | Clinical isolate | 31 |
| NU2-25 | Clinical isolate | 31 |
| NU2-38 | Clinical isolate | 31 |
| NU2-39 | Clinical isolate | 31 |
| AAEC189 | YMC9 Δfim operon Δlac recA endA | 5 |
| DH5α | supE44 ΔlacU169(lacZΔM15) hsdR17 endA1 gyrA96 thi-1 relA1 | Gibco/BRL |
| MC4100 | F− Δ(lac)U169 araD139 rpsL relA thiA flbB | Linda Kenney |
| MH1160 | MC4100 ompB101 (ompR101) | Linda Kenney |
| RR1 | supE44 lacY1 hsdS20 ara-19 proA2 galK2 rpsL20 xyl-5 mtl-1 | Staffan Normark |
| RR1 bglY | RR1 bglY (hns) | Staffan Normark |
| AAEC189-4fimB | AAEC189 chromosomal fimB-lacZ integration | This study |
| AAEC189-2fimE | AAEC189 chromosomal fimE-lacZ integration | This study |
| Plasmids | ||
| pPR274 | Mini-F replicon Cmr | 48 |
| pPP2-6 | pPR274 with MCSa | James Duncan (Brenda Bloom) |
| pBB2-1 | fimA-lacZYA on pPR274 | James Duncan (Magaret Pearle) |
| pUTE1 | fimE-lacZYA on pUT-Tc | James Duncan (Margaret Pearle) |
| pP5-48 | fimB-lacZYA on pUJ8 | James Duncan (Margaret Pearle) |
| pAON-1 | fimA-lacZYA locked on on pUJ8 | James Duncan (Margaret Pearle) |
| pUJ8 | trp′-′lacZ phoA Apr | 10 |
| pUT-Tc | Mini-Tn5 on pUT Tcr | 10 |
| pJB5A | fimB-lacZYA on pPP2-6 | This study |
| pJLE4-3 | fimE-lacZYA on pPP2-6 | This study |
| pUTE4 | fimB-lacZYA on pUT-Tc | James Duncan (Margaret Pearle) |
| pWS124-17 | fimA-lacZYA locked on on pPP2-6 | This study |
Multiple cloning sites.
Construction of the fim-lacZYA fusions.
The fimA-lacZYA fusion on single-copy plasmid pPR274 was constructed by Brenda Bloom (Northwestern University) by using the pORN129 plasmid, which contains a lacZYA operon fused to the promoter of fimA (55). The entire fim operon containing this fusion was ligated to the single-copy pPR274 plasmid, creating pBB2-1, and was transformed into E. coli strain DH5α. Plasmid DNA from this strain was isolated with a commercial kit (Qiagen, Valencia, Calif.) and was used to transform E. coli strain AAEC189 (59). Transformants were selected on LB agar containing chloramphenicol. Transformants were confirmed by plasmid isolation and restriction endonuclease digestion (59). Since plasmid pBB2-1 has a functional invertible element and intact fimB and fimE genes, the invertible element can flip from the phase-on position to the phase-off position and vice versa. Transcription of fimA fused to lacZYA reflects the position of the invertible element.
For construction of the fimB promoter-containing plasmid, we used the pP5-48 plasmid generated by Margaret Pearle, Northwestern University, as a template. Plasmid DNA was isolated from DH5α/pP5-48 cells as described above and was digested with restriction endonuclease NotI. The proper fragment was separated on a low-melting-point agarose gel and processed as previously described (64). This fragment contained the fimB promoter linked to a promoterless lacZ gene from pUJ8. Using this fimB promoter fragment, we ligated (62) this fragment and NotI-digested pPP2-6, a single-copy plasmid derived from pPR274 that contains a multiple cloning site (48). The ligated DNAs were used to transform DH5α cells, and the transformants were selected on LB agar plates containing chloramphenicol and were screened for the fimB-lacZ fusion by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The colonies that were Cmr and blue were processed further. After confirmatory tests were performed, plasmid pJB5A (fimB-lacZ) was used for the remaining tests. DNA from pJB5A was used to transform E. coli strain AAEC189.
To create the fimE-lacZ fusion on a single-copy plasmid, the pUTE1 plasmid constructed by Margaret Pearle was used. This plasmid has a fimE-lacZ fusion inserted into a mini-Tn5 transposon on a multicopy plasmid. Plasmid DNA was isolated from DH5α/pUTE1 cells and cut with NotI. The correct DNA fragment was isolated and processed as desribed above. This DNA fragment contained the fimE promoter linked to a promoterless lacZ gene from pUJ8. With this DNA fragment, ligation to NotI-digested pPP2-6 DNA was carried out, and then DH5α cells were transformed with the ligated material. Transformants were selected on LB agar containing chloramphenicol and X-Gal. Any blue colonies that arose on the plates were processed further. From this screening process plasmid pJLE4-3 (fimE-lacZYA) was selected and used for the remaining tests. DNA from pJLE4-3 was used to transform E. coli strain AAEC189.
Construction of a recombinant plasmid with the promoter locked in the phase-on position began with plasmid DNA from pAON-1 generated by Margaret Pearle. Plasmid DNA was isolated as described above, digested with NotI, and ligated to NotI-cut pPP2-6 DNA. DNA was transformed into DH5α cells, and transformants were selected for and screened as described above. From this screening process plasmid pWS124-17 (fimA-lacZ locked in the phase-on position) was selected and used for the remaining tests. DNA from pWS124-17 was used to transform E. coli strain AAEC189.
The fimB-lacZ and fimE-lacZ fusions integrated into the chromosome of strain AAEC189 were constructed as follows. Plasmid pP5-48 (fimB-lacZ) DNA was digested with NotI and ligated to NotI-digested pUT-Tc (10) DNA. The resulting plasmid, pUTE4, was transformed into S17.1λpir cells. Then S17.1λpir/pUTE4 cells and AAEC189 cells were conjugated. Transconjugants that were tetracycline resistant and blue on X-Gal-containing media were selected. One of these transconjugants, AAEC189-4fimB, was used to test environmental conditions. To construct the fimE-lacZ fusion in the genome of strain AAEC189, plasmid DNA from pUTE1 was processed as the fimB-lacZ fusion was processed. The resulting plasmid, pUTE4, was transformed into S17.1λpir cells and then mated with strain AAEC189 cells. From this conjugation strain AAEC189-2fimE was chosen and used for further analysis.
Environmental conditions.
To obtain variations in pH in vitro, the pH of LB medium was adjusted by using 0.1 M Na2HPO4-NaH2PO4 buffer combined with 1% (vol/vol) glycerol. We prepared a series of LB media with pHs between 5.0 and 8.0 at 0.5-pH unit increments, and the pH values were confirmed with a pH meter (Orion Research). To each aliquot of LB medium 100 μl of a culture of AAEC189 carrying fim-lacZ recombinant plasmids was added. The cultures were incubated overnight at 37°C with shaking. The next day the pHs of the cultures were checked, and a 100-μl aliquot of each culture was transferred to another portion of the same LB medium. The resulting cultures were then incubated with shaking at 37°C, β-galactosidase assays were performed, and the final pHs were determined.
The osmolarity of Luria broth was adjusted by adding NaCl to final concentrations of 100, 200, 400, and 800 mM. The incubation conditions were the same as those described above. Combinations of different osmolarities and different pHs were also tested by using pHs 5.5 and 7.0 with the same NaCl concentrations.
Bacterial cultures grown in LB medium supplemented with human urine or in pure human urine were also tested. Clean-catch midstream urine samples were collected at different times from a healthy male, and the pH was monitored with a pH meter. Aliquots of the urine were plated onto brain heart infusion agar (Difco) to check for sterility. The urine was added to pH 5.5 LB medium at concentrations of 10, 20, 40, and 80%. Recombinant bacterial cultures were inoculated into the broth media and incubated as described above. To test pure human urine, the pHs of aliquots of fresh human urine obtained from one urine specimen were adjusted to 5.5, 6.0, 6.5, and 7.0 by using the sodium phosphate buffer mentioned above without added glycerol. The urine samples having different pHs were each divided into two portions in order to perform tests on two separate days.
β-Galactosidase assays.
The β-galactosidase activity of mid-logarithmic-phase bacteria permeabilized with sodium dodecyl sulfate and CHCl3 was determined by using the method of Miller (47). Measurements were obtained after 20 min of exposure to the substrate. Most assays were performed at least three times on different days, and the data were expressed as means ± standard deviations based on the values obtained; the only exception was the analysis of the recombinant strains in pure human urine, for which only two assays were performed on separate days.
Extraction of total RNAs and conversion to cDNAs.
Total RNAs were extracted from NU149 cells grown in pH 7.0 low-osmolarity LB medium, in pH 5.5 low-osmolarity LB medium, and in pH 5.5 high-osmolarity (400 mM NaCl) LB medium by using the hot phenol extraction procedure utilized previously (63). The cDNAs used for PCR amplification were each synthesized from 6 μg of total RNA as previously described (62) by using the random hexamer primer from a reverse transcription (RT)-PCR kit (Stratagene, La Jolla, Calif.).
Limiting-dilution PCR analyses.
Chromosomal DNAs from E. coli strain NU149 grown in pH 7.0 LB medium containing no added NaCl, in pH 5.5 LB medium containing no added NaCl, and in pH 5.5 LB medium containing 400 mM NaCl were extracted by using a commercial DNA extraction kit (AGTC, Gaithersburg, Md.). The DNA concentrations were standardized, and the preparations were used for PCR amplification as described by Schwan et al. (63) with the INV-FIMA primer pair to amplify the phase-on orientation of the invertible element and the FIME-INV primer pair to amplify the phase-off orientation of the invertible element. The chromosomal DNAs were each serially twofold diluted in double-distilled H2O to a dilution of 1/256, and an aliquot of each dilution was then amplified. PCR amplification was performed at least three times with at least two separate chromosomal DNA preparations for each type of growth conditions.
LD-RT-PCR analyses.
Limiting-dilution RT-PCR (LD-RT-PCR) amplification was performed as previously described (66), with the following modifications. A Perkin-Elmer 2400 thermocycler was used under the following PCR conditions: initial denaturation at 94°C for 5 min; 33 cycles consisting of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and elongation at 72°C for 1 min; and 10 min of elongation at 72°C after the last cycle. The following primers were used: FimB1 (5′ CGAATCACTCCTTAAAGCAGC 3′), FimB2 (5′ GGCGTAACATGTGCGGATGAA 3′), FimE1 (5′ CTGTTGGCATATCGGCATGGG 3′), FimE2 (5′ TCGATGCCCGAGATAATCCTGA 3′), EcFtsZ1 (5′ TAGCGGTATCACCAAAGGACT 3′), and EcFtsZ2 (5′ GTGATCAGAGAGTTCACATGCT 3′). The fimB and fimE primers were derived from the sequence described by Klemm (38), and the ftsZ primer pair was derived from the E. coli genome sequencing project (4). LD-RT-PCR amplification was performed at least three times with cDNAs derived from at least two separate total RNA extractions for each type of culture conditions tested.
EIAs.
Bacterial strains were transferred to the appropriate broth cultures and incubated overnight at 37°C with shaking. Then each of the strains was passaged a second time in the same medium overnight at 37°C with shaking. We used agar-grown cultures from serially agar-passaged cultures that have been shown previously to become nonpiliated (31). For all cultures, the optical density at 600 nm was adjusted to 0.800, and 500-μl aliquots of each culture were transferred to 1.5-ml microcentrifuge tubes. The enzyme immunoassay (EIA) procedure described by Hultgren et al. (29) was used. EIAs were performed at least twice for each type of conditions tested, and the values given below are means.
HA assays.
Hemagglutination (HA) assays were performed for each culture as previously described (31). These assays were repeated three or four times.
Statistics.
Student's t test was used for statistical analyses. P values of ≤0.05 were considered significant.
RESULTS
Examination of the fim-lacZYA fusions at different pHs.
To address the question of whether pH affects the transcription of fim genes, several fim-lacZYA fusions were generated. All of the fim-lacZYA fusions (fimA-, fimB-, and fimE-lacZYA) were successfully cloned onto single-copy plasmids pPR274 and pPP2-6 and then transformed into E. coli strains DH5α and AAEC189 (Δfim operon). Single-copy plasmid pPR274 has been used previously for this purpose (63). Strain AAEC189 was chosen because it lacks the fim and lac genes.
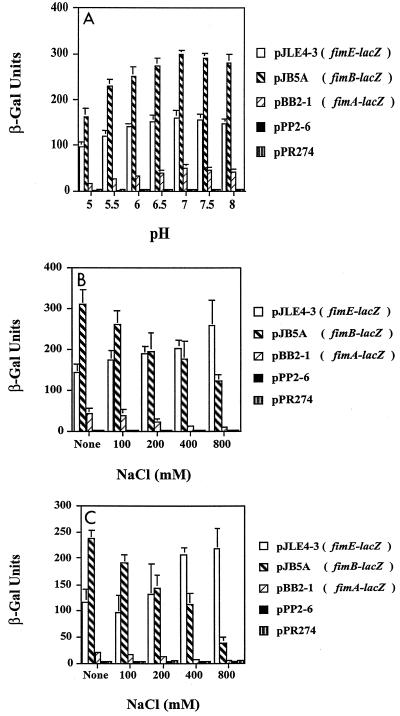
First, the pH of LB medium was adjusted to 5.0 to 8.0 by using NaPO4 buffering and glycerol to maintain the pH. The resulting media were inoculated with strain AAEC189 containing the recombinant single-copy plasmids, and the β-galactosidase activities of mid-logarithmic-phase cells were determined. The pH of each medium was measured before and after growth to ensure that the pH did not change. We found that the pH before growth and the pH after growth differed by no more than 0.04 unit (data not shown). The optimal pH for expression from all of the fim promoters was found to be pH 7.0 (fimA, 50 Miller units; fimB, 300 Miller units; fimE, 161 Miller units), but the values varied less than threefold over the pH range (Fig. 1A). Incremental increases at pHs greater than 7.0 resulted in slight decreases in activity for all three fim promoters, which were not significant. Moreover, a shift to an acidic pH resulted in decreases in activity for all three promoters, with the greatest decrease occurring as the pH fell to 5.0 (fimA, 16 Miller units; fimB, 162 Miller units; fimE, 95 Miller units). For all three promoters, these changes were significant (fimA, P < 0.003; fimB, P < 0.0003; fimE, P < 0.0004). However, the cultures grown at pH 5.0 grew poorly. The pH 5.5 cultures were able to reach almost the same turbidity as the pH 7.0 cultures. The β-galactosidase activities were significantly lower at pH 5.5 than at pH 7.0 (fimA, 25 Miller units [P < 0.008]; fimB, 162 Miller units [P < 0.002]; fimE, 95 Miller units [P < 0.03]). These results indicate that pH alone had a minor effect on expression of the fim promoters.
FIG. 1.
Effects of pH and osmolarity on fimA, fimB, and fimE expression as determined with lacZYA transcriptional fusions in strain AAEC189. The β-galactosidase (β-Gal) activity (expressed in Miller units) was determined after 20 min; means ± standard deviations are indicated. Osmolarity effects were tested by using NaCl as the osmolyte. (A) Different pHs; (B) different osmolarities in pH 7.0 media; (C) different osmlarities in pH 5.5 media.
Examination of the fim-lacZYA fusions under different osmotic conditions.
Another growth factor that was tested with all of the fim-lacZYA fusions in strain AAEC189 was osmolarity. In pH 7.0 medium, the highest osmolarity resulted in reduced expression from the fimA promoter (P < 0.01) and the fimB promoter (P < 0.006) in the AAEC189 background (Fig. 1B). Surprisingly, the level of fimE expression in strain AAEC189 increased slightly less than twofold (similar results were obtained for strain DH5α [data not shown]), which was significant (P < 0.0007). Even with added 400 mM NaCl the fimA promoter (P < 0.009) and fimB promoter (P < 0.009) activities were significantly lower than the activities observed in LB medium without added NaCl. Moreover, the fimE activity was significantly higher (P < 0.0007) in LB medium containing 400 mM NaCl.
Effects of pH and osmotic conditions together on expression of the fim genes.
As described above, changing the pH or osmolarity had modest effects on expression of the fim genes. Because there are environments in which fluctuations in both pH and osmolarity can occur, such as the human or murine urinary tract, both pH and osmolarity were changed in some experiments. At pH 5.5, the NaCl concentration was changed to values up to 800 mM. In strain AAEC189, the β-galactosidase activity for fimB (238 Miller units) was approximately twice the β-galactosidase activity for fimE (116 Miller units) under low-osmolarity conditions (Fig. 1C). When the NaCl concentration was increased by 400 mM (1.2 mol/kg), fimB expression decreased significantly to 112 Miller units (P < 0.0006) and fimE expression increased to 208 Miller units (P < 0.003). When the NaCl concentration was increased to 800 mM (1.8 mol/kg), fimB expression decreased considerably (39 Miller units) (P < 0.00003), whereas fimE expression nearly doubled (218 Miller units) (P < 0.01) compared to the values obtained at pH 7.0 under low-osmolarity conditions. The level of fimA activity decreased almost to the background level in medium containing 400 mM NaCl and to the background level (5 Miller units) under very-high-osmolarity conditions (P < 0.0004). Similar changes were observed for the fim-lacZYA fusions in strain DH5α (data not shown). These results suggest that a low-pH high-osmolarity environment may favor expression of fimE.
Chromosomally integrated fimB-lacZ and fimE-lacZ fusion variants of AAEC189 were also tested under different pH and osmolarity conditions. Strain AAEC189-2fimE grown at pH 7.0 under low-osmolarity conditions had an enzyme activity of 72.6 Miller units; the activity increased slightly to 73.4 Miller units when the NaCl concentration was increased to 400 mM (Table 2). When the pH was 5.5, the activity was 56.6 Miller units, but the activity increased to 62 Miller units when the osmolarity was increased. There was no statistically significnt variation in β-galactosidase expression in AAEC189 cells grown under these conditions. However, when AAEC189-4fimB was used, there was a significant difference in expression of β-galactosidase between cultures grown at pH 7.0 (90.6 Miller units) and cultures grown at pH 7.0 with high osmolarity (65.6 Miller units) (P < 0.05). Growth in pH 5.5 medium resulted in 58 Miller units (P < 0.008), and there was an additional decrease in expression in pH 5.5 high-osmolarity medium (49.6 Miller units) (P < 0.002).
TABLE 2.
Effects of pH and osmolarity on chromosomal fimB-lacZ and fimE-lacZ fusions in strain AAEC189
| Strain | Fusion | pH | Added NaCl concn (mM) | β-Galactosidase activity (Miller units) |
|---|---|---|---|---|
| AAEC189-2fimE | fimE-lacZ | 7.0 | 0 | 72.6 ± 7.4 |
| AAEC189-2fimE | fimE-lacZ | 7.0 | 400 | 73.4 ± 9.5 |
| AAEC189-2fimE | fimE-lacZ | 5.5 | 0 | 56.6 ± 14.3 |
| AAEC189-2fimE | fimE-lacZ | 5.5 | 400 | 62.2 ± 7.0 |
| AAEC189-4fimB | fimB-lacZ | 7.0 | 0 | 90.6 ± 14.7 |
| AAEC189-4fimB | fimB-lacZ | 7.0 | 400 | 65.6 ± 11.1 |
| AAEC189-4fimB | fimB-lacZ | 5.5 | 0 | 58.0 ± 6.7 |
| AAEC189-4fimB | fimB-lacZ | 5.5 | 400 | 49.6 ± 5.7 |
Examination of fimA promoter activity in a phase-on orientation in bacteria grown in media at different pHs under different osmotic conditions.
Expression of fimA in a low-pH high-osmolarity environment could be affected either directly by something that represses fimA promoter activity or indirectly by something that affects the position of the invertible element that contains the fimA promoter. To assess whether repression of the fimA promoter was directly responsible for less fimA expression, a construct was made on a single-copy plasmid containing the fimA promoter with the invertible element locked in the phase-on position (pWS124-17), which prevented any switching. When recombinant AAEC189 cells with the pWS124-17 plasmid were grown in pH 7.0 medium with no added NaCl (201 Miller units) and in pH 7.0 medium with 400 mM added NaCl (high osmolarity) (209 Miller units), the β-galactosidase activities were not significantly different statistically. When the pH was 5.5, the fimA activity (169 Miller units) was slightly lower than the activity at pH 7.0 (201 Miller units) in low-osmolarity media, but the difference was not significant. In media containing 400 mM NaCl, the levels of β-galactosidase activity did not differ significantly when pH 5.5 medium (202 Miller units) was compared to pH 7.0 medium (209 Miller units). This level of fimA expression was substantially higher than the levels obtained with the pBB2-1 plasmid, which had a functional invertible element and intact fimB and fimE genes.
Urine-containing media and pure human urine affect fim::lacZ expression.
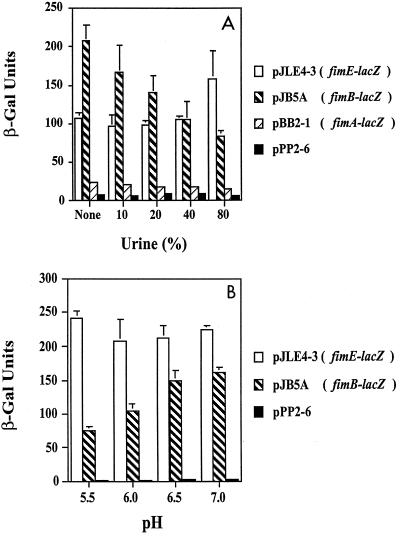
Since the fim gene constructs were derived from the J96 UPEC strain (29), we wanted to determine whether growth in human urine could affect expression of the fimA, fimB, and fimE promoters. To elucidate whether human urine itself could have an effect, recombinant AAEC189 bacteria were initially grown in LB media adjusted to pH 5.5 containing different concentrations of filter-sterilized normal male urine; the final pHs were 5.68 to 5.76, and the osmolalities were between 0.78 and 0.90 mol/kg.
For fimA expression, the levels of β-galactosidase activity were 22.5 Miller units without urine and 14.5 Miller units in the presence of 80% urine (P < 0.09) (Fig. 2A). When the strain was grown in 100% urine, the level of activity was 9 Miller units (data not shown). The results showed that fimB expression decreased approximately twofold (P < 0.0004) when the concentration of urine in the medium was increased to 80% and that fimE expression was constant until 80% urine was present; an approximately 50% increase in fimE expression occurred in the presence of 80% urine (Fig. 2A) (P < 0.04). To further assess the effects of human urine on expression of the fimB and fimE genes, the pH of filtered human urine was adjusted to various values with sodium phosphate buffer, and then the urine was inoculated with strain AAEC189 containing either pJB5A (fimB-lacZYA fusion) or pJLE4-3 (fimE-lacZYA fusion). β-Galactosidase assays were performed, and the results showed that the fimB levels decreased approximately twofold when the pH was changed from 7.0 to 5.5, whereas the fimE levels remained constant at all pH values (Fig. 2B). These analyses showed that human urine can have effects on fim expression similar to those of low-pH high-osmolarity LB medium.
FIG. 2.
Effects of growth in human urine on fimA, fimB, and fimE expression as determined by using lacZYA transcriptional fusions in strain AAEC189. (A) Different percentages of human urine mixed with LB medium; (B) different pHs of human urine, adjusted with sodium phosphate buffer. The β-galactosidase (β-Gal) activity is expressed in Miller units; means ± standard deviations are indicated.
Examination of fim transcript levels in a clinical isolate grown under the same environmental conditions
The fim-lacZYA fusion data suggested that transcription of fimA, transcription of fimB, and transcription of fimE were affected by growth in a low-pH high-osmolarity environment. To further examine these possibilities, wild-type clinical isolate NU149 was grown in LB media prepared so that the pH was either 5.5 or 7.0 and the osmolarity was either low (no added NaCl) or high (400 mM NaCl). Total RNAs from bacterial cells grown under these conditions were isolated. Using fimB- and fimE-specific oligonucleotide primers, we performed LD-RT-PCR. The level of fimB transcripts in strain NU149 decreased twofold in pH 5.5 low-osmolarity medium and then another fourfold to a barely detectable level in low-pH high-osmolarity medium (Fig. 3). Although the level of fimE transcripts also decreased about twofold in a low-pH low-osmolarity growth environment, the level of fimE transcripts increased in cells grown under low-pH high-osmolarity conditions. Oligonucleotide primers specific for ftsZ, encoding a cell division protein, were used to standardize the results obtained under the different conditions. The results validated the fim-lacZ fusion results, showing that low-pH high-osmolarity conditions favor expression of fimE over expression of fimB.
FIG. 3.
Quantitative determination of mRNA regulation by LD-RT-PCR analysis of cDNAs of strain NU149 cells grown in pH 7.0 LB medium with low osmolarity, in pH 5.5 LB medium with low osmolarity, and in pH 5.5 LB medium with high osmolarity. The FimB1-FimB2, FimE1-FimE2, and EcFtsZ1-EcFtsZ2 primer pairs were used to amplify serially twofold diluted cDNAs and targeted fimB (379-bp product), fimE (392-bp product), and ftsZ (302-bp product) transcripts, respectively. All PCR products were electrophoresed on 1.5% agarose gels. The folowing dilutions of cDNAs were used: undiluted (lanes 1), 1/2 (lanes 2), 1/4 (lanes 3), 1/8 (lanes 4), 1/16 (lanes 5), 1/32 (lanes 6), 1/64 (lanes 7), 1/128 (lanes 8), and 1/256 (lanes 9).
Measurement of invertible element switching.
Growth of the clinical isolate in a low-pH high-osmolarity environment showed that there was a distinct shift that favored transcription of fimE over transcription of fimB. Since FimE is responsible for switching the invertible element from the phase-on position to the phase-off position, we predicted that the position of the invertible element would shift accordingly. To determine whether the position of the invertible element did shift in the low-pH high-osmolarity environment compared to the neutral-pH low-osmolarity environment, PCR amplification was performed by using chromosomal DNAs extracted from NU149 cells grown in pH 7.0 LB medium with no added NaCl (low osmolarity), in pH 5.5 LB medium with no added NaCl (low osmolarity), and in pH 5.5 LB medium containing 400 mM NaCl (high osmolarity). These DNA populations were serially twofold diluted and PCR amplified. Oligonucleotide primers specific for the phase-on and phase-off orientations of the invertible element were used (63). The orientation of the invertible element containing the fimA promoter changed markedly when the growth medium was switched from pH 7.0 low-osmolarity medium to pH 5.5 high-osmolarity (400 mM NaCl) medium. There was a twofold decrease in phase-on positioning when the medium was switched from pH 7.0 low-osmolarity medium to pH 5.5 low-osmolarity medium, and there was an additional fourfold decline when the medium was switched to pH 5.5 high-osmolarity medium (Fig. 4B). Phase-off positioning also changed, increasing fourfold when pH 5.5 low-osmolarity medium was compared to pH 7.0 low-osmolarity medium and increasing an additional fourfold in pH 5.5 high-osmolarity medium.
FIG. 4.
Determination of the invertible element orientation in strain NU149 grown under different environmental conditions by PCR analysis. The PCR analysis was performed with chromosomal DNA isolated from NU149 cells grown in pH 7.0 LB medium with no added NaCl (low osmolarity), in pH 5.5 LB medium with no added NaCl (low osmolarity), and in pH 5.5 LB medium with 400 mM NaCl (high osmolarity), using the INV and FIMA primers to amplify phase-on-oriented DNA (450-bp product) and the FIME and INV primers to amplify phase-off-oriented DNA (750-bp product). The following dilutions were used for the PCR: undiluted (lanes 1), 1/2 (lanes2), 1/4 (lanes 3), 1/8 (lanes 4), 1/16 (lanes 5), 1/32 (lanes 6), 1/64 (lanes 7), 1/128 (lanes 8), and 1/256 (lanes 9). All PCR products were electrophoresed on 1.5% agarose gels.
Examination of expression of type 1 pili under different environmental conditions.
The transcriptional analyses described above suggested that neutral-pH low-osmolarity conditions favor fimB expression and concomitant expression of type 1 pili, whereas under low-pH high-osmolarity conditions transcription of fimE is favored. Furthermore, the shift of the invertible element to the phase-off position also suggested that type 1 pilus expression is shut down. To ascertain if the results of the transcriptional studies and switching of the invertible element were reflected at the protein level, a series of EIAs were performed with polyclonal anti-149 type 1 pilus antibody and UPEC strains NU149 and J96. The NU149 strain was chosen because of the extensive transcriptional analyses and type 1 pilus detection assays that have been performed with it (63), and the J96 strain (28) was chosen because the fim gene promoters used in the β-galactosidase assays described above were derived from sequences of this strain (63). EIAs showed that there were high levels of expression of type 1 pili in both E. coli strains at pH 7.0 under low-osmolarity conditions and that there were slight declines in the levels of type 1 pili as the osmolarities of the broth cultures increased (Fig. 5A). Agar-grown cells were used as negative controls because both of the strains used have been shown by electron microscopy to be nonpiliated after five serial passages on Luria agar (unpublished observations). A more marked decline in type 1 pilus expression was observed when the pH was 5.5 and the osmolarity was adjusted. At very high osmolarity (800 mM), type 1 pilus production decreased in both strains to levels that were near the level in nonpiliated agar-grown cells (Fig. 5B) (NU149, P < 0.00009; J96, P < 0.002).
FIG. 5.
EIA analyses of strains NU149 and J96 grown in media with different osmolarities. Optical densities at 492 nm (O.D.492) were determined, and means ± standard deviations are indicated. a, agar grown; None, no NaCl added. (A) Bacteria grown in pH 7.0 media; (B) bacteria grown in pH 5.5 media.
Additional UPEC strains obtained from the Northwestern University collection (31) were then tested with EIAs, as well as with the more classical HA assays. All of the strains were grown at either pH 7.0 or pH 5.5 under low-osmolarity conditions (no additional NaCl) or high-osmolarity conditions (400 mM NaCl). Agar-grown cells of strains NU149 and J96 were used as negative controls in these assays. As described previously (31), all of the strains except NU14 became nonpiliated after five serial passages on agar, as determined by the HA assays (Table 3). Strain NU14, a phase-variable strain of E. coli that expresses type 1 pili when it is grown on agar (31), had an HA titer. The base levels for the EIAs were also set by using the measurements obtained with the agar-grown cells. Again, all of the agar-grown strains except NU14 had background levels that indicated that they were nonpiliated, whereas NU14 exhibited a low level of type 1 pili on the surfaces of the cells. When the bacteria were passaged in broth media at pH 7.0 under low-osmolarity conditions, both the HA titer (range, 128 to 1,024) and the EIA values (range, 1.88 to 2.76) were high for all of the strains. An increase in osmolarity resulted in slight declines in the HA titer (range, 64 to 256) and the EIA value (range, 0.89 to 2.05) for all of the strains. A shift to pH 5.5 had an even more dramatic effect on type 1 pilus expression. With no added NaCl, the EIA values ranged from 0.63 to 1.77, and there was a similar decrease in the HA titers (range, 12 to 96). Even more striking were the results obtained after the NaCl concentration was increased to 400 mM. The EIA values were 0.24 to 0.86, and the HA titers varied from 48 to “±” (weak reaction in the first well). Strain NU2-39 had barely detectable levels of type 1 pili, as shown by the low EIA value (0.24) and HA titer (±) for this strain. These findings demonstrate that the pH and osmolarity of the growth medium also affect expression of type 1 pili at the protein level.
TABLE 3.
EIAs and HA assays with type 1 piliated bacteria grown in pH 7.0 and 5.5 broth media under low- or high-osmolarity conditions
| Strain | Agar
|
pH 7.0 broth
|
pH 5.5 broth
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No NaCl added
|
400 mM NaCl added
|
No NaCl added
|
400 mM NaCl added
|
|||||||||
| EIAa | HA assayb | EIA | HA assay | EIA | HA assay | EIA | HA assay | EIA | HA assay | |||
| NU149 | 0.15 ± 0.01 | 0 | 2.56 ± 0.23 | 512 | 1.84 ± 0.16 | 128 | 1.40 ± 0.01 | 64 | 0.60 ± 0.04 | 24 | ||
| J96 | 0.16 ± 0.02 | 0 | 2.76 ± 0.18 | 1,024 | 2.03 ± 0.08 | 256 | 1.77 ± 0.52 | 96 | 0.86 ± 0.06 | 48 | ||
| NU14 | 0.67 ± 0.01 | 32 | 2.52 ± 0.17 | 512 | 1.81 ± 0.27 | 196 | 1.47 ± 0.20 | 96 | 0.76 ± 0.06 | 48 | ||
| NU2-25 | 0.16 ± 0.01 | 0 | 2.11 ± 0.15 | 196 | 1.68 ± 0.09 | 128 | 1.60 ± 0.12 | 64 | 0.51 ± 0.04 | 16 | ||
| NU2-38 | 0.22 ± 0.01 | 0 | 2.33 ± 0.16 | 512 | 2.05 ± 0.06 | 196 | 1.60 ± 0.08 | 64 | 0.57 ± 0.06 | 24 | ||
| NU2-39 | 0.17 ± 0.02 | 0 | 1.88 ± 0.05 | 128 | 0.89 ± 0.04 | 64 | 0.63 ± 0.07 | 12 | 0.24 ± 0.14 | ±c | ||
EIA data are expressed in optical density at 492 nm; the values are means ± standard deviations based on at least two separate experiments.
The HA assay data are average HA titers based on at least three separate experiments.
±, weak reaction; the first well showed some agglutination.
Role of hns and ompR in expression of fimB and fimE genes in bacteria grown in low-pH high-osmolarity media.
Low-pH high-osmolarity growth conditions favored fimE expression, so we wondered what mechanism or mechanisms are responsible for the changes in type 1 pilus expression. Previous work has shown that the nucleoid-associated protein H-NS can repress fimB expression (12), so fimB expression was tested by using an hns mutant strain and media that had different pH values and different osmolarities. hns mutant strain RR1 bglY containing either the fimB-lacZ recombinant plasmid or the fimE-lacZ recombinant plasmid was grown in LB medium with or without 400 mM NaCl at pH 5.5 or 7.0 and compared to wild-type strain RR1 grown under the same conditions. Derepression of fimB expression occurred in the hns mutant background under all growth conditions tested (Table 4). In pH 7.0 low-osmolarity growth medium, the levels of fimB expression were 352 Miller units for the wild-type strain and 453 Miller units for the hns mutant. More interesting was the difference observed in pH 7.0 high-osmolarity medium, in which there was more than a twofold difference in fimB expression (307 versus 698 Miller units). Although the hns mutation resulted in an increase in overall fimB expression, the level of fimB expression when cells were grown in pH 5.5 medium containing 400 mM NaCl (486 Miller units) was slightly lower than the level of fimB expression when cells were grown in pH 5.5 medium without added NaCl (534 Miller units); the difference was similar to the difference observed for wild-type strain RR1 (258 versus 236 Miller units). At pH 5.5 there was a significant twofold difference in fimB expression in the hns background in low-osmolarity medium (P < 0.008), as well as high-osmolarity medium (P < 0.0002), compared to growth in the wild-type background. Both the RR1 strain and the RR1 bglY strain transformed with the pPP2-6 plasmid exhibited mean levels of β-galactosidase activity of 4 to 8 Miller units under the growth conditions tested, showing that there was minimal background β-galactosidase activity in these strains containing the plasmid backbone (data not shown).
TABLE 4.
Effect of an hns mutation on fimB and fimE expression compared to expression in a wild-type strain
| Strain | Plasmid | pH | Added NaCl concn (mM) | β-Galactosidase activity (Miller units) |
|---|---|---|---|---|
| RR1 | pJB5A (fimB-lacZ) | 5.5 | 0 | 258 ± 30 |
| RR1 | pJB5A | 5.5 | 400 | 236 ± 26 |
| RR1 | pJB5A | 7.0 | 0 | 352 ± 12 |
| RR1 | pJB5A | 7.0 | 400 | 307 ± 90 |
| RR1 bglY (hns) | pJB5A | 5.5 | 0 | 534 ± 18 |
| RR1 bglY | pJB5A | 5.5 | 400 | 486 ± 15 |
| RR1 bglY | pJB5A | 7.0 | 0 | 453 ± 123 |
| RR1 bglY | pJB5A | 7.0 | 400 | 698 ± 156 |
| RR1 | pJLE4-3 (fimE-lacZ) | 5.5 | 0 | 139 ± 13 |
| RR1 | pJLE4-3 | 5.5 | 400 | 176 ± 26 |
| RR1 | pJLE4-3 | 7.0 | 0 | 294 ± 40 |
| RR1 | pJLE4-3 | 7.0 | 400 | 318 ± 37 |
| RR1 bglY (hns) | pJLE4-3 | 5.5 | 0 | 151 ± 27 |
| RR1 bglY | pJLE4-3 | 5.5 | 400 | 190 ± 13 |
| RR1 bglY | pJLE4-3 | 7.0 | 0 | 313 ± 34 |
| RR1 bglY | pJLE4-3 | 7.0 | 400 | 336 ± 27 |
The effect of the hns mutation on fimE expression was more subtle. In pH 7.0 low-osmolarity medium, the level of fimE expression was slightly higher in the hns mutant strain than in the wild type (245 Miller units for the wild type and 303 Miller units for the hns mutant) (P < 0.02) (Table 4). When the NaCl concentration in pH 7.0 medium was increased to 400 mM, slight increases in the levels of fimE expression were also observed (308 Miller units for the wild type and 342 Miller units for the hns mutant); these increases were not significant (P < 0.06). A decrease in the pH to 5.5 resulted in no significant difference in fimE expression when wild-type bacteria were compared to the hns mutant regardless of the osmolarity.
Osmotic changes in the environment affect supercoiling of the DNA, and they also trigger the EnvZ-OmpR two-component regulatory system in E. coli. To ascertain if the EnvZ-OmpR system is also involved somehow in osmotic regulation of fimB and fimE, an unmarked ompR mutant (26) was transformed with the recombinant fimB-lacZ and fimE-lacZ plasmids and grown in the media described above. Derepression of fimB expression occurred in the ompR mutant strain grown in all of the media tested (Table 5). In the pH 7.0 low-osmolarity medium fimB expression changed from 208 Miller units in the wild-type strain to 283 Miller units in the ompR mutant (P < 0.03). When the NaCl concentration in pH 7.0 medium was increased by 400 mM, the wild-type strain expressed 163 Miller units of activity from the fimB promoter, whereas the ompR mutant expressed 252 Miller units (P < 0.03). As observed for the hns mutant strain, pH had a slight effect on fimB expression in the ompR mutant strain grown at pH 5.5. When the pH of low-osmolarity medium was 5.5, the fimB-lacZ plasmid in wild-type bacteria expressed 119 Miller units of activity, compared to the 151 Miller units of activity observed for the ompR mutant (P < 0.03). After growth in pH 5.5 high-osmolarity medium the wild-type strain exhibited threefold less fimB activity (103 Miller units) than the ompR mutant strain (312 Miller units) (P < 0.0007).
TABLE 5.
Effect of an ompR mutation on fimB and fimE expression compared to expression in a wild-type strain
| Strain | Plasmid | pH | Added NaCl concn (mM) | β-Galactosidase activity (Miller units) |
|---|---|---|---|---|
| MC4100 | pJB5A (fimB-lacZ) | 5.5 | 0 | 119 ± 22 |
| MC4100 | pJB5A | 5.5 | 400 | 103 ± 6 |
| MC4100 | pJB5A | 7.0 | 0 | 208 ± 36 |
| MC4100 | pJB5A | 7.0 | 400 | 163 ± 30 |
| MH1160 (ompR) | pJB5A | 5.5 | 0 | 151 ± 12 |
| MH1160 | pJB5A | 5.5 | 400 | 312 ± 50 |
| MH1160 | pJB5A | 7.0 | 0 | 283 ± 53 |
| MH1160 | pJB5A | 7.0 | 400 | 252 ± 64 |
| MC4100 | pJLE4-3 (fimE-lacZ) | 5.5 | 0 | 101 ± 23 |
| MC4100 | pJLE4-3 | 5.5 | 400 | 158 ± 21 |
| MC4100 | pJLE4-3 | 7.0 | 0 | 148 ± 21 |
| MC4100 | pJLE4-3 | 7.0 | 400 | 195 ± 42 |
| MH1160 (ompR) | pJLE4-3 | 5.5 | 0 | 126 ± 18 |
| MH1160 | pJLE4-3 | 5.5 | 400 | 124 ± 30 |
| MH1160 | pJLE4-3 | 7.0 | 0 | 212 ± 30 |
| MH1160 | pJLE4-3 | 7.0 | 400 | 213 ± 57 |
The results of the analysis with the fimE-lacZ plasmid were unclear. Growth in pH 7.0 medium resulted in a slight increase in fimE expression in the wild-type strain from 148 to 195 Miller units (P < 0.23) as the osmolarity increased (Table 5). No increase was observed for the ompR mutant strain (212 versus 213 Miller units). Even in pH 5.5 medium, fimE expression in the wild-type background increased from 101 to 158 Miller units (P < 0.03) when the osmolarity increased, but the levels of fimE expression in the ompR mutant background were not significantly different after growth in low-osmolarity medium (126 Miller units) and growth in high-osmolarity medium (124 Miller units). In the ompR mutant background, there was no osmotic activation of fimE expression compared to the wild-type bacteria. Both the MC4100 strain and the MH1160 strain transformed with pPP2-6 exhibited 2 to 4 Miller units of β-galactosidase activity under the growth conditions tested, indicating that there was minimal background β-galactosidase activity when the plasmid backbone was used (data not shown).
DISCUSSION
In this study, we examined how pH, osmolarity, and human urine affect transcription of several fim genes involved in type 1 pilus expression in UPEC. We found that an environment with a low pH combined with moderate to high osmolarity, whether it is LB medium or human urine, not only affects transcription of the fim genes but also alters the position of the invertible element containing the fimA promoter and ultimately the amount of type 1 pili expressed on the surfaces of the UPEC cells. Both H-NS and OmpR may play roles in transcriptional regulation of the fim genes in a low-pH high-osmolarity environment. Our results suggest that regulation of fim gene transcription is multifaceted and depends on the environmental stimuli received by the UPEC cells.
Transcription of all of the fim genes tested (fimA, fimB, and fimE) was maximal at pH 7.0. Alkaline environments had a slight repressing effect on transcription of these fim genes, like the effect observed for the E. coli pap and fan operons (72). A pH of 5.5 or lower also reduced transcription of all of the fim promoters compared to transcription in pH 7.0 medium. In a recent study White-Ziegler et al. (72) demonstrated that growth in pH 5.5 media downregulated pap, daa, and fan pilus operons found in E. coli, suggesting that an acid response global regulator may be involved. The acid tolerance response systems in E. coli include a σs-cyclic AMP receptor-protein system, a glutamate-gamma-aminobutyric acid antiporter, and an arginine-dependent system that requires arginine decarboxylase (22). One or more of these systems may repress transcription of the fim genes. Besides limiting transcription of the fim genes, low-pH environments, particularly human urine (2, 36), may also inhibit the growth of E. coli, and this could help explain the poor growth results obtained for cultures grown at pH 5.0.
Osmolarity was another environmental factor which we examined. Expression of fimA and expression of fimB were maximal in pH 7.0 LB medium without added NaCl, whereas fimE expression was maximal in pH 7.0 LB medium containing 800 mM NaCl. This indicated that single parameters, such as osmotic conditions and pH, had relatively minor effects on transcription of the fim genes. On the other hand, a combination of low pH and moderate to high osmolarity had a profound synergistic effect on fim gene expression, the position of the invertible element, and ultimately expression of type 1 pili on the surfaces of the bacteria. The level of fimB transcription in pH 5.5 medium with 400 mM NaCl was considerably lower than the level of fimB transcription in pH 7.0 medium with low osmolarity, as demonstrated with the fimB-lacZ fusion assays and LD-RT-PCR analysis. Differences between the LD-RT-PCR analysis results and the fim-lacZ reporter fusion analysis results may have been due to the fact that reporter fusions measured transcription over a defined period of time and to the fact that the results did not take into account RNA stability and accumulation that was measured by the LD-RT-PCR. Nevertheless, similar trends were observed when we compared the LD-RT-PCR results to the β-galactosidase activity results for bacteria grown in the various LB media. A low-pH high-osmolarity environment caused fimE levels to rise and fimB levels to decline. The results obtained with fimB-lacZ and fimE-lacZ fusions integrated into the chromosome of strain AAEC189 supported the single-copy plasmid results, indicating that the use of single-copy plasmids can solve some of the problems in interpretation encountered when multicopy plasmids are used (45, 46).
Since FimB and FimE affect the position of the 314-bp invertible element containing the fimA promoter (45, 46), we predicted that this would have an effect on fimA transcription. Indeed, fimA transcription appeared to be affected based on a shift in the invertible element to the phase-off position in low-pH high-osmolarity conditions rather than on a direct effect on the fimA promoter itself. Ultimately, this led to a decrease in type 1 pilus expression on the bacterial cells as determined by the EIA.
Using different in vitro conditions provided clues about what was happening, but we wanted to know what relevance the results might have for in vivo growth and survival. Experiments performed with urine specimens collected from a single source (pH, around 5.5; osmolarity, 250 to 300 mM [0.8 to 0.9 mol/kg]) substantiated the changes in transcription of the key fim genes when the results were compared to the results obtained with LB media whose pH values and osmolarities were adjusted to various values. Although the use of a single source of urine was a limitation in this study, we found that urine from other individuals did not support the growth of UPEC strains, possibly due to inhibitory agents in the urine samples (data not shown). Thus, there are also inherent dangers in using pooled urine from many individuals. Nonetheless, the bacteria displayed results similar to those obtained with LB medium.
The UPEC strains in the human intestinal tract and in patients with urinary tract infections have been shown to originate from E. coli strains of fecal origin (34). Unsupplemented LB medium has an osmolality similar to the osmolality of the small intestines of mammals (21, 72). Piliated bacteria enter the urethra and move into the bladder, sometimes ascending to the kidneys of an infected host. We used LB medium with 400 mM NaCl (1.2 mol/kg) as the high-osmolarity medium for most of our studies, and this osmolarity was in the normal physiological range for urine in the collecting ducts and Henle's loop within the renal medulla of the human kidney (58) and certainly was in the normal physiological range for the murine kidney (43). Even 800 mM NaCl (1.6 mol/kg) is moderate osmolarity in murine kidneys, in which an osmolality of 3 mol/kg can be reached (43, 61). However, in the human urinary tract, an NaCl concentration of 800 mM is encountered only in a severely dehydrated person on a specific diet, so we performed no further analysis after the initial test under these very high-osmolarity conditions. Instead, we used 400 mM as the highest osmolarity in most of this study.
Osmolarity may affect two systems, the OmpR-EnvZ and H-NS systems. Hyperosmotic conditions trigger the EnvZ protein, an inner membrane histidine kinase osmosensor, which in turn phosphorylates the cytoplasmic DNA-binding protein OmpR (33). At a high osmolarity, there is an increase in the level of phosphorylated OmpR, which in turn binds to DNA sequences to either activate or repress transcription (56). We found that an ompR mutant exhibited threefold derepression of fimB expression in low-pH high-osmolarity growth medium compared to fimB expression in neutral-pH low-osmolarity medium (Table 5); however, fimE expression in the ompR mutant background was not as clear-cut. Wild-type bacteria grown in pH 7.0 low-osmolarity medium had virtually the same β-galactosidase activity with a fimE-lacZ fusion as bacteria grown in pH 5.5 high-osmolarity medium. This result contrasted with the significantly lower value obtained with cells grown in a pH 5.5 low-osmolarity environment. The divergent effects of OmpR (repressing fimB and possibly neutralizing the repressive effect of an acid response regulator on fimE) favor fimE expression. Based on our data, it appears that FimB is more favored in a neutral-pH low-osmolarity environment and that FimE predominates in a low-pH high-osmolarity environment.
An increase in osmolarity also increases DNA supercoiling (27), providing more curvature to the DNA and more template for the H-NS histone-like DNA-binding protein to bind to (3). The H-NS protein, encoded by the hns gene, is involved in global regulatory events (27, 32, 44). The hns or osmZ gene is allelic to bglY, pilG, and virR (44). Mutations in the hns (pilG) gene affect type 1 pilus expression by causing a higher rate of inversion of the invertible element (35, 66), and fimB transcription is repressed by H-NS (11, 12, 15, 50). Our results support the hypothesis that fimB is derepressed in an hns mutant strain, but there was only a slight effect on fimE expression. Strain differences could explain variations in fimE expression.
Observations made in this study may also help explain murine models of infection that have shown that many E. coli strains become nonpiliated after 5 days in the mouse kidney but remain piliated in the mouse bladder (30, 60). Human or murine kidneys are bathed in urine with higher osmolarity (up to 1.4 mol/kg in humans and 3 mol/kg in mice) and slightly lower pH than the urine in the bladder (43, 58, 61). Moreover, the abundance of mannose receptors in the bladder (51, 71) and the paucity of such receptors in the kidneys (69, 70) may have an effect on expression of both fimB and fimE.
Our hierarchical model for type 1 pilus regulation involves several mechanisms. Within a human or murine kidney, one or more acid adaptation systems and an increase in DNA supercoiling may be triggered in a low-pH high-osmolarity environment, leading to an acid response protein and more H-NS binding to the supercoiled fimB promoter DNA, repressing fimB expression. The EnvZ-OmpR osmolarity sensing system may also be activated, causing either phosphorylated OmpR or an intermediary to bind and repress fimB transcription. If this happens, the level of fimB plummets, so there is limited switching from the phase-off position to the phase-on position. In turn, an acid response system triggers downregulation of fimE that may be countered by slight fimE transcriptional activation by phosphorylated OmpR binding directly to fimE or OmpR acting through an intermediary to activate fimE transcription. More fimE transcription combined with a drastic reduction in the level of fimB transcripts switches the invertible element from the phase-on position to the phase-off position. In this series of events, the invertible element may eventually be locked in the phase-off position. Transcription of fimA would be severely reduced or aborted in this environment, and a nonpiliated phenotype would predominate in time in the kidney.
In the human or murine bladder, an abundance of mannose receptors may circumvent some of the regulatory changes described above. Some recent work has shown that binding of type 1 piliated bacteria to their mannose receptors triggers downregulation of fimE and slight activation of fimB transcription (W. R. Schwan, J. Pinkner, S. J. Hultgren, and M. T. Beck, unpublished data). Urine is slightly less acidic in the bladder than in the kidneys, and the osmolality in the bladder is lower than the osmolality in the kidneys. More FimB and less FimE in the bacteria in the bladder mean that more of the invertible element is in the phase-on position. Recent studies have shown that strains of UPEC from patients are more apt to have the invertible element in the phase-on position when the strains colonize the mouse bladder than when they are grown in vitro (25, 42, 68). Ultimately, fim-reporter fusions in UPEC strains will need to be tested in vivo to ascertain if the observations described above truly reflect what occurs in the murine or human urinary tract.
Acknowledgments
We thank Kenneth Timmis, James Duncan, Margaret Pearle, Hank Seifert, and Brenda Bloom for constructing and/or providing plasmids used in this study. We also thank Richard Hull, Ian Blomfield, Linda Kenney, Staffan Normark, and James Duncan for providing strains that were used in this study.
This work was supported in part by an undergraduate research grant from University of Wisconsin-La Crosse to F.A.L. and by a University of Wisconsin-La Crosse faculty research grant to W.R.S.
REFERENCES
- 1.Abraham, J. M., C. S. Freitag, J. R. Clements, and B. I. Eisenstein. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:5724-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asscher, A. W., M. Sussman, W. E. Waters, R. H. Davis, and S. Chick. 1966. Urine as a medium for bacterial growth. Lancet ii:1037-1041. [DOI] [PubMed]
- 3.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Blomfield, I. C., M. S. McClain, J. A. Princ, P. J. Calie, and B. I. Eisenstein. 1991. Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J. Bacteriol. 173:5298-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield, I. C., P. J. Calie, K. J. Eberhardt, M. S. McClain, and B. I. Eisenstein. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinton, C. C. 1959. Non-flagellar appendages of bacteria. Nature 183:782-786. [DOI] [PubMed] [Google Scholar]
- 8.Buchanan, K., S. Falkow, R. A. Hull, and S. I. Hull. 1985. Frequency among Enterobacteriaceae of the DNA sequences encoding type 1 pili. J. Bacteriol. 162:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers, S., and C. M. Kunin. 1985. The osmoprotective properties of urine for bacteria: the protective effect of betaine and human urine against low pH and high concentrations of electrolytes, sugars, and urea. J. Infect. Dis. 152:1308-1316. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donato, G. M., and T. H. Kawula. 1999. Phenotypic analysis of random hns mutations differentiate DNA-binding activity from properties of fimA promoter inversion modulation and bacterial motility. J. Bacteriol. 181:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donato, G. M., M. J. Lelivelt, and T. H. Kawula. 1997. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol. 179:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman, C. J., and C. F. Higgins. 1987. Fimbrial phase variation in Escherichia coli: dependence of integration host factor and homologies with other site-specific recombinases. J. Bacteriol. 169:3840-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman, C. J., and N. N. Bhriain. 1992. Thermal regulation of fimA, the Escherichia coli gene coding for the type 1 fimbrial subunit protein. FEMS Microbiol. Lett. 99:125-130. [DOI] [PubMed] [Google Scholar]
- 15.Dove, S. L., and C. J. Dorman. 1994. The site-specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Mol. Microbiol. 14:975-988. [DOI] [PubMed] [Google Scholar]
- 16.Dove, S. L., S. G. J. Smith, and C. J. Dorman. 1997. Control of Escherichia coli type 1 fimbrial gene expression in stationary phase: a negative role for RpoS. Mol. Gen. Genet. 254:13-20. [DOI] [PubMed] [Google Scholar]
- 17.Duguid, J. P., W. Smith, G. Dempster, and P. N. Edmunds. 1955. Non-flagellar filamentous appendages (‘fimbriae’) and haemagglutinating activity in Bacterium coli. J. Pathol. Bacteriol. 70:335-348. [DOI] [PubMed] [Google Scholar]
- 18.Eisenstein, B. I. 1981. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214:347-349. [DOI] [PubMed] [Google Scholar]
- 19.Eisenstein, B. I., and D. C. Dodd. 1982. Pseudocatabolite repression of type 1 fimbriae of Escherichia coli. J. Bacteriol. 151:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenstein, B. I., D. S. Sweet, V. Vaughn, and D. I. Friedman. 1987. Integration host factor is required for the DNA inversion that controls phase variation in Escherichia coli. Proc. Natl. Acad. Sci. USA 84:6505-6510. [DOI] [PMC free article] [PubMed]
- 21.Ferraris, R. P., S. Yasharpour, K. C. Lloyd, R. Mirzayan, and J. M. Diamond. 1990. Luminal glucose concentrations in the gut under normal conditions. Am. J. Physiol. 259:G822-G837. [DOI] [PubMed]
- 22.Foster, J. W. 1999. When protons attack: microbial strategies of acid adaptation. Curr. Opin. Microbiol. 2:170-174. [DOI] [PubMed]
- 23.Gally, D. L., T. J. Rucker, and I. C. Blomfield. 1994. The leucine-responsive regulatory protein binds to the fim switch to control phase variation of type 1 fimbrial expression in Escherichia coli K-12. J. Bacteriol. 176:5665-5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunther, N. W., V. Lockatell, D. E. Johnson, and H. L. T. Mobley. 2001. In vivo dynamics of type 1 fimbria regulation in uropathogenic Escherichia coli during experimental urinary tract infection. Infect. Immun. 69:2838-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall, M. N., and T. J. Silhavy. 1981. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J. Mol. Biol. 146:23-43. [DOI] [PubMed] [Google Scholar]
- 27.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569-584. [DOI] [PubMed]
- 28.Hull, R. A., R. E. Gill, P. Hsu, B. H. Minshew, and S. Falkow. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect. Immun. 33:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hultgren, S. J., J. L. Duncan, A. J. Schaeffer, and S. K. Amundsen. 1990. Mannose-sensitive haemagglutination in the absence of piliation in Escherichia coli. Mol. Microbiol 4:1311-1318. [DOI] [PubMed] [Google Scholar]
- 30.Hultgren, S. J., T. N. Porter, A. J. Schaeffer, and J. L. Duncan. 1985. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect. Immun. 50:370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hultgren, S. J., W. R. Schwan, A. J. Schaeffer, and J. L. Duncan. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect. Immun. 54:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hulton, C. S., A. Seirafi, J. C. Hinton, J. M. Sidebotham, L. Waddell, G. D. Pavitt, T. Owen-Hughes, A. Spassky, H. Buc, and C. F. Higgins. 1990. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell 63:631-642. [DOI] [PubMed] [Google Scholar]
- 33.Igo, M. M., A. J. Ninfa, and T. J. Silhavy. 1989. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 3:598-605. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawula, T. H., and P. E. Orndorff. 1991. Rapid site-specific DNA inversion in Escherichia coli mutants lacking the histone-like protein H-NS. J. Bacteriol. 173:4116-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaye, D. 1968. Antibacterial activity of human urine. J. Clin. Investig. 47:2374-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klemm, P. 1985. Fimbrial adhesions of Escherichia coli. Rev. Infect. Dis. 7:321-340. [DOI] [PubMed] [Google Scholar]
- 38.Klemm, P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunin, C. M., and S. T. Chambers. 1989. Osmoprotective properties for bacteria of renal papilla and urine: role of betaines as osmoprotectant molecules, p. 327-332. In E. Kass and C. Svanborg Eden (ed.), Host-parasite interactions in urinary tract infections. University of Chicago Press, Chicago, Ill.
- 40.Kunin, C. M., T. H. Hua, L. Van Arsdale White, and M. Villarejo. 1992. Growth of Escherichia coli in human urine: role of salt tolerance and accumulation of glycine betaine. J. Infect. Dis. 166:1311-1315. [DOI] [PubMed] [Google Scholar]
- 41.Kunin, C. M., T. H. Hua, R. L. Guerrant, and L. O. Bakaletz. 1994. Effect of salicylate, bismuth, osmoloytes, and tetracycline resistance on expression of fimbriae by Escherichia coli. Infect. Immun. 62:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim, J. K., N. W. Gunther IV, H. Zhao, D. E. Johnson, S. K. Keay, and H. L. T. Mobley. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect. Immun. 66:3303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loeb, W. F., and F. W. Quimby. 1989. The clinical chemistry of laboratory animals. Pergamon Press, New York, N.Y.
- 44.May, G., P. Dersch, M. Haardt, A. Middendorf, and E. Bremer. 1990. The osmZ (bglY) gene encodes the DNA-binding protein H-NS (H1a), a component of the Escherichia coli K12 nucleoid. Mol. Gen. Genet. 224:81-90. [DOI] [PubMed] [Google Scholar]
- 45.McClain, M. S., I. C. Blomfield, and B. I. Eisenstein. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClain, M. S., I. C. Blomfield, K. J. Eberhardt, and B. I. Eisenstein. 1993. Inversion-dependent phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 175:4335-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Misra, R., and P. Reeves. 1987. Role of micF in the tolC-mediated regulation of ompF, a major outer membrane protein of Escherichia coli K-12. J. Bacteriol. 169:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ofek, I., S. Hadar, R. Heiber, D. Zafiri, and M. Maayan. 1989. Effect of urea and urine dialysate on the growth of type-1 fimbriated and nonfimbriated phenotypes of isolates of Escherichia coli, p. 122-126. In E. Kass and C. Svanborg Eden (ed.), Host-parasite interactions in urinary tract infections. University of Chicago Press, Chicago, Ill.
- 50.O'Gara, J. P., and C. J. Dorman. 2000. Effects of local transcription and H-NS on inversion of the fim switch of Escherichia coli. Mol. Microbiol. 36:457-466. [DOI] [PubMed] [Google Scholar]
- 51.O'Hanley, P., D. Lark, S. Falkow, and G. Schoolnik. 1985. Molecular basis of Escherichia coli colonization of the upper urinary tract in BALB/c mice. J. Clin. Investig. 75:347-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Old, D. C., and J. P. Duguid. 1970. Selective outgrowth of fimbriate bacteria in static liquid medium. J. Bacteriol. 103:447-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen, P. B., and P. Klemm. 1994. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 116:95-100. [DOI] [PubMed] [Google Scholar]
- 54.Olsen, P. B., M. A. Schembri, D. L. Gally, and P. Klemm. 1998. Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett. 162:17-23. [DOI] [PubMed] [Google Scholar]
- 55.Orndorff, P., P. Spears, D. Schauer, and S. Falkow. 1985. Two modes of control of pilA, the gene encoding type 1 pilin in Escherichia coli. J. Bacteriol. 164:321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 57.Roesch, P. L., and I. C. Blomfield. 1998. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol. 27:751-761. [DOI] [PubMed] [Google Scholar]
- 58.Ross, D. L., and A. E. Neely. 1983. Textbook of urinalysis and bodily fluids. Appleton-Century-Crofts, Norwalk, Conn.
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Schaeffer, A. J., W. R. Schwan, S. J. Hultgren, and J. L. Duncan. 1987. Relationship of type 1 pilus expression in Escherichia coli to ascendingurinary tract infections in mice. Infect. Immun. 55:373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt-Nielsen, B., B. Graves, and J. Roth. 1983. Water removal and solute additions determining increases in renal medullary osmolarity. Am. J. Physiol. 244:F472-F482. [DOI] [PubMed]
- 62.Schwan, W. R., and W. Goebel. 1994. Host cell responses to Listeria monocytogenes infection include differential transcription of host cell stress genes involved in signal transduction. Proc. Natl. Acad. Sci. USA 91:6428-6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwan, W. R., H. S. Seifert, and J. L. Duncan. 1992. Growth conditions mediate differential transcription of fim genes involved in phase variation of type 1 pili. J. Bacteriol. 174:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwan, W. R., H. S. Seifert, and J. L. Duncan. 1994. Analysis of the fimB promoter region involved in type 1 pilus phase variation in Escherichia coli. Mol. Gen. Genet. 242:623-629. [DOI] [PubMed] [Google Scholar]
- 65.Smith, S. G. J., and C. J. Dorman. 1999. Functional analysis of the FimE integrase of Escherichia coli K-12: isolation of mutant derivatives with altered DNA inversion preferences. Mol. Microbiol. 34:965-979. [DOI] [PubMed] [Google Scholar]
- 66.Spears, P. A., D. Schauer, and P. E. Orndorff. 1986. Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J. Bacteriol. 168:179-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stentebjerg-Olesen, B., T. Chakraborty, and P. Klemm. 2000. FimE-catalyzed off-to-on inversion of the type 1 fimbrial phase switch and insertion sequence recruitment in an Escherichia coli K-12 fimB strain. FEMS Microbiol. Lett. 182:319-325. [DOI] [PubMed] [Google Scholar]
- 68.Struve, C., and K. A. Krogfelt. 1999. In vivo detection of Escherichia coli type 1 fimbrial expression and phase variation during experimental urinary tract infection. Microbiology 145:2683-2690. [DOI] [PubMed] [Google Scholar]
- 69.Vaisanen-Rhen, V., M. Rhen, E. Linder, and T. K. Korhonen. 1985. Adhesion of Escherichia coli to human kidney cryostat sections. FEMS Microbiol. Lett. 27:179-182. [Google Scholar]
- 70.Virkola, R. 1987. Binding characteristics of Escherichia coli type 1 fimbriae to human kidney cryostat sections. FEMS Microbiol. Lett. 40:257-262. [Google Scholar]
- 71.Virkola, R., B. Westerlund, H. Holthofer, J. Parkkinen, M. Kekomaki, and T. K. Korhonen. 1988. Binding characteristics of Escherichia coli adhesins in human urinary bladder. Infect. Immun. 56:2615-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White-Ziegler, C. A., A. Villapakkam, K. Ronaszeki, and S. Young. 2000. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J. Bacteriol. 182:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]