Abstract
Pertussis toxin is secreted from Bordetella pertussis with the assistance of the Ptl transport system, a member of the type IV family of macromolecular transporters. The S1 subunit and the B oligomer combine to form the holotoxin prior to export from the bacterial cell, although the site of assembly is not known. To better understand the pathway of pertussis toxin assembly and secretion, we examined the subcellular location of the S1 subunit, expressed with or without the B oligomer and the Ptl proteins. In wild-type B. pertussis, the majority of the S1 subunit that remained cell associated localized to the bacterial membranes. In mutants of B. pertussis that do not express pertussis toxin and/or the Ptl proteins, full-length S1, expressed from a plasmid, partitioned almost entirely to the bacterial membranes. Several lines of evidence strongly suggest that the S1 subunit localizes to the outer membrane of B. pertussis. First, we found that membrane-bound full-length S1 was almost completely insoluble in Triton X-100. Second, recombinant S1 previously has been shown to localize to the outer membrane of Escherichia coli (J. T. Barbieri, M. Pizza, G. Cortina, and R. Rappuoli, Infect. Immun. 58:999-1003, 1990). Third, the S1 subunit possesses a distinctive amino acid motif at its carboxy terminus, including a terminal phenylalanine, which is highly conserved among bacterial outer membrane proteins. By using site-directed mutagenesis, we determined that the terminal phenylalanine is critical for stable expression of the S1 subunit. Our findings provide evidence that prior to assembly with the B oligomer and independent of the Ptl proteins, the S1 subunit localizes to the outer membrane of B. pertussis. Thus, outer membrane-bound S1 may serve as a nucleation site for assembly with the B oligomer and for interactions with the Ptl transport system.
Many bacteria manifest important pathogenic effects by releasing proteins, such as degradative enzymes, toxins, and invasins, that function outside the bacterial cell. In order to secrete these virulence factors into the extracellular environment, bacteria have developed complex protein secretion mechanisms. One example of this is the pathway by which pertussis toxin is secreted from Bordetella pertussis.
Pertussis toxin is an oligomeric protein composed of five subunits, S1 to S5, which assemble into two functionally distinct moieties (40). The enzymatically active A component consists of the S1 subunit, which ADP ribosylates a family of GTP-binding regulatory proteins involved in signal transduction in eukaryotic cells (18). The B oligomer binds a eukaryotic cell and mediates translocation of the S1 subunit into the cell (24, 40). Structurally, the subunits comprising the B oligomer (one copy each of subunits S2, S3, and S5 and two copies of S4) form a pentameric ring that is penetrated by the carboxy terminus of the S1 subunit (37). Residues of the S1 subunit between positions 219 and 235 are involved in an interaction with the B oligomer (29).
Secretion of pertussis toxin is thought to involve at least two distinct steps. The DNA sequence of each of the pertussis toxin subunits indicates that the subunit is synthesized as a precursor containing a secretory leader peptide (32, 33). Presumably, the individual subunits cross the inner membrane through a Sec-like pathway and enter the periplasm, where the leader peptides are removed. In the second step, pertussis toxin traverses the outer membrane with the assistance of a set of nine accessory transport proteins, the Ptl proteins (41). The Ptl proteins belong to the type IV family of secretion systems that are involved in the transport of proteins and/or DNA from bacterial cells (9, 10, 41, 42). Recently, we showed that the individual subunits of pertussis toxin assemble into the holotoxin while they are still cell associated and that the Ptl transport system functions by exporting the assembled holotoxin (16).
In a previous study, Nicosia and Rappouli detected biologically active pertussis toxin in the periplasm of a B. pertussis mutant that is deficient in pertussis toxin secretion, indicating that at least to some extent, holotoxin can be found in the periplasmic compartment of the cell (34). However, two lines of evidence have led us to examine whether the S1 subunit may localize to the bacterial outer membrane prior to assembly into the secretion-competent holotoxin. First, Barbieri and colleagues have shown that when expressed in Escherichia coli, a recombinant pertussis toxin S1 subunit localized almost exclusively to the bacterial outer membrane (2). Second, we noted that the S1 subunit of pertussis toxin possesses a distinctive amino acid motif at its carboxy terminus (VYYESIAYSF) that is characteristic of outer membrane proteins of gram-negative bacteria (Table 1) (20, 39). This motif consists of hydrophobic residues at positions 1, 3, 5, 7, and 9 from the carboxy terminus, with a preference for tyrosine at position 3 and a highly conserved aromatic amino acid, usually phenylalanine but occasionally tryptophan, as the carboxy-terminal residue (20, 39). Studies of some outer membrane proteins have shown that removal of the carboxy-terminal segment or, more specifically, deletion or replacement of the terminal phenylalanine results in decreased outer membrane insertion and/or reduced protein stability (12, 21, 39).
TABLE 1.
Carboxy-terminal motifs of the S1 subunit of pertussis toxin, selected bacterial outer membrane proteins, and selected β-domains of autotransporter proteinsa
| Protein | Organism | Carboxy-terminal amino acids |
|---|---|---|
| S1 | B. pertussis | VYYESIAYSF |
| LamB | E. coli | TFGAQMEIWW |
| OmpC | E. coli | IVALGLVYQF |
| OmpF | E. coli | TVAVGIVYQF |
| PhoE | E. coli | IVAVGMTYQF |
| BrkA | B. pertussis | SFHAGYRYSF |
| Hap | H. influenzae | NVGVKLGYRW |
| Immunoglobulin A protease | Neisseria gonorrhoeae | SGQIKIQIRF |
| Pertactin | B. pertussis | TFHAGYRYSW |
The carboxy-terminal amino acid sequences of the outer membrane proteins LamB, OmpC, OmpF, and PhoE were obtained from reference 39. The carboxy-terminal amino acid sequences of the β-domains of the autotransporters BrkA, Hap, immunoglobulin A protease, and pertactin were obtained from reference 20. The carboxy termini (β-domains) of the autotransporters insert into the bacterial outer membrane and mediate secretion of the mature protein (20).
To better understand the events involved in the assembly and secretion of pertussis toxin, we examined the subcellular location of the S1 subunit in B. pertussis and studied whether the B oligomer and the Ptl transport system affect its localization. We also examined the importance of the carboxy-terminal phenylalanine for the stability of the S1 subunit.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The strains of B. pertussis and E. coli and the plasmids used in this study are listed in Table 2. B. pertussis strains were grown at 37°C on Bordet Gengou agar for 48 to 72 h or in Stainer-Scholte liquid medium. For liquid cultures, cells that had been grown on agar plates were resuspended in 20 ml of Stainer-Scholte medium at an A550 of 0.6. The strains were then grown with shaking for 48 h to an A550 of 1.3.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44λ−thi-1 gyrA96 relA1 | GIBCO BRL |
| B. pertussis strains | ||
| BP536 | Wild-type, nalidixic acid-resistant, streptomycin-resistant derivative of Tohama I | 38 |
| BP536Δptx | BP536 with a 2.6-kb deletion in the ptx genes from nucleotide 936 to nucleotide 3514 | 16 |
| BP536Δptxptl | BP536 with an 11.5-kb deletion in the ptx-ptl genes from nucleotide 425 to nucleotide 11971 | 16 |
| Plasmids | ||
| pTH18 | pUFR047 containing ptxS2, ptxS4,ptxS5, and ptxS3 under lacZ promoter control | 16 |
| pTH19 | pRK415 containing the ptx-ptl promoter region and ptxS1 | 16 |
| pDMC36 | pUFR047 containing the ptx-ptl promoter region and ptxS1 | 16 |
| pAMC61 | pUFR047 containing the ptx-ptl promoter region and mutant ptxS1 with a deletion in the coding sequence for Phe235 | This study |
| pSZH4 | pUC19 containing the entire ptx-ptl region | 19 |
| pUFR047 | Broad-host-range (IncW) vector, Mob+lacZα+, gentamicin resistant | 13 |
| pRK415 | Broad-host-range (IncP1) vector, tetracycline resistant | 25 |
| pRK2013 | ColE1 Tra+ kanamycin-resistant helper plasmid | 17 |
| pGEM11Zf(+) | Ampicillin resistant cloning vector | Promega |
| pAlterEx1 | Tetracycline-resistant cloning vector | Promega |
| pUC19 | Ampicillin-resistant cloning vector | GIBCO BRL |
B. pertussis mutants.
Construction of BP536Δptx and BP536Δptxptl has been described previously (16). Briefly, BP536Δptx has an in-frame deletion in the pertussis toxin structural genes from nucleotide 936 to nucleotide 3514, corresponding to the second half of ptxS1, all of ptxS2, ptxS4, and ptxS5, and 84% of ptxS3 (Fig. 1). This strain is expected to produce the Ptl proteins but not the pertussis toxin subunits, except possibly a truncated S1 protein or a mutant protein consisting of the N-terminal half of S1 and the C-terminal end of S3. Previously, we have demonstrated that this strain produces no detectable forms of S1 (as determined by immunoblot analysis) but, as expected, produces a functional Ptl transport system (16).
FIG. 1.
ptx-ptl region. The structure and nucleotide numbering system of the ptx-ptl region (41) are shown. Pr indicates the location of the ptx-ptl promoter. The position of the stop codon for ptlH, the most downstream of the nine ptl genes, is indicated by an asterisk. The DNA segments deleted in BP536Δptx and BP536Δptxptl are indicated.
In BP536Δptxptl, virtually the entire ptx-ptl region, extending from nucleotide 425 through nucleotide 11971, has been deleted from the B. pertussis chromosome (16) (Fig. 1). The 11.5-kb deletion in this strain encompasses the ptx-ptl promoter, all of the ptx genes, and the ptl genes except for the extreme 3′ end of ptlH (Fig. 1). Thus, BP536Δptxptl does not produce any of the pertussis toxin subunits or a functional Ptl transport system.
Plasmid construction.
Plasmid pTH18, which carries the genes encoding the subunits of the B oligomer (ptxS2, ptxS4, ptxS5, and ptxS3) behind the lacZ promoter of pUFR047 has been described previously (16). Construction of plasmids pDMC36 and pTH19, which carry the ptx-ptl promoter and ptxS1, in pUFR047 and pRK415, respectively, also has been described previously (16).
Plasmid pAMC61 encodes a mutant S1 subunit with a deletion of the carboxy-terminal phenylalanine, Phe235. This residue is encoded by nucleotides 1311 to 1313 of the ptx region (33). Plasmid pAMC61 was constructed as follows. First, using PCR as described previously (23), a DNA fragment which included nucleotides 921 to 1310 of the ptx region, followed by three stop codons, one nucleotide, and an XbaI site, was generated. The upstream primer was 5′-TTCGAATACGTCGACACTTATGGC (nucleotides 921 to 944 of ptx region; SalI site underlined). The downstream primer was 5′-CTATTATCTAGATCTATCATTACGAATACGCGATGCTTTCGTAGTA (six nucleotides, followed by an XbaI site, one nucleotide, three stop codons, and nucleotides 1310 to 1287 of the ptx region; XbaI site underlined). The DNA fragment generated by PCR was digested with SalI and XbaI and inserted into pGEM-11Zf(+) for DNA sequencing, which indicated that only the desired nucleotide changes had been incorporated. Next, nucleotides 1 to 4570 of the ptx-ptl region were excised as an EcoRI-BamHI fragment from pSZH4, a plasmid that contains the entire ptx-ptl region (19). The EcoRI-BamHI fragment from pSZH4 was inserted into pAlterExI. The SalI-XbaI fragment from the resultant vector, corresponding to nucleotides 930 to 1316 of the ptx region, was excised and replaced with the DNA fragment generated by PCR. The resultant vector was digested with EcoRI and XbaI, which generated a fragment containing the ptx-ptl promoter, followed by mutant ptxS1. This fragment was inserted into the EcoRI-XbaI site of pUC19. The EcoRI-HindIII fragment from the resultant vector was inserted into pUFR047, generating pAMC61.
Introduction of plasmids into B. pertussis.
Introduction of pDMC36 (pUFR047 containing the ptx-ptl promoter and wild-type ptxS1) into strains BP536Δptx and BP536Δptxptl has been described previously (16). Similarly, pAMC61 (pUFR047 containing the ptx-ptl promoter and the DNA sequence encoding S1ΔPhe235) was introduced into BP536Δptx by triparental conjugation by using E. coli DH5α containing the helper plasmid pRK2013, as described previously (3). Exconjugants were selected on Bordet Gengou agar containing gentamicin (10 μg/ml) and streptomycin (100 μg/ml). Introduction of pTH19 (pRK415 containing the ptx-ptl promoter and wild-type ptxS1) and pTH18 (pUFR047 containing the genes encoding the B oligomer) together into BP536Δptxptl has been described previously (16). The resultant strain, BP536Δptxptl(pTH18)(pTH19), contains ptxS1 and the genes encoding the B-oligomer subunits on two separate plasmids.
Preparation and fractionation of cell extracts.
A cell extract of B. pertussis whole cells that had been grown on Bordet Gengou agar was prepared by suspending cells in 5 ml of phosphate-buffered saline to an A550 of 2. A sample containing whole cells was removed. The remaining cell suspension was centrifuged at 17,000 × g for 5 min. The washed cells were suspended in 1.25 ml of 10 mM Tris-HCl (pH 8.0) containing 20% sucrose, 1 mM EDTA, and 0.1 mg of lysozyme per ml and incubated on ice for 10 min. After incubation, the volume was adjusted to 2.5 ml by adding 10 mM Tris-HCl (pH 8.0). The mixture was frozen in a dry ice-ethanol bath, thawed in cold water, and sonicated (model W225; Heat Systems Ultrasonics, Inc.) by using the 50% pulsed mode and a power setting of 4 for 5 min on ice. The mixture was centrifuged for 10 min at 1,300 × g to remove unbroken cells. The supernatant (2.5 ml) was centrifuged for 1 h at 190,000 × g (at Rmax). After centrifugation, the soluble fraction that contained cytoplasmic and periplasmic proteins was collected. The pellet containing the membrane fraction was suspended in a volume of phosphate-buffered saline equal to the volume of the soluble fraction. Portions of the whole-cell extract (50 μl), the soluble fraction (150 μl), and the membrane fraction (150 μl) were precipitated with an equal volume of 20% trichloroacetic acid. The precipitates were collected after centrifugation and suspended in 20 μl of the sample buffer used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Thus, the amounts of the membrane fraction and the soluble fraction loaded were six times the amount of the whole-cell extract loaded.
We used alkaline phosphatase, an enzyme that is active in the periplasm, and pertactin, a protein that localizes to the outer membrane of B. pertussis, as markers to assess the effectiveness of our cell fractionation procedure. In a B. pertussis strain expressing a ptxS2::phoA fusion, 94% of the total alkaline phosphatase activity was in the soluble (cytoplasmic-periplasmic) fraction, and only 6% of the total activity was in the membrane fraction. The alkaline phosphatase assay was performed as described previously (6, 16). When immunoblot analysis and monoclonal antibody BPE3 (5) were used, pertactin could not be detected in the soluble fraction, but this molecule was readily detected in the B. pertussis membrane fraction (data not shown).
Detergent solubilization of the membrane fraction of BP536Δptx(pDMC36) with Triton X-100 was performed essentially as described previously (23). The pellet obtained after centrifugation of the sonicated cell extract at 190,000 × g for 1 h, as described above, was suspended in 1.5 ml of 10 mM Tris-HCl (pH 8.0) containing 2% Triton X-100 and 7.5 mM MgCl2. The preparation was incubated at room temperature for 30 min and was then centrifuged at 190,000 × g for 1 h. The pellet (Triton X-100-insoluble fraction) was suspended in 1.5 ml of 10 mM Tris-HCl (pH 8.0). Portions of the Triton X-100-insoluble and Triton X-100-soluble fractions (100 μl each) were precipitated with 2 volumes of ice-cold 100% ethanol and stored for 30 min in a dry ice-ethanol bath. The precipitates were collected after centrifugation, suspended in 20 μl of SDS-PAGE sample buffer, and subjected to electrophoresis.
SDS-PAGE and immunoblot analysis.
SDS-PAGE was performed essentially as described by Laemmli (30) by using 4 to 20% gradient polyacrylamide gels obtained from Bio-Rad Laboratories, Hercules, Calif. Immunoblot analysis was performed essentially as previously described (7); monoclonal antibody 3CX4 (26, 27) or monoclonal antibody X2X5 (26, 27) was used to visualize the S1 subunit, and polyclonal antibody preparations were used to visualize PtlE and PtlF (23).
RESULTS
Cellular localization of the S1 subunit of pertussis toxin in wild-type B. pertussis.
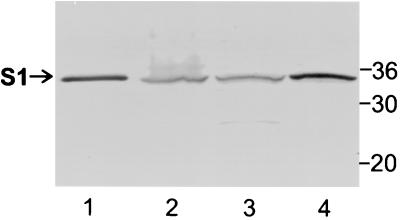
While pertussis toxin is secreted from B. pertussis, a significant amount remains cell associated (16). Using immunoblot analysis and monoclonal antibody 3CX4, we found that the S1 subunit was readily visualized in whole cells of the wild-type strain B. pertussis BP536 (Fig. 2, lane 2). Cells of this strain were then fractionated into the soluble fraction containing the cytoplasmic and periplasmic compartments and the total membrane fraction containing the inner and outer membranes. In BP536 preparations, the S1 subunit partitioned to both the soluble fraction and the total membrane fraction (Fig. 2, lanes 3 and 4). Since equivalent amounts of the two fractions were loaded, substantially more of the S1 subunit partitioned to the membranes than to the cytoplasmic-periplasmic fraction (Fig. 2, lanes 3 and 4). Virtually all of the S1 observed in BP536 was full-length S1; only a very minute amount was a lower-molecular-weight form that probably represented a commonly observed degradation product (8) detected in the soluble fraction (Fig. 2, lane 3). The full-length form of S1 represented the mature protein, in which the signal peptide had been removed. Since cleavage of the signal peptide occurs in the periplasm (36), any full-length S1 observed must have at least traversed the inner membrane and reached the periplasmic space. Thus, full-length S1 in the cytoplasmic-periplasmic fraction was probably located in the periplasm. Membrane-bound full-length S1 may have been located on the periplasmic face of the inner membrane or may have been associated with the outer membrane.
FIG. 2.
Immunoblot analysis of the S1 subunit in cell fractions of wild-type strain B. pertussis BP536. The cell fractionation procedures used and the methods used to prepare samples are described in Materials and Methods. Samples were subjected to SDS-PAGE and immunoblot analysis using monoclonal antibody 3CX4 to visualize the S1 subunit of pertussis toxin. Equivalent amounts of the cytoplasmic-periplasmic fraction and the total membrane fraction (inner and outer membranes) were loaded. Six times more of each of these fractions than whole-cell extract was loaded. The positions of molecular mass markers (in kilodaltons) are indicated on the right. The arrow indicates the protein band corresponding to full-length S1. Lane 1, purified pertussis toxin (0.1 μg); lane 2, whole-cell extract; lane 3, cytoplasmic-periplasmic fraction; lane 4, total membrane fraction.
Cellular localization of the S1 subunit of pertussis toxin in B. pertussis in the absence of the Ptl proteins.
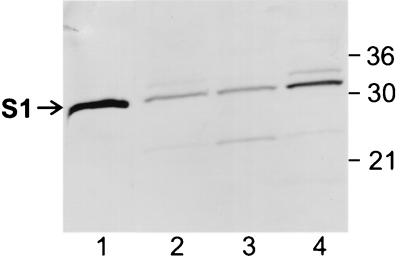
The Ptl proteins, several of which localize to the membranes of B. pertussis (11, 15, 23), are critical for export of the S1 subunit when it is part of the holotoxin (28). Therefore, we wanted to assess whether the finding that a substantial amount of S1 partitioned to the membranes of wild-type B. pertussis could be explained by S1 associating with the Ptl proteins. To examine this possibility, we used strain BP536Δptxptl(pTH18)(pTH19), which lacks the Ptl transport system and contains ptxS1 and the genes encoding the B-oligomer subunits on two separate plasmids. Full-length S1 was readily detected in a whole-cell extract of BP536Δptxptl(pTH18)(pTH19) (Fig. 3, lane 2). Minute amounts of two other forms of S1, a lower-molecular-weight degradation product and a slightly higher-molecular-weight form, were observed in whole cells of BP536Δptxptl(pTH18)(pTH19) (Fig. 3, lane 2). The higher-molecular-weight form migrated slightly slower than full-length S1. This species has been observed previously in strains of B. pertussis, including a wild-type strain carrying a plasmid encoding the S1 subunit, and is thought to represent a form of S1 in which the signal peptide is not removed because high-level production from the plasmid may have exceeded the normal processing capability of the cell (16, 28).
FIG. 3.
Immunoblot analysis of the S1 subunit in cell fractions of BP536Δptxptl(pTH18)(pTH19). The cell fractionation procedures used and the methods used to prepare samples are described in Materials and Methods. Samples were subjected to SDS-PAGE and immunoblot analysis using monoclonal antibody 3CX4 to visualize the S1 subunit of pertussis toxin. Equivalent amounts of the cytoplasmic-periplasmic fraction and the total membrane fraction (inner and outer membranes) were loaded. Six times more of each of these fractions than whole-cell extract was loaded. The positions of molecular mass markers (in kilodaltons) are indicated on the right. The arrow indicates the protein band corresponding to full-length S1. Lane 1, purified pertussis toxin (0.1 μg); lane 2, whole-cell extract; lane 3, cytoplasmic-periplasmic fraction; lane 4, total membrane fraction.
As in wild-type B. pertussis, full-length S1 was observed in both the cytoplasmic-periplasmic fraction and the total membrane fraction of BP536Δptxptl(pTH18)(pTH19), and relatively more partitioned to the membranes (Fig. 3, lanes 3 and 4). This finding indicated that in B. pertussis, S1 localizes to the membranes independent of the Ptl transport system. In BP536Δptxptl(pTH18)(pTH19), the lower-molecular-weight degradation product of S1 partitioned primarily to the cytoplasmic-periplasmic fraction (Fig. 3, lane 3), and unprocessed S1 partitioned primarily to the membrane fraction (Fig. 3, lane 4).
Cellular localization of the S1 subunit of pertussis toxin in B. pertussis in the absence of the B oligomer and the Ptl proteins.
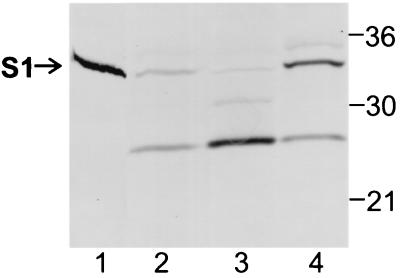
Previously, we have shown that the S1 subunit and the B oligomer must assemble into the holotoxin to be efficiently released from the bacterial cell by the Ptl transport system (16). To assess whether the association with the B oligomer affects the subcellular localization of the S1 subunit in B. pertussis, we examined strain BP536Δptxptl(pDMC36). This strain lacks both the genes encoding the Ptl proteins and the genes encoding the B-oligomer subunits. In this strain, the S1 subunit is expressed from a plasmid that contains the coding sequence for the ptx-ptl promoter region, followed by ptxS1. We should note that we have previously shown that the S1 subunit expressed from a plasmid can associate with the B oligomer and can subsequently be secreted by a Ptl-dependent mechanism in a strain of B. pertussis that also produces the B oligomer (expressed from a second plasmid) and a functional Ptl system (expressed from genes located on the chromosome) (16). Thus, the location of ptxS1 (i.e., whether it is located on a plasmid or on the chromosome) does not seem to affect the biogenesis pathway. In BP536Δptxptl(pDMC36) (which expresses the S1 subunit but no B oligomer and no Ptl proteins), substantial degradation of S1 was observed (Fig. 4, lanes 2 to 4), which was not surprising because S1 is known to be susceptible to proteolysis, particularly when it is not associated with the B oligomer (8). The S1 subunit partitioned to both the cytoplasmic-periplasmic fraction and the membrane fraction of BP536Δptxptl(pDMC36) (Fig. 4, lanes 3 and 4). However, the patterns of S1 and its proteolytic fragments were markedly different in the two fractions. Full-length S1 was found almost exclusively in the membranes (Fig. 4, lane 4); very little was observed in the cytoplasmic-periplasmic fraction (Fig. 4, lane 3). The membrane fraction contained predominantly full-length S1, along with smaller amounts of degraded S1 and unprocessed S1 (Fig. 4, lane 4). Almost all of the S1 in the cytoplasmic-periplasmic fraction had been proteolytically degraded into a major degradation product and a minor species that migrated approximately midway between full-length S1 and the predominant degradation product (Fig. 4, lane 3). We also examined the subcellular localization of the S1 subunit in BP536Δptx(pDMC36), a strain that differs from BP536Δptxptl(pDMC36) only in that it produces a functional Ptl transport system. The pattern of subcellular localization of S1 and its proteolytic fragments in BP536Δptx(pDMC36) was essentially identical to that observed in BP536Δptxptl(pDMC36) (compare Fig. 4 and Fig. 5, lanes 2 and 4).
FIG. 4.
Immunoblot analysis of the S1 subunit in cell fractions of BP536Δptxptl(pDMC36). The cell fractionation procedures used and the methods used to prepare samples are described in Materials and Methods. Samples were subjected to SDS-PAGE and immunoblot analysis using monoclonal antibody 3CX4 to visualize the S1 subunit of pertussis toxin. Equivalent amounts of the cytoplasmic-periplasmic fraction and the total membrane fraction (inner and outer membranes) were loaded. Six times more of each of these fractions than whole-cell extract was loaded. The positions of molecular mass markers (in kilodaltons) are indicated on the right. The arrow indicates the protein band corresponding to full-length S1. Lane 1, purified pertussis toxin (0.1 μg); lane 2, whole-cell extract; lane 3, cytoplasmic-periplasmic fraction; lane 4, total membrane fraction.
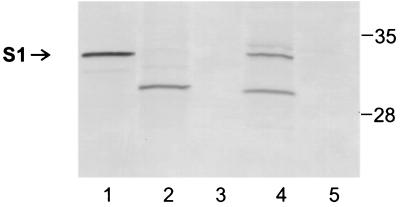
FIG. 5.
Immunoblot analysis of the S1 subunit in cell fractions of BP536Δptx(pDMC36) and BP536Δptx(pAMC61). The cell fractionation procedures used and the methods used to prepare samples are described in Materials and Methods. Samples were subjected to SDS-PAGE and immunoblot analysis using monoclonal antibody 3CX4 to visualize the S1 subunit of pertussis toxin. Equivalent amounts of the cytoplasmic-periplasmic fractions and the total membrane fractions (inner and outer membranes) were loaded. The positions of molecular mass markers (in kilodaltons) are indicated on the right. The arrow indicates the protein band corresponding to full-length S1. Lane 1, purified pertussis toxin (0.2 μg); lane 2, BP536Δptx(pDMC36) cytoplasmic-periplasmic fraction; lane 3, BP536Δptx(pAMC61) cytoplasmic-periplasmic fraction; lane 4, BP536Δptx(pDMC36) total membrane fraction; lane 5, BP536Δptx(pAMC61) total membrane fraction.
In BP536Δptxptl(pDMC36), in the absence of the B oligomer, almost all of the full-length S1 and unprocessed S1 partitioned to the membrane fraction. Thus, in the absence of the B oligomer, if S1 survives proteolytic degradation, almost all of it localizes to the membranes. Taken together, our findings indicate that the propensity for full-length S1 to localize to the membranes of B. pertussis is independent of the association of S1 with the B oligomer or the Ptl proteins.
Triton X-100 solubility of the S1 subunit.
Whereas isopycnic centrifugation cannot separate the membranes of B. pertussis into distinguishable fractions corresponding to the inner and outer membranes (14), it is thought that Triton X-100 preferentially solubilizes inner membrane proteins of B. pertussis (1, 14, 23). Using strain BP536Δptx(pDMC36), we used differential solubilization of the S1 subunit in Triton X-100 as an indicator of the inner or outer membrane location. We found that while a significant fraction of the degraded form of the S1 subunit was soluble in Triton X-100, membrane-bound, full-length S1 was almost completely insoluble in Triton X-100 (Fig. 6A), suggesting an outer membrane location. The lack of solubility of the membrane-bound, full-length S1 subunit in Triton X-100 was similar to the lack of solubility of PtlF (Fig. 6B), a protein which is believed to localize to the outer membrane of B. pertussis (23), and to the lack of solubility of the major porin protein of B. pertussis (data not shown), which is known to localize to the outer membrane (31). In contrast, a significant fraction of PtlE, a protein believed to localize to the inner membrane (23), was soluble in Triton X-100 (Fig. 6C).
FIG. 6.
Triton X-100 solubility of the S1 subunit. The cell fractionation procedures used and the methods used to prepare samples are described in Materials and Methods. The total membrane fraction of BP536Δptx(pDMC36) was treated with 2% Triton X-100 as described in Materials and Methods. Samples were subjected to SDS-PAGE and immunoblot analysis using monoclonal antibody 3CX4 to visualize the S1 subunit of pertussis toxin (A) and polyclonal antibody preparations to visualize PtlF (B) and PtlE (C). The positions of protein bands corresponding to full-length S1, the degraded form of S1 (S1*), PtlF, and PtlE are indicated on the left. The positions of molecular mass markers (in kilodaltons) are indicated on the right. The immunoblots used to visualize PtlF (B) and PtlE (C) were developed with polyclonal antibody preparations that exhibit binding to either PtlF or PtlE but also exhibit some nonspecific binding to other protein bands. Several bands are present in these immunoblots; the positions of bands identified previously as either PtlF or PtlE (23) are indicated. Lanes 1, Triton X-100-soluble fraction; lanes 2, Triton X-100-insoluble fraction.
Effect of the carboxy-terminal phenylalanine on the stability of the S1 subunit of pertussis toxin.
Next, we wanted to examine the role, if any, of the carboxy-terminal phenylalanine in membrane localization and/or stability of the S1 subunit. We introduced plasmid pAMC61, which encodes a mutant S1 subunit with a deletion of the carboxy-terminal phenylalanine, Phe235, into BP536Δptx, a strain that does not produce any of the pertussis toxin subunits. The backbone used for pAMC61 was the broad-host-range vector pUFR047, the backbone that was used for pDMC36, a plasmid that contains wild-type ptxS1. In pAMC61 and pDMC36, mutant ptxS1 and wild-type ptxS1, respectively, follow the ptx-ptl promoter region. We examined the cytoplasmic-periplasmic fraction and the membranes of BP536Δptx(pDMC36) and BP536Δptx(pAMC61) for the presence of S1 by immunoblot analysis. In BP536Δptx(pDMC36), S1 was found in both the cytoplasmic-periplasmic fraction and the membranes (Fig. 5, lanes 2 and 4). Essentially all of the full-length S1 partitioned to the membranes (Fig. 5, lane 4), whereas more of the degraded forms of S1 were observed in the cytoplasmic-periplasmic fraction (Fig. 5, lane 2). In contrast, in BP536Δptx(pAMC61), the mutant S1 subunit lacking the terminal phenylalanine could be detected only barely, if at all, by immunoblot analysis with monoclonal antibody 3CX4 (27), which recognizes a conformational epitope on the S1 subunit (Fig. 5), or monoclonal antibody X2X5, which recognizes a linear epitope (27) located near the N terminus of S1 (data not shown). To determine whether any mutant S1 that might have been produced by BP536Δptx(pAMC61) was secreted from the bacterial cells, we grew this strain in Stainer-Sholte liquid medium and examined the cellular material and culture supernatant for the presence of S1 by immunoblot analysis, using methods described previously (16). Mutant S1 could not be detected in either the cellular material or the culture supernatant (data not shown). Thus, our findings indicate that deletion of the carboxy-terminal phenylalanine of S1 results in markedly reduced protein stability. The mutant construct apparently could be transcribed and translated since we detected minute amounts of the mutant S1 in cell lysates when monoclonal antibody X2X5 was used to visualize the protein (data not shown). In view of the importance of this highly conserved amino acid residue in outer membrane insertion and/or stability of other proteins known to localize to bacterial outer membranes (12, 21, 39), these findings support the hypothesis that S1 is localized in the outer membrane in B. pertussis.
DISCUSSION
Secretion of pertussis toxin from B. pertussis is thought to involve at least two distinct steps. First, the individual subunits of the toxin, each of which is synthesized with an amino-terminal signal peptide (32, 33), are probably translocated across the inner membrane in an unfolded state by a Sec-like pathway. Second, the Ptl proteins, which are required for pertussis toxin secretion (41), are thought to facilitate transport of the toxin across the outer membrane. Recently, we have shown that the individual components of the toxin assemble prior to release from the bacterial cell and that only the assembled holotoxin is efficiently released from B. pertussis by the Ptl transport system (16). The site of holotoxin assembly in the bacterial cell is not known. Previously, it was thought that pertussis toxin assembly occurs in the periplasmic space (34), in a manner similar to cholera toxin assembly (22). This view was based on the results of one study in which some biologically active pertussis toxin was found in the periplasmic fraction of a mutant of B. pertussis which does not secrete pertussis toxin (34). Because the S1 subunit must combine with the B oligomer to manifest biological activity, this finding indicated that at least to some extent holotoxin could be detected in the periplasmic space. However, the possibility that individual subunits could also reach the outer membrane prior to assembly was not addressed. In another study, pertussis toxin subunits were detected by immunoblot analysis in the periplasmic fraction of wild-type cells; however, membrane fractions were not examined, so the proportion of subunits that localized to the periplasmic space and the proportion of subunits that localized to the membrane could not be determined (35). In the present study, to better understand the pathway of pertussis toxin assembly and secretion, we examined the subcellular location of the S1 subunit when it was expressed with and without the B oligomer. In contrast to the previously held view that holotoxin assembly is completed in the periplasmic space, we obtained evidence that the S1 subunit localizes to the outer membrane of B. pertussis before it combines with the B oligomer to form the secretion-competent holotoxin. Our results also indicate that the ability of the S1 subunit to localize to the outer membrane is independent of the Ptl transport system.
We found that in a wild-type strain of B. pertussis, the majority of the S1 of the cell partitioned to the membranes, and a much smaller amount was present in the cytoplasmic-periplasmic fraction (Fig. 2). The Ptl transport system is critical for export of S1 that is part of the holotoxin (28), and several of the Ptl proteins have been shown to associate with the bacterial membranes (11, 15, 23). While it was possible that the membrane location of the S1 subunit was due to an association with the Ptl proteins, we determined that this was not the case since the relative amounts of S1 observed in the membrane and cytoplasmic-periplasmic fractions were similar for strains that produced the Ptl proteins and strains that lacked the Ptl proteins.
We also wanted to determine if S1 localized to the membranes in the absence of the B oligomer. As expected, in strains that did not express the B oligomer, S1 was susceptible to proteolytic degradation. Degraded forms of S1 were observed primarily in the cytoplasmic-periplasmic fraction (Fig. 4). Nevertheless, a substantial amount of full-length S1 was observed in these strains, and virtually all of it localized to the membranes (Fig. 4). Thus, it appears that localization of full-length S1 to the membranes of B. pertussis occurs independent of the association of S1 with the B oligomer.
Several lines of evidence from this study and from the work of other researchers strongly suggest that a substantial amount of the S1 subunit observed in the membranes of B. pertussis is associated with the bacterial outer membrane. First, we found that membrane-bound, full-length S1 was almost completely insoluble in Triton X-100. Since Triton X-100 has been shown to preferentially solubilize inner membrane proteins of B. pertussis, these results are consistent with an outer membrane location for the S1 subunit (1, 14, 23).
Second, Barbieri and colleagues expressed a recombinant S1 subunit in a protease-deficient strain of E. coli and found that the major product, which corresponded to authentic full-length S1, localized to the bacterial membranes (2). These investigators found that when the membrane fraction of E. coli was separated into inner and outer membranes by sucrose gradient centrifugation, more than 90% of the recombinant S1 was associated with the outer membrane. They detected minor proteolytic fragments in the periplasm. In our study, the pattern of partitioning of full-length S1 and its degradation products to the membranes and cytoplasmic-periplasmic fractions of B. pertussis strains that do not express the B oligomer was remarkably similar to the pattern observed by Barbieri et al. when the S1 subunit was expressed in E. coli.
Another line of evidence that supports the hypothesis that the S1 subunit is located in the outer membrane is the presence of a carboxy-terminal motif that is highly characteristic of bacterial outer membrane proteins and β-domains of autotransporter proteins (20, 39). The carboxy-terminal β-domains of autotransporters insert into the bacterial outer membrane and mediate secretion of the mature protein (20). The last 10 amino acids of bacterial outer membrane proteins, but not periplasmic proteins, comprise a highly conserved segment consisting of hydrophobic residues at positions 1, 3, 5, 7, and 9 from the carboxy terminus (20, 39). The most pronounced similarity involves the presence of a phenylalanine or tryptophan residue at the carboxy-terminal position in the vast majority of outer membrane proteins (20, 39) (Table 1). Except for a hydrophilic residue at position 7 from the carboxy terminus, the carboxy-terminal segment of the S1 subunit, VYYESIAYSF, conforms remarkably with the consensus sequence found in bacterial outer membrane proteins.
The carboxy-terminal amino acid motif of bacterial outer membrane proteins is thought to form a membrane-spanning segment that contains information important for outer membrane insertion and retention and/or inherent protein stability (4, 12, 20, 21, 39). The role of the carboxy-terminal segment, or the carboxy-terminal phenylalanine itself, in outer membrane localization of PhoE, a porin of E. coli, has been studied in detail (4, 12, 39). Mutant PhoE proteins lacking either the carboxy-terminal 10 amino acids or only the carboxy-terminal phenylalanine were not efficiently incorporated into the outer membrane (4, 39). The terminal phenylalanine appears to be essential for the stability of the protein (12). Similarly, deletion of the last three amino acids of Hap, an autotransporter of Haemophilus influenzae, drastically affected the stability of this protein, and it was suggested that the instability observed was due to improper localization of the protein (21).
In our study, we wanted to examine the role of the carboxy-terminal phenylalanine itself, irrespective of the B oligomer, in the membrane location and/or stability of the S1 subunit. Whereas full-length S1 and its major proteolytic degradation product were readily detected in BP536Δptx(pDMC36), a strain expressing wild-type S1, virtually no S1 was detected in BP536Δptx(pAMC61), a strain that is expected to produce the mutant S1 subunit lacking the terminal phenylalanine residue (Fig. 5). Thus, the carboxy-terminal phenylalanine itself appears to play a major role in the stability of S1. The fact that the carboxy-terminal phenylalanine alone has such a profound effect on the stability of the S1 monomer (in the absence of the B oligomer) is not immediately apparent from the crystal structure of pertussis toxin (37). It is possible that the carboxy-terminal phenylalanine is important for outer membrane targeting or for interaction with outer membrane components that promote proper folding of the S1 subunit and its stabilization.
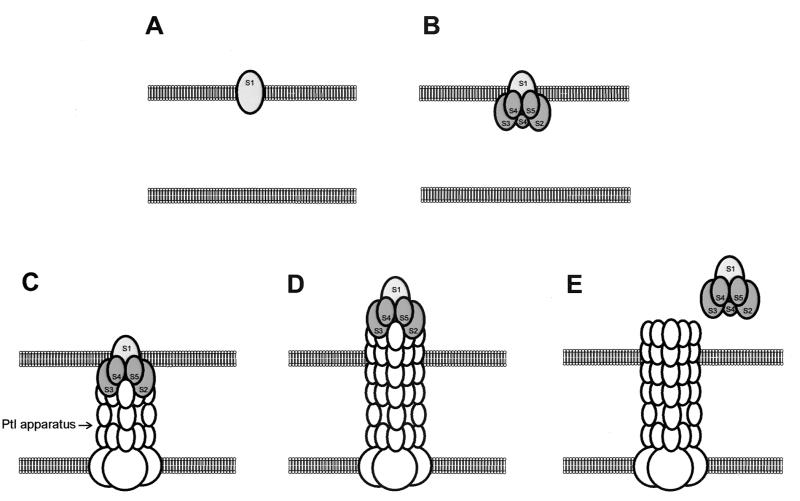
Our results indicate that in the pertussis toxin assembly and secretion pathway, the S1 subunit associates with the outer membrane before it joins with the B oligomer to form the secretion-competent holotoxin. Our findings are in contrast to the previously widely held view that assembly into the holotoxin is completed in the periplasmic space. The outer-membrane-bound S1 subunit may serve as a nucleation site for assembly with the B oligomer and subsequent interactions with the Ptl transport system. While it has been hypothesized that the Ptl system acts as a channel through which the assembled holotoxin is transported, an equally plausible model is that the Ptl system acts as a piston which pushes the toxin molecule through the outer membrane. Type IV transport systems often are associated with pilus structures (10). If the toxin subunits transiently associate with the tip of a pilus-like structure formed by the Ptl proteins, then elongation and extrusion of this pilus-like structure through the outer membrane would result in export of the toxin. Localization of the S1 subunit to the outer membrane is consistent with such a model, as shown in Fig. 7. Further studies are needed to understand the structure of the Ptl transport system and the mechanism by which it enables pertussis toxin to traverse the outer membrane.
FIG. 7.
Model for secretion of pertussis toxin from B. pertussis. S1 first localizes to the outer membrane (A). The B oligomer then associates with the S1 subunit (B), and this is followed by association of the toxin with Ptl proteins (C). Formation a pilus-like structure consisting of Ptl proteins and elongation of this structure result in extrusion of the toxin through the outer membrane (D). Dissociation of the toxin from the Ptl apparatus results in release of the toxin into the extracellular space (E).
REFERENCES
- 1.Armstrong, S. K., and C. D. Parker. 1986. Heat-modifiable envelope proteins of Bordetella pertussis. Infect. Immun. 54:109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbieri, J. T., M. Pizza, G. Cortina, and R. Rappuoli. 1990. Biochemical and biological activities of recombinant S1 subunit of pertussis toxin. Infect. Immun. 58:999-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, E. M., A. A. Weiss, I. E. Ehrmann, M. C. Gray, E. L. Hewlett, and M. S. M. Goodwin. 1991. Bordetella pertussis adenylate cyclase toxin and hemolytic activities require a second gene, cyaC, for activation. J. Bacteriol. 173:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, D., M. Scholten, C. Verhagen, and J. Tommassen. 1989. The role of the carboxy-terminal membrane-spanning fragment in the biogenesis of Escherichia coli K12 outer membrane protein PhoE. Mol. Gen. Genet. 216:144-148. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, M. J., Z. M. Li, J. L. Cowell, M. E. Bisher, A. C. Steven, P. Novotny, and C. R. Manclark. 1988. Identification of a 69-kilodalton nonfimbrial protein as an agglutinogen of Bordetella pertussis. Infect. Immun. 56:3189-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase using deletions and 80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 7.Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 8.Burns, D. L., S. Z. Hausman, W. Lindner, F. A. Robey, and C. R. Manclark. 1987. Structural characterization of pertussis toxin A subunit. J. Biol. Chem. 262:17677-17682. [PubMed] [Google Scholar]
- 9.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook, D. M., K. M. Farizo, and D. L. Burns. 1999. Identification and characterization of PtlC, an essential component of the pertussis toxin secretion system. Infect. Immun. 67:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Cock, H., M. Struyve, M. Kleerebezem, T. van der Krift, and J. Tommassen. 1997. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Esherichia coli K-12. J. Mol. Biol. 269:473-478. [DOI] [PubMed] [Google Scholar]
- 13.DeFeyter, R., Y. Yang, and D. W. Gabriel. 1993. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant-Microbe Interact. 6:225-237. [DOI] [PubMed] [Google Scholar]
- 14.Ezzel, J. W., W. J. Dobrogosz, W. E. Kloos, and C. R. Manclark. 1981. Phase shift markers in Bordetella: alterations in envelope proteins. J. Infect. Dis. 143:562-569. [DOI] [PubMed] [Google Scholar]
- 15.Farizo, K. M., T. G. Cafarella, and D. L. Burns. 1996. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 271:31643-31649. [DOI] [PubMed] [Google Scholar]
- 16.Farizo, K. M., T. Huang, and D. L. Burns. 2000. Importance of holotoxin assembly in Ptl-mediated secretion of pertussis toxin from Bordetella pertussis. Infect. Immun. 68:4049-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilman, A. G. 1987. G proteins: transducers of receptor generated signals. Annu. Rev. Biochem. 56:615-649. [DOI] [PubMed] [Google Scholar]
- 19.Hausman, S. Z., J. D. Cherry, U. Heininger, C. H. Wirsing von Konig, and D. L. Burns. 1996. Analysis of proteins encoded by the ptx and ptl genes of Bordetella bronchiseptica and Bordetella parapertussis. Infect. Immun. 64:4020-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 21.Hendrixson, D. R., M. L. de la Morena, C. Stathopoulos, and J. W. St. Geme III. 1997. Structural determinants of processing and secretion of the Haemophilus influenzae Hap protein. Mol. Microbiol. 26:505-518. [DOI] [PubMed] [Google Scholar]
- 22.Hirst, T. R., J. Sanchez, J. B. Kaper, S. J. S. Hardy, and J. Holgren. 1984. Mechanism of toxin secretion by Vibrio cholerae investigated in strains harboring plasmids that encode heat labile enterotoxins of Escherichia coli. Proc. Natl. Acad. Sci. USA 81:7752-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, F. D., and D. L. Burns. 1994. Detection and subcellular localization of three Ptl proteins involved in the secretion of pertussis toxin from Bordetella pertussis. J. Bacteriol. 176:5350-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katada, T., M. Tamura, and M. Ui. 1983. The A protomer of islet-activating protein, pertussis toxin, as an active peptide catalyzing ADP-ribosylation of a membrane protein. Arch. Biochem. Biophys. 224:290-298. [DOI] [PubMed] [Google Scholar]
- 25.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 26.Kenimer, J. G., K. J. Kim, P. G. Probst, C. R. Manclark, D. G. Burstyn, and J. L. Cowell. 1989. Monoclonal antibodies to pertussis toxin: utilization as probes of toxin function. Hybridoma 8:37-51. [DOI] [PubMed] [Google Scholar]
- 27.Kim, K. J., W. N. Burnette, R. D. Sublett, C. R. Manclark, and J. G. Kenimer. 1989. Epitopes on the S1 subunit of pertussis toxin recognized by monoclonal antibodies. Infect. Immun. 57:944-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotob, S. I., and D. L. Burns. 1997. Essential role of the consensus nucleotide-binding site of PtlH in secretion of pertussis toxin from Bordetella pertussis. J. Bacteriol. 179:7577-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krueger, K. M., and J. T. Barbieri. 1994. Assignment of functional domains involved in ADP-ribosylation and B-oligomer binding with the carboxyl terminus of the S1 subunit of pertussis toxin. Infect. Immun. 62:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Li, Z. M., J. H. Hannah, S. Stibitz, N. Y. Nguyen, C. R. Manclark, and M. J. Brennan. 1991. Cloning and sequencing of the structural gene for the porin protein of Bordetella pertussis. Mol. Microbiol. 5:1649-1656. [DOI] [PubMed] [Google Scholar]
- 32.Locht, C., and J. M. Keith. 1986. Pertussis toxin gene: nucleotide sequence and genetic organization. Science 232:1258-1264. [DOI] [PubMed] [Google Scholar]
- 33.Nicosia, A., M. Perugini, C. Franzini, M. C. Casagli, M. G. Borri, G. Antoni, M. Almoni, P. Neri, G. Ratti, and R. Rappuoli. 1986. Cloning and sequencing of the pertussis toxin genes: operon structure and gene duplication. Proc. Natl. Acad. Sci. USA 83:4631-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicosia, A., and R. Rappuoli. 1987. Promoter of the pertussis toxin operon and production of pertussis toxin. J. Bacteriol. 169:2843-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pizza, M., M. Bugnoli, R. Manetti, A. Covacci, and R. Rappuoli. 1990. The subunit S1 is important for pertussis toxin secretion. J. Biol. Chem. 265:17759-17763. [PubMed] [Google Scholar]
- 36.Randall, L. L., S. J. S. Hardy, and J. R. Thom. 1987. Export of protein: a biochemical view. Annu. Rev. Biochem. 41:507-541. [DOI] [PubMed] [Google Scholar]
- 37.Stein, P. E., A. Boodhoo, G. D. Armstrong, S. A. Cockle, M. H. Klein, and R. J. Read. 1994. The crystal structure of pertussis toxin. Structure 2:45-57. [DOI] [PubMed] [Google Scholar]
- 38.Stibitz, S., and M.-S. Yang. 1991. Subcellular localization and immunochemical detection of proteins encoded by the vir locus of Bordetella pertussis. J. Bacteriol. 173:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 40.Tamura, M., K. Nogimori, S. Murai, M. Yajima, K. Ito, T. Katada, M. Ui, and S. Ishii. 1982. Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry 21:5516-5522. [DOI] [PubMed] [Google Scholar]
- 41.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]