Abstract
The Bacillus subtilis LexA protein represses the SOS response to DNA damage by binding as a dimer to the consensus operator sequence 5′-CGAACN4GTTCG-3′. To characterize the requirements for LexA binding to SOS operators, we determined the operator bases needed for site-specific binding as well as the LexA amino acids required for operator recognition. Using mobility shift assays to determine equilibrium constants for B.subtilis LexA binding to recA operator mutants, we found that several single base substitutions within the 14 bp recA operator sequence destabilized binding enough to abolish site-specific binding. Our results show that the AT base pairs at the third and fourth positions from the 5′ end of a 7 bp half-site are essential and that the preferred binding site for a LexA dimer is 5′-CGAACATATGTTCG-3′. Binding studies with LexA mutants, in which the solvent accessible amino acid residues in the putative DNA binding domain were mutated, indicate that Arg-49 and His-46 are essential for binding and that Lys-53 and Ala-48 are also involved in operator recognition. Guided by our mutational analyses as well as hydroxyl radical footprinting studies of the dinC and recA operators we docked a computer model of B.subtilis LexA on the preferred operator sequence in silico. Our model suggests that binding by a LexA dimer involves bending of the DNA helix within the internal 4 bp of the operator.
INTRODUCTION
The LexA protein is the transcriptional repressor of the bacterial SOS DNA repair system, which comprises a set of DNA repair and cellular survival genes that are induced in response to DNA damage [reviewed in (1–3)]. First characterized in Escherichia coli, LexA binds as a dimer to operator sequences preceding more than 30 SOS genes (3–6). DNA damage generates single-stranded DNA, which activates the RecA protein to stimulate the autocatalytic cleavage activity of LexA (7,8). The site-specific autodigestion of LexA separates the N-terminal DNA binding domain from the C-terminal dimerization domain, which in turn causes a decrease in its binding affinity for SOS operators. This regulatory mechanism has been highly conserved among gram-negative and gram-positive bacteria (2). Correspondingly, bacterial LexA proteins share a significant degree of homology in the dimerization domain, which contains the catalytic site for cleavage. In contrast, the DNA binding domains of gram-positive LexA proteins have diverged considerably from their counterparts in gram-negative bacteria, consistent with the fact that the two classes of repressor bind to completely different sequences (2,9).
The Bacillus subtilis LexA protein was first identified as the product of the dinR gene, a damage-inducible gene whose product is homologous with E.coli LexA (10). Its role as the SOS repressor was confirmed by its ability to bind specifically to sequences overlapping the promoters of din genes (9,11) and its ability to inhibit transcription (11). The consensus SOS operator sequence, 5′-CGAACN4GTTCG-3′, was first identified based on visual inspection of known din promoter sequences (12) and later confirmed by deletion analyses (13) and DNA binding studies (9,11,14,15). It is significantly different from the SOS operator of E.coli and other gram-negative bacteria, which is TACTGTATATATATACAGTA (3).
The B.subtilis SOS operators characterized thus far include the recA, lexA, uvrB (formerly dinA), dinB and dinC (also called tagC) operators (12,16). We report here the B.subtilis SOS operator sequence requirements, which should facilitate the identification of other B.subtilis SOS genes. To assess the sequence requirements, we determined equilibrium dissociation constants for LexA binding to SOS operators and to recA operator mutants in which bases within the 14 bp operator were individually changed. We also identified LexA amino acid residues that contribute to SOS operator binding and we used computational docking of LexA to the operator sequence to visualize possible interactions between these residues and the operator.
MATERIALS AND METHODS
The B.subtilis LexA protein was purified as described previously (11). Oligonucleotide primers were purchased from Sigma Genosys. Pfu polymerase and the Quick change mutagenesis kit purchased from Stratagene, and T4 kinase, purchased from Promega Corp., were used as recommended by the manufacturers.
Preparation of promoter regions for mobility shift assays
DNA fragments corresponding to the recA(−110 to +170), uvrB(−200 to +75), dinB(−155 to +70) and dinC(−85 to +110) promoters were prepared by PCR amplification of YB886 (17) DNA (10 ng/ml) using synthetic oligonucleotide primers (2 µM) with a Peltier PTC-200 Thermal Cycler (MJ Research). The recA operator mutants were made by annealing complementary oligonucleotides containing variants of the 14 bp operator sequence flanked by six Gs on the 5′ end and six Cs on the 3′ end. DNA samples were radiolabeled with [γ-32P]ATP using T4 kinase and purified by electrophoresis on an 8% non-denaturing polyacrylamide gel. DNA concentration was determined by fluorometry using the Hoescht 33258 fluorochrome dye and/or by densitometric analysis using known mass standards (18).
Mobility shift assays
Purifed LexA or recA4 crude extract, prepared as described previously (14), was incubated with radiolabeled promoter DNA (5.0 nM) for 30 min at 25°C in the following incubation buffer: 12 mM HEPES–NaOH (pH 7.9), 4 mM Tris–HCl (pH 7.9), 12% glycerol, 60 mM KCl, 1 mM EDTA, 1 mM DTT, 2 µg poly(dI–dC) poly(dI–dC) and 0.3 mg/ml BSA. This incubation mixture (10 µl) was loaded on a 4% (acrylamide: bisacrylamide ratio of 80:1) non-denaturing polyacrylamide gel and electrophoresis was begun immediately. The buffer within the gel and the running buffer were both 25 mM Tris–HCl (pH 8.5), 250 mM glycine and 1 mM EDTA. Samples were electrophoresed and the dried gel was subjected to densitometric analysis using a BioRad Molecular Imager FX phosphoimaging system. To determine equilibrium dissociation constants, the fraction of DNA bound was plotted against the concentration of unbound LexA, which was determined by subtracting the amount of bound LexA from the amount of total LexA.
Molecular modeling
We built a homology model of the B.subtilis LexA protein using the Protein Local Optimization and Prediction (PLOP) program developed by Jacobson et al. (19) and maintained by the Jacobson laboratory at the University of California, San Francisco. We aligned the B.subtilis LexA sequence with the sequences of E.coli LexA, E.coli UmuD, pKM101 MucA and repressors from 434 and lambda bacteriophages using the ClustalW alignment program (20), which provided identical results to those shown by Luo et al. (21). This alignment assured that we preserved the spatial orientation of important residues. Using PLOP, we created a final model of the B.subtilis LexA protein using the crystal structure of the E.coli LexA protein (21) as a template (PDB ID: 1JHF). Refinement of the model included energy minimization and side chain optimization, using PLOP with a surface implementation of the generalized born implicit solvent model (22). As a control, energy minimization and side chain optimization was also performed on the crystal structure of E.coli LexA resulting in minimal movement of both side chains and the backbone of this protein. We visualized our models and generated molecular graphics images using the Chimera package from the Computer Graphics Laboratory at UCSF [(23) supported by NIH P41 RR-01081].
Docking LexA on the SOS operator consensus sequence
We produced a structural model for the interaction of the 203 amino acid LexA polypeptide with DNA containing the sequence 5-CGAACATATGTTCG-3′ using the Chimera visualization application combined with PLOP's ability to manipulate amino acid dihedral angles. The orientation of helix-III within the half-site major groove was guided by hydroxyl radical footprinting analyses of LexA binding to the dinC (11) and recA (15) operators and by the crystal structure of the HNF3/forkhead domain complexed with DNA (24). The HNF3/forkhead–DNA structure was used as a reference to dock the E.coli LexA DNA binding domain on the SOS operator because the helix–turn–helix motif of HNF3/forkhead closely resembles that of the NMR structure of the LexA DNA binding domain (25). Correspondingly, our homology model of the B.subtilis LexA protein contains a helix–turn–helix DNA binding motif resembling the HNF3/forkhead DNA binding domain. Unfortunately, the coordinates for the HNF3/forkhead complex were never deposited in the PDB and they are not available (S. K. Burley, personal communication). Thus, instead of using a strict superposition of the B.subtilis LexA binding domain onto the HNF3/forkhead–DNA complex, we used Chimera to align helix-III in the half-site major groove. A comprehensive dihedral angle search suggested possible positions of Arg-49 in the DNA interaction.
Mutational analysis of the DNA binding domain
We constructed the following LexA mutants using the quick change mutagenesis kit as described by the manufacturer: Ser41Ala, His44Ala, His46Ala, Ala48Gly, Arg49Ala, Thr52Ala and Lys53Ala. The mutant proteins were purified as described previously (11) and used in mobility shift assays with the recA promoter region.
RESULTS
Measurement of dissociation constants for LexA binding to SOS operators
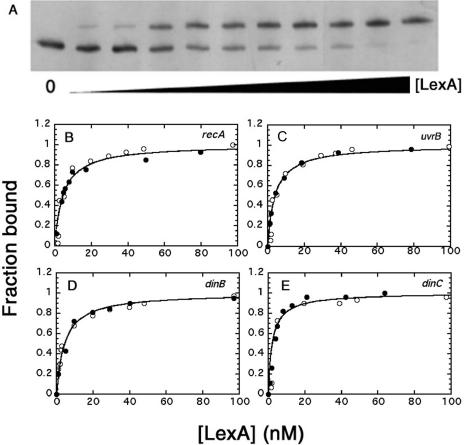
We used mobility shift assays to determine the equilibrium dissociation constants for B.subtilis LexA binding to the SOS operators of four genes, recA, uvrB, dinB and dinC (Table 1). Assays were conducted with purified protein and with a crude extract from B.subtilis cells to assess whether other cellular proteins affect binding. The amount of active LexA in purified samples and in crude extracts was determined by titrating with excess dinB promoter DNA at concentrations of both DNA and repressor that ensured stoichiometric binding (data not shown). Figure 1A shows a typical mobility shift assay with the recA promoter region. Fractional saturation of binding sites was determined by densitometric analysis of unbound and bound bands. For dinC, which shows two bound bands at sub-saturating LexA concentration (14,15), the higher mobility band was assumed to correspond to two sites bound and the lower mobility band to one occupied site. Figure 1B–E shows plots of fractional saturation versus the concentration of unbound LexA for the four promoter regions. The apparent Kd values for the four operators were determined by curve fitting the ligand binding equation, fraction bound = [LexA]/(Kd + [LexA]), to be 4.6, 4.1, 3.6 and 2.3 nM for recA, uvrB, dinB and dinC promoters, respectively. These values are the averages of at least three determinations with standard deviations of less than ±0.5 nM. The dissociation constants for LexA binding to each of the four distinct SOS promoters were not affected by other proteins in a crude extract suggesting that LexA acts alone in the repression of the B.subtilis SOS regulon.
Table 1.
Binding constants for SOS operators of recA, uvrB, dinB and dinC genes
| Gene | Operator | Positiona | Kd (nM) | ΔG (kJ/mol) |
|---|---|---|---|---|
| recA | 5′-CGAATATGCGTTCG-3′ | −73 (−44) | 4.6 | −47.6 |
| uvrB | 5′-CGAACTTTAGTTCG-3′ | −79 (−42) | 4.1 | −47.9 |
| dinB | 5′-AGAACTCATGTTCG-3′ | −42 (−14) | 3.6 | −48.2 |
| dinC (1) | 5′-AGAACAAGTGTTCT-3′ | −85 (−44) | 2.3 | −49.3 |
| dinC (2) | 5′-CGAACGTATGTTTG-3′ | −55 (−14) |
aLocation of 3′ end relative to the ATG codon of the respective gene and (relative to 3′ end of −10 region of canonical promoter sequence).
Figure 1.
Binding of B.subtilis LexA to SOS operators. Graphical analyses of mobility shift titrations of 32P-labelled recA (B), uvrB (C), dinB (D) and dinC (E) promoter DNA (5.0 nM) incubated with increasing concentrations of purified LexA (black circles) or crude extract (open circles). (A) A typical mobility shift assay with the recA promoter as described in Materials and Methods. (B–E) Typical plots of fractional saturation of promoter DNA versus the concentration of 'unbound LexA.
Inhibition studies to assess binding of LexA to recA operator mutants
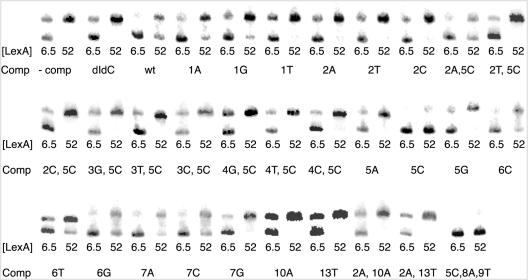
We tested the ability of 26 bp DNA fragments containing recA operator variants to inhibit LexA binding to the wild-type recA operator. Figure 2 shows the results of mobility shift experiments in which LexA was incubated with a radiolabeled 232 bp region of the recA promoter and a 100-fold molar excess of unlabeled recA operator mutant DNA at saturating (52 nM) and subsaturating (6.5 nM) concentrations of LexA. At 52 nM LexA, a 10% inhibition of binding (which is easily detectable in our assay) by a 100-fold molar excess of competitor DNA would correspond to an inhibition constant in the micromolar range—well beyond the range for specific binding (see below). Thus, if no unbound DNA was detected in the presence of competitor DNA at 52 nM LexA, we inferred that the altered operator sequence is not bound specifically by LexA. Moreover, binding at 6.5 nM LexA, in the presence of sequences that did not detectably reduce binding at 52 nM LexA, was similar to that in the presence of poly(dI–dC).
Figure 2.
Inhibition of LexA binding to the recA operator by recA operator mutants. Mobility shift assays were conducted as described in Materials and Methods with purified LexA, radiolabeled recA promoter DNA (5.0 nM) and a 100-fold molar excess of competitor recA operator mutant. Base changes are indicated by position from the 5′ end of the 14 bp operator, and the base substitution relative to the recA operator sequence. Lower and upper bands correspond to unbound and LexA-bound recA promoter DNA, respectively. Lanes corresponding to no addition of competitor DNA and non-specific DNA [poly(dI–dC)] are indicated. LexA concentrations given are for the total amount of LexA.
Identification of recA operator base substitutions that prevent LexA binding
Several single- and double-base substitutions within the 14 bp recA operator sequence did not inhibit LexA binding to the recA operator (Table 2). We further tested the ability of LexA to bind to these sites by titrating radiolabeled mutant fragments directly with LexA. In every case, we did not detect any shift at LexA concentrations under 400 nM. In the absence of excess poly(dI–dC) DNA, LexA concentrations above 400 nM produce a diffuse supershifted band, which we attribute to non-specific LexA binding. A similar supershift is observed at comparably high LexA concentrations with any DNA fragments and the supershifted band can be eliminated by the addition of excess non-specific DNA.
Table 2.
Thermodynamics of LexA binding to recA operator mutants
| Inhibitor sequencea | Kd(app) (nM) | Kd(calc) (nM)b | ΔG (kJ/mol) | |ΔΔG|c (kJ/mol) |
|---|---|---|---|---|
| 5′-CGAACATATGTTCG-3′ | 993 | 0.17 | −34.2 | 0 |
| 5′-CGAACATGCGTTCG-3′ | 767 | 0.22 | −34.9 | 0.7 |
| 5′-AGAACATGCGTTCG-3′ | 391 | 0.42 | −36.6 | 2.4 |
| 5′-CGAACTTGCGTTCG-3′ | 337 | 0.49 | −36.9 | 2.7 |
| 5′-CGAACCTGCGTTCG-3′ | 299 | 0.56 | −37.2 | 3.3 |
| 5′-CGAACAAGCGTTCG-3′ | 268 | 0.62 | −37.5 | 3.4 |
| 5′-GGAACATGCGTTCG-3′ | 166 | 1.0 | −38.6 | 4.4 |
| 5′-CGAATATGCGTTCG-3′(wt) | 40.3 | 4.6 | −42.2 | 8.0 |
| 5′-CAAACATGCGTTCG-3′ | 36.3 | 5.2 | −42.4 | 8.2 |
| 5′-CGAATGTGCGTTCG-3′ | 30.4 | 6.4 | −42.9 | 8.7 |
| 5′-CGAATACGCGTTCG-3′ | 25.6 | 7.8 | −43.4 | 9.1 |
| 5′-AGAATATGCGTTCG-3′ | 19.0 | 11.4 | −44.0 | 9.8 |
| 5′-CGAATTTGCGTTCG-3′ | 18.9 | 11.5 | −44.1 | 9.9 |
| 5′-CGAATAAGCGTTCG-3′ | 14.2 | 17.1 | −44.8 | 10.6 |
| 5′-CGAATCTGCGTTCG-3′ | 12.8 | 20.0 | −45.0 | 10.8 |
| 5′-GGAATATGCGTTCG-3′ | 7.7 | 53.0 | −46.3 | 12.1 |
| recA operator base substitutions that prevent LexA binding | ||||
| 5′-TGAATATGCGTTCG-3′ | 5′-CTAATATGCGTTCG-3′ | |||
| 5′-CCAATATGCGTTCG-3′ | 5′-CGTATATGCGTTCG-3′ | |||
| 5′-CGCATATGCGTTCG-3′ | 5′-CGGATATGCGTTCG-3′ | |||
| 5′-CGATTATGCGTTCG-3′ | 5′-CGACTATGCGTTCG-3′ | |||
| 5′-CGAGTATGCGTTCG-3′ | 5′-CGAAGATGCGTTCG-3′ | |||
| 5′-CGAAAATGCGTTCG-3′ | 5′-CGAATAGGCGTTCG-3′ | |||
| 5′-CTAACATGCGTTCG-3′ | 5′-CCAACATGCGTTCG-3′ | |||
| 5′-CGAATATGCATTCG-3′ | 5′-CGAATATGCGTTTG-3′ | |||
| 5′-CAAACATGCATTCG-3′ | 5′-CAAACATGCGTTTG-3′ | |||
aBases in boldface are changes relative to the wild-type recA operator.
bKd values were determined by calculating KI from the relationship Kd(app) = Kd(recA) (1 + [I]/KI) and then multiplying the KI values by the ratio Kd(recA)/KI (wt).
cΔΔG values are given as absolute values.
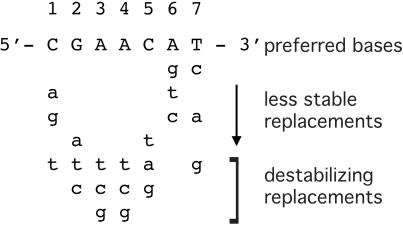
Our results indicate that the AT base pairs at the third and fourth positions within a half-site, or monomer binding site, are essential for operator binding because replacement with any other base pairs eliminates competition with the labeled recA DNA for LexA binding. The G and C at the second and fifth half-site positions of the consensus sequence 5′-CGAAC-3′ are also critical because two mismatches at these sites, in either half-site, eliminates competition with the labeled recA DNA for LexA binding. They are not essential because substitution with A at the second site (as in the dinC operator) and T at the fifth site (as in the recA operator) do not eliminate site-specific operator binding. Other bases that destabilize LexA binding to the recA operator are T at the first site and G at the seventh site.
Identification of recA operator base substitutions that do not prevent LexA binding
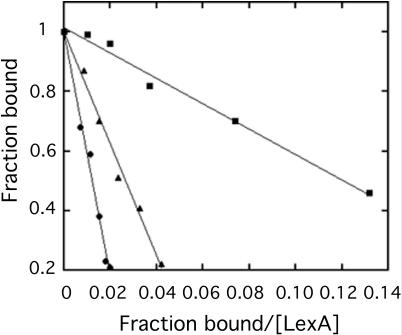
The recA operator mutants that inhibited LexA binding to the recA operator are listed in Table 2. Relative binding affinities of LexA for the operator mutants were assessed by determining the apparent Kd for binding to the recA operator in the presence of competitor DNA. Figure 3 shows representative Scatchard plots for two DNA fragments that displaced LexA from the recA operator to varying degrees. The sequence that was most effective at inhibiting LexA binding was 5′-CGAACATATGTTCG-3′, which corresponds to the defined consensus sequence plus an internal ATAT (also found in the center of the E.coli SOS operator consensus sequence). Comparison of the KI values, calculated from the standard relationship for Kd in the presence of a competitive inhibitor, Kd(app) = Kd (1 + [I]/KI), shows that this preferred sequence binds LexA ∼30 times tighter than the recA operator (Table 3). The sequence 5′-CGAACATGCGTTCG-3′, corresponding to the recA operator with a single mutation at position 5 (changing it to the consensus sequence 5′-CGAAC-3′), was nearly as effective. ΔG values were determined from the apparent binding constants using the Gibbs free energy equation, ΔG = −RTlnKd(app), and ΔΔG values were calculated relative to the free energy change for the sequence 5′-CGAACATATGTTCG-3′. The degrees to which specific base substitutions destabilize binding relative to the preferred sequence are summarized qualitatively in Figure 4.
Figure 3.
Representative Scatchard plots for quantifying inhibition of LexA binding to the recA operator by operator mutants. Mobility shift assays were conducted as described in Materials and Methods with purified LexA, radiolabeled recA promoter DNA (5.0 nM), and a 100-fold molar excess of wild-type recA operator (black circles), recA operator with A substituted for C at the first position (black triangles) and no competitor (black squares). LexA concentrations used were for unbound LexA.
Table 3.
Thermodynamics of LexA mutants binding to the recA operator
| LexA | Kd (nM) | ΔΔG (kJ/mol) |
|---|---|---|
| Wild-type | 4.6 | |
| Ser41Ala | 6.0 | 0.7 |
| His44Ala | 3.7 | 0.5 |
| His46Ala | >400 | |
| Ala48Gly | 22 | 3.9 |
| Arg49Ala | >400 | |
| Thr52Ala | 6.6 | 0.9 |
| Lys53Ala | 58 | 6.3 |
Figure 4.
Sequence requirements for LexA binding. The preferred half-site sequence based on thermodynamic analysis of LexA binding to recA operator mutants. Base substitutions labeled as destabilizing abolish LexA binding to the recA operator.
In contrast with an earlier study where recA operator mutants were assayed for their ability to inhibit LexA binding in vivo (15), our results indicate some sequence preference for the internal 4 bp by LexA. Footprinting analyses of LexA binding to the recA (15) and dinC (11) operators show that LexA interacts with the CGAAC sequence in the major groove and that any interactions with the four internal base pairs would likely be in the minor groove. The fact that LexA interacts with a GC sequence at the sixth and seventh position nearly as well as an AT base pair suggests that the interactions are not specific, but rather due to a distortion or bending of the DNA. To test this hypothesis, we used computer modeling to construct a model of LexA bound to the recA operator.
Modeling the B.subtilis LexA DNA binding domain
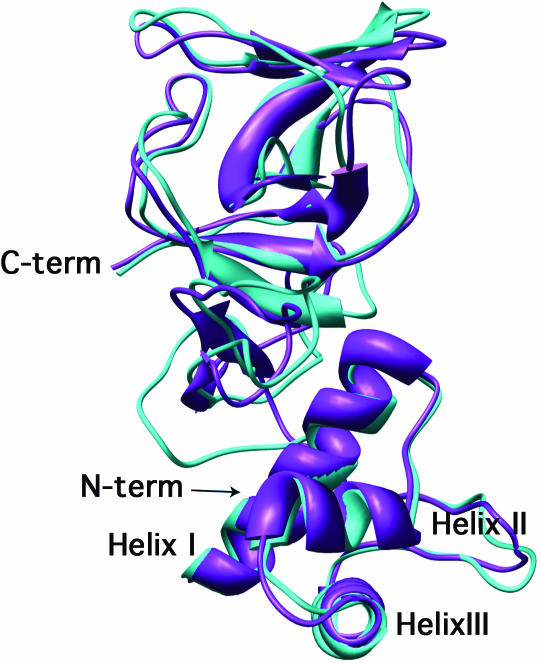
The E.coli LexA DNA binding domain is contained within the first 84 amino acid residues (26) and shares 32% sequence identity and an additional 21% amino acid similarity with the corresponding region of B.subtilis LexA. Based on the crystal structure of the E.coli LexA repressor (21), we constructed a model of B.subtilis LexA as described in Materials and Methods. Figure 5 shows a backbone trace of the E.coli LexA crystal structure overlayed with our model of B.subtilis LexA. The energy minimization and side chain optimization techniques used to construct the B.subtilis repressor structure were also applied to the E.coli LexA crystal structure and did not perturb the overall structure of the E.coli protein.
Figure 5.
Computer model of B.subtilis LexA superimposed on the structure of E.coli LexA. A homology model of the B.subtilis LexA protein (turquoise) was superimposed on the crystal structure of the E.coli LexA protein (purple) as described in Materials and Methods.
Similar to its E.coli counterpart, our model of the B.subtilis LexA DNA binding domain has a helix–turn–helix motif. The tertiary structure contains three α-helices: helix I (residues 5–20), helix II (residues 29–34) and helix III (residues 39–54). Helices I and II are separated by a turn of seven amino acid residues and helices II and III are separated by a turn of five residues. A β-sheet spans residues 55–69 and residues 70–75 correspond to part of the hinge region that connects the N-terminal DNA binding domain and the C-terminal dimerization domain of E.coli LexA.
Mutational analysis of the LexA DNA binding domain
The E.coli LexA DNA binding site primarily comprises helix III (25,27,28), which is believed to interact with an operator half-site in the major groove (25,29). According to our model, the hydrophobic residues of helix III of B.subtilis LexA (Val-43, Leu-47 and Leu-50) are packed into the interior of the protein, whereas the remaining residues are on the helix surface that is accessible to solvent. Although, in theory, all residues on helix III that are accessible to solvent can potentially interact with the DNA, spatial constraints suggest that only seven of these residues have the potential to make contacts with operator DNA in the major groove: Ser-41, His-44, His-46, Ala-48, Arg-49, Thr-52 and Lys-53. We used site-specific mutagenesis to replace each of these solvent accessible residues with alanine (except Ala-48, which was replaced by glycine). Table 3 shows the equilibrium dissociation constants for each of the LexA mutants with the recA operator. Only four of the seven mutants showed significant binding deficiencies: H46A, A48G, R49A and K53A. The R49A and H46A mutants are essential for binding because they abolish operator binding. The A48G and K53A mutants reduce binding by 5- and 13-fold, respectively.
Docking the DNA binding domain model with operator DNA
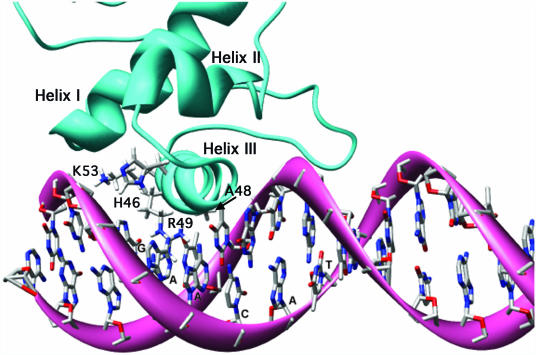
For docking studies we constructed a model of the recA promoter with the operator replaced by the preferred SOS operator, 5′-CGAACATATGTTCG-3′. Docking helix III of a LexA monomer to an operator half-site was guided by (i) the mutational analyses described above; (ii) hydroxyl-radical footprinting results for B.subtilis LexA binding the dinC (11) and recA (15) operators; and (iii) the crystal structure of HNF3/forkhead–DNA complex (24), which was used to model the E.coli LexA–operator complex because the helix–turn–helix motif of HNF3/forkhead closely resembles that of the NMR structure of the LexA DNA binding domain (25). The result is shown in Figure 6.
Figure 6.
Computer model of the putative B.subtilis LexA DNA binding domain docked at one half-site of the preferred operator sequence CGAACATATGTTCG. The homology model depicted in Figure 5 was docked on the preferred operator sequence in silico as described in Materials and Methods.
According to the model and consistent with our mutational analyses, the amino acids best poised for interactions with the operator within the major groove are His-46, Ala-48, Arg-49 and Lys-53. The docking analysis places Arg-49 in a position where it could form hydrogen bonds with one or both of the two essential AT base pairs. Ala-48 appears to contribute to the interactions with either or both AT base pairs by providing favorable hydrophobic contacts. The model indicates the potential for His-46 and Lys-53 to form either non-specific interactions with the phosphate backbone, which could help to orient and anchor the binding domain, or specific interactions with the GC base pair in the second position. The specific base pair interactions could be facilitated by bridging water molecules or helix rotation, which may accompany bending of the DNA (see below). Such a rotation would orient Arg-49 to interact with both adenines without a change in the overall conformation of the arginine side chain and would bring His-46 and Lys-53 closer to the GC base pair.
There are no obvious potential interactions between helix III residues and the first and fifth positions of the half-site. However, our model suggests other regions of the protein that may be capable of interacting with these base pairs. The alpha carbon of Arg-58 is located on the portion of the protein that overhangs the internal ATAT region between the two half-sites, but the guanidinium group of this residue is located next to the binding helix and could make contact with the cytosine at the fifth position. Our model also shows a hydrogen bond between the epsilon nitrogen of His-46 and the guanidinium group of Arg-7. Although not part of helix III, this arginine appears well positioned to interact with the phosphate backbone when hydrogen bonded to His-46. On the other hand, if there is no hydrogen bond between these residues and the side chains are free to rotate, it would be possible for Arg-7 to interact with the CG base pair at the first position. Thus, the only part of the operator that does not appear to be capable of specific interactions between base pairs and LexA residues is the ATAT internal region.
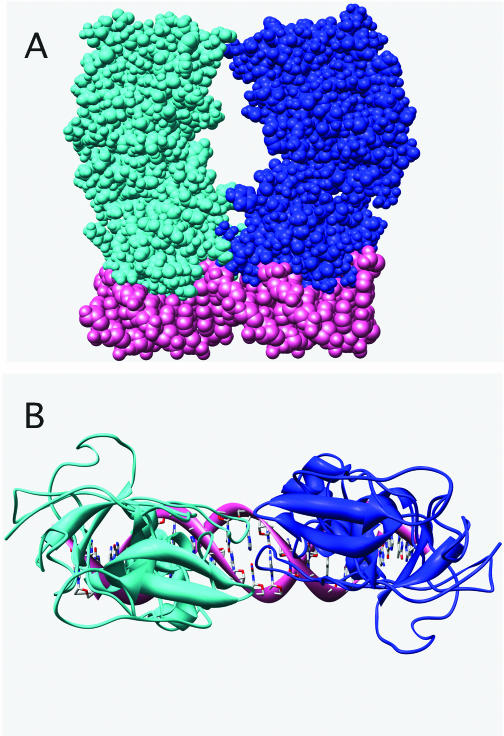
When a second monomer is similarly docked at the other half-site (Figure 7), the C-terminal domains have little contact with each other. This seems highly unlikely given the evidence implicating the C-terminal domain of E.coli LexA in the dimerization that stabilizes site-specific binding (29). In order for the C-terminal domains of our model to interact effectively, LexA must undergo a substantial conformation change and/or the DNA must bend. It is easy to imagine from Figure 7 that bending of the operator DNA could facilitate effective interactions between the C-terminal domains of the two monomers. Such bending would likely occur in the internal region between the two half-sites and is consistent with the sequence preference in this region, despite no apparent interactions between the base pairs and LexA.
Figure 7.
Computer model of LexA dimer bound to the preferred operator sequence CGAACATATGTTCG. A second LexA monomer was docked on the preferred operator sequence in the same orientation as shown in Figure 6. Side view (A) and top view (B) of the model of the LexA dimer bound to DNA show that distortion of the DNA and/or LexA must occur for the C-terminal domains to interact effectively.
DISCUSSION
We analyzed the binding affinity of B.subtilis LexA for four known SOS operators and recA operator mutants using mobility shift assays to determine equilibrium binding constants. We found that binding affinity varies slightly for the four different operator sequences ranging from 2.3 nM for dinC to 4.6 nM for recA, consistent with a qualitative analysis of binding to these operators that suggested similar affinities (15). In contrast, E.coli LexA has a wide range of binding constants for SOS operators (27).
Our characterization of LexA binding to recA operator mutants has revealed a preference for the sequence, 5′-CGAACATATGTTCG-3′. This differs from the preferred sequence reported by Winterling et al. 5′-t/c GAA t/c g/a NNCGT g/t C g/t-3′, based on an analysis of β-galactosidase activity in recA-lacZ fusion strains containing recA operator mutants (15). One explanation for this difference is that sequences outside the operator region may affect the interaction of LexA with operator bases in vivo. This would also explain why the preferred site for a homodimer in vivo is not symmetrical. However, their reported consensus sequence, 5′-CGAACRNRYGTTCG-3′, based on computational analyses of putative SOS boxes in other gram-positive organisms, is identical to our preferred sequence except for the four internal base pairs, which are consistent with our results showing similar affinity with either ATAT or GCGC in these positions.
Although our model of the LexA–DNA complex indicates that the repressor does not make sequence-specific contacts with the internal 5′-ATAT-3′ region, our binding results suggest that this region contributes to binding affinity. Thus, the effects of this region on binding may be due to a distortion in the DNA that facilitates interactions between monomers. The effects of non-contacted bases between operator half-sites have been well studied for the bacteriophage 434 and P22 repressors, for which operators with central T-A base pairs bind repressor more strongly than those containing C-G pairs (30,31). Crystallographic studies have shown that the non-contacted operator bases are overwound and distorted when bound to the 434 repressor (32) and recent evidence indicates that the presence of the N2-NH2 group of guanine may oppose repressor induced overwindng (33). Our results with G and C at the sixth and seventh half-site positions argue against a similar effect of this amino group on LexA binding.
Our model of two LexA monomers docked at the two half-sites supports the hypothesis that distortion of the DNA contributes to LexA binding affinity because, unless the DNA bends (or LexA undergoes a substantial conformational change), there is not enough contact between the C-terminal dimerization domains for them to interact effectively. Extensive interaction between dimerization domains is supported by the fact that SOS genes are derepressed in vivo by proteolytic removal of the C-terminal domain and by biochemical analyses of the E.coli LexA protein; the E.coli repressor has a dimer dissociation constant in the picomolar range and dimerization involves the burial of a large surface area (34).
Assuming that LexA does not undergo a substantial conformation change, our computer model of the bound complex is consistent with our mutational analysis of the helix III amino acids and the orientation of the dimer is entirely consistent with footprinting analyses of the recA and dinC promoters (11,15). Although it is not possible to know the exact positions of amino acid side chains, we believe the position of the helix itself is accurate because a slight rotation or translation of the helix relative to the DNA either puts the essential amino acids too far away from the major groove or results in steric hindrance between the protein and the DNA. Of course, our model assumes that the 3D structure of B.subtilis LexA is similar to that of its E.coli counterpart and that, similar to its counterpart, helix III is the part of the DNA binding domain that interacts in the major groove. It is noteworthy that two of the four amino acid residues that contribute to binding, His-46 and Lys-53, are conserved in the E.coli LexA protein, which recognizes a completely different operator sequence. However, the computer model of E.coli LexA bound to its operator (25) indicates that the amino acids involved in specific interactions—Ser-39, Asn-41, Ala-42, Glu-44 and Glu-45—are closer to the N-terminal end of the helix than the interacting B.subtilis LexA residues suggested by our model. The difference in the helix positions relative to the DNA may reflect the fact that the operators are different lengths; according to the models, there are four non-contacted base pairs between the two half-sites in B.subtilis and eight in E.coli operators.
Considering that only five LexA binding sites have been identified in B.subtilis, a significant outcome of this report is our determination of the minimum sequence requirements for LexA binding as summarized in Figure 4, i.e. our results should provide a guide for identifying other potential SOS genes. In fact, we report elsewhere that these sequence requirements have served as a basis for identifying 40 potential binding sites of which 33 are bound specifically by LexA (35). In contrast, a search for a 14mer with up to five mismatches relative to our preferred sequence gives ∼18 000 possible binding sites.
Acknowledgments
We thank Matthew Jacobson for helpful discussions about computer modeling. This work was supported by NSF Grants MCB-9601398 and MCB-0135899. Funding to pay the Open Access publication charges for this article was provided by Williams College.
Conflict of interest statement. None declared.
REFERENCES
- 1.Little J.W., Mount D.W. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E.C., Walker G.C., Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. pp. 407–522. [Google Scholar]
- 3.Walker G.C. The SOS response of Escherichia coli. In: Neidhardt F.C., editor. Escherichia coli and Salmonella. Washington, DC: ASM Press; 1996. [Google Scholar]
- 4.Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc. Natl Acad. Sci. USA. 1981;78:4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little J.W., Mount D.W., Yanisch-Perron C.R. Purified lexA protein is a repressor of the recA and lexA genes. Proc. Natl Acad. Sci. USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez De Henestrosa A.R., Ogi T., Aoyagi S., Chafin D., Hayes J.J., Ohmori H., Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 7.Sassanfar M., Roberts J.W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 8.Little J.W. Autodigestion of lexA and phage lambda repressors. Proc. Natl Acad. Sci. USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winterling K.W., Levine A.S., Yasbin R.E., Woodgate R. Characterization of DinR, the Bacillus subtilis SOS repressor. J. Bacteriol. 1997;179:1698–1703. doi: 10.1128/jb.179.5.1698-1703.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raymond-Denise A., Guillen N. Expression of the Bacillus subtilis dinR and recA genes after DNA damage and during competence. J. Bacteriol. 1991;173:7084–7091. doi: 10.1128/jb.174.10.3171-3176.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller M.C., Resnick J.B., Smith B.T., Lovett C.M. The Bacillus subtilis dinR gene codes for the analogue of Escherichia coli LexA. Purification and characterization of the DinR protein. J. Biol. Chem. 1996;271:33502–33508. [PubMed] [Google Scholar]
- 12.Cheo D.L., Bayles K.W., Yasbin R.E. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J. Bacteriol. 1991;173:1696–1703. doi: 10.1128/jb.173.5.1696-1703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheo D.L., Bayles K.W., Yasbin R.E. Elucidation of regulatory elements that control damage induction and competence induction of the Bacillus subtilis SOS system. J. Bacteriol. 1993;175:5907–5915. doi: 10.1128/jb.175.18.5907-5915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovett C.M., Cho K.C., O'Gara T.M. Analysis of the SOS inducing signal in Bacillus subtilis using Escherichia coli LexA as a probe. J. Bacteriol. 1993;175:6842–6849. doi: 10.1128/jb.176.16.4914-4923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winterling K.W., Chafin D., Hayes J.J., Sun J., Levine A.S., Yasbin R.E., Woodgate R. The Bacillus subtilis DinR binding site: redefinition of the consensus sequence. J. Bacteriol. 1998;180:2201–2211. doi: 10.1128/jb.180.8.2201-2211.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love P.E., Lyle M.J., Yasbin R.E. DNA-damage-inducible (din) loci are transcriptionally activated in competent Bacillus subtilis. Proc. Natl Acad. Sci. USA. 1985;82:6201–6205. doi: 10.1073/pnas.82.18.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yasbin R.E., Fields P.I., Anderson B.J. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene. 1980;12:155–159. doi: 10.1016/0378-1119(80)90026-8. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidham J.G., Smith J.A., Struhl K. Current Protocols in Molecular Biology. NY: John Wiley & Sons; 1998. [Google Scholar]
- 19.Jacobson M.P., Kaminski G.A., Friesner R.A., Rapp C.S. Force field validation using protein side chain prediction. J. Phys. Chem. B. 2002;106:11673–11680. [Google Scholar]
- 20.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Y., Pfuetzner R.A., Mosimann S., Paetzel M., Frey E.A., Cherney M., Kim B., Little J.W., Strynadka N.C. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh A., Rapp C.S., Friesner R.A. Generalized born model based on a surface integral formulation. J. Phys. Chem. B. 1998;102:10983–10990. [Google Scholar]
- 23.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 24.Clark K.L., Halay E.D., Lai E., Burley S.K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 25.Knegtel R.M., Fogh R.H., Ottleben G., Ruterjans H., Dumoulin P., Schnarr M., Boelens R., Kaptein R. A model for the LexA repressor DNA complex. Proteins. 1995;21:226–236. doi: 10.1002/prot.340210305. [DOI] [PubMed] [Google Scholar]
- 26.Hurstel S., Granger-Schnarr M., Daune M., Schnarr M. In vitro binding of LexA repressor to DNA: evidence for the involvement of the amino-terminal domain. EMBO J. 1986;5:793–798. doi: 10.1002/j.1460-2075.1986.tb04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnarr M., Oertel-Buchheit P., Kazmaier M., Granger-Schnarr M. DNA binding properties of the LexA repressor. Biochimie. 1991;73:423–431. doi: 10.1016/0300-9084(91)90109-e. [DOI] [PubMed] [Google Scholar]
- 28.Fogh R.H., Ottleben G., Ruterjans H., Schnarr M., Boelens R., Kaptein R. Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy. EMBO J. 1994;13:3936–3944. doi: 10.1002/j.1460-2075.1994.tb06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumoulin P., Ebright R.H., Knegtel R., Kaptein R., Granger-Schnarr M., Schnarr M. Structure of the LexA repressor–DNA complex probed by affinity cleavage and affinity photo-cross-linking. Biochemistry. 1996;35:4279–4286. doi: 10.1021/bi9529162. [DOI] [PubMed] [Google Scholar]
- 30.Koudelka G.B., Carlson P. DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature. 1992;355:89–91. doi: 10.1038/355089a0. [DOI] [PubMed] [Google Scholar]
- 31.Wu L., Vertino A., Koudelka G.B. Non-contacted bases affect the affinity of synthetic P22 operators for P22 repressor. J. Biol. Chem. 1992;267:9134–9139. [PubMed] [Google Scholar]
- 32.Aggarwal A., Rodgers D.W., Drottar M., Ptashne M., Harrison S.C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 33.Mauro S.A., Pawlowski D., Koudelka G.B. The role of the minor groove substituents in indirect readout of DNA sequence by 434 repressor. J. Biol. Chem. 2003;278:12955–12960. doi: 10.1074/jbc.M212667200. [DOI] [PubMed] [Google Scholar]
- 34.Mohana-Borges R., Pacheco A.B., Sousa F.J., Foguel D., Almeida D.F., Silva J.L. LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein–protein interactions. J. Biol. Chem. 2000;275:4708–4712. doi: 10.1074/jbc.275.7.4708. [DOI] [PubMed] [Google Scholar]
- 35.Au N., Kuester-Schoeck E., Mandava V., Bothwell L.E., Canny S.P., Chachu K., Colavito, Fuller S.A., Groban E.S., Hensley L.A., et al. The genetic composition of the Bacillus subtilis SOS system. J. Bacteriol. 2005;187 doi: 10.1128/JB.187.22.7655-7666.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]