Abstract
Escherichia coli polynucleotide phosphorylase (PNPase), a protein that has both ribonucleolytic and synthetic capabilities, binds, along with the 48-kDa glycolytic enzyme enolase, the 50-kDa DEAD-box protein RhlB helicase and other cellular proteins to the C-terminal “scaffold” region of RNase E to form a complex termed the RNA degradosome. PNPase itself has been reported to exist as a complex (α3β2) containing trimers of a catalytic subunit (α) and dimers of another subunit (β). The β-subunit has been believed to be enolase; we report here that it is instead the RhlB helicase. Whereas interaction between PNPase-α and enolase was observed in bacteria that synthesize RNase E having a scaffold region, immunoprecipitates from cells expressing PNPase-α, RhlB, and enolase from single-copy chromosomal loci, plus a mutant RNase E protein lacking its C-terminal half, showed direct association of PNPase-α only with RhlB. Using affinity chromatography, we found that PNPase-α and RhlB form a ribonucleolytically active complex corresponding to the mass calculated previously for α3β2 (i.e., 377–380 kDa), whereas no association between PNPase-α and enolase was detected. Chromosomal deletion of the eno gene had no effect on the ability of PNPase to degrade either single- or double-stranded RNAs. Collectively, our findings show that direct interaction between PNPase-α and RhlB occurs physiologically in the absence of the RNase E C-terminal region, that enolase association with PNPase-α is a consequence of the interaction of both proteins with RNase E, and that, contrary to current notions, enolase is not the β-subunit of E. coli PNPase complex.
Keywords: degradosome, RNase E
Polynucleotide phosphorylase (PNPase) is a major 3′ to 5′ exoribonuclease of Escherichia coli and functions both in the degradation of mRNA and stable RNA species and as a poly(A) polymerase (1, 2). The enzyme initially was discovered by Grunberg-Manago et al. in 1955 (3) in Azotobacter vinelandii. Subsequently, its enzymatic activity was detected among eubacteria (4, 5), Archea (6), eukaryotic microbes (7), plants (8), and animal cells (9). “Degradosome” complexes containing PNPase, RNase E, the RhlB helicase, enolase, and other proteins have been isolated from E. coli and other bacteria (10–13) and recently have been shown to exist also in vivo (14) and to function as ribonucleolytic machines (15, 16). In yeast and animal cells, several PNPase-related 3′ to 5′ exonucleases have been identified and shown, along with an RNA helicase, to form an “exosome” complex (17–19). Because of its occurrence in a broad spectrum of organisms and its involvement in a variety of ribonucleolytic complexes (for recent reviews, see refs. 20 and 21), the identification and characterization of protein complexes containing PNPase continue to be of general interest.
In crude cell extracts of E. coli, PNPase displays heterogeneity (22). Early in its history, PNPase was shown by Portier (23) to exist in two active forms, A or B, having molecular masses of ≈252 and 365 kDa, respectively. The A form contains three identical catalytic (α) subunits and is present as a homotrimeric exoribonuclease (i.e., the α3 type structure), whereas the B form consists of two types of chains, α and β, and has been assigned a structure of α3β2 (23). The molecular mass of the β chain was determined by Portier to range between 48 and 50 kDa (23). Although it was concluded by Carpousis et al. (24) and has been generally accepted (e.g., refs. 11, 25, and 26) that the PNPase β-subunit is the 48-kDa protein enolase, direct association of PNPase-α with enolase has not been detected (refs. 25, 27, and 28; see Discussion), and whether PNPase-β is in fact enolase has remained unclear.
The experiments reported here demonstrate that direct interaction between PNPase-α and RhlB occurs when these proteins are expressed in vivo at physiological levels from single-copy chromosomal genes in bacterial strains that lack the C-terminal region of RNase E, and that consequently are unable to assemble degradosomes. Using M2 affinity column purified overexpressed FLAG-tagged protein complexes from the above bacterial strains, we found no detectable association between PNPase-α and enolase but instead detected a ribonuclease-insensitive complex consisting of PNPase-α and RhlB helicase. The PNPase protein complex isolated from bacteria lacking enolase showed no difference in its ability to degrade either single-stranded or duplex RNAs, whereas RhlB is necessary for PNPase-α to efficiently degrade structured mRNAs (refs. 11 and 27 and our current data). Biochemical reconstitution of PNPase-α–RhlB protein complex in vitro yielded an enzymatically active product having a molecular mass of 377–380 kDa, consistent with the α3β2 configuration. Collectively, the genetic and biochemical experiments we report prove that enolase is not the β-subunit of the B form PNPase complex, but rather that the 50-kDa RhlB RNA helicase is PNPase-β. The association of an RNA helicase and PNPase-α in an exoribonucleolytic protein complex of E. coli has thus been evolutionally conserved in eukaryotic exosome complexes.
Materials and Methods
Bacterial Strains and Culture Conditions. Bacterial strains BL(DE3) rne131 (16) and BL21(DE3) rne131ΔrhlB (this work; see below) were cultured in LB media (29) supplemented with the antibiotics kanamycin (50 μg/ml) or ampicillin (100 μg/ml). BL21(DE3) rne131ΔrhlB mutant was constructed by P1 transduction by using the ΔrhlB mutant SU02 (15) as a donor and BL(DE3) rne131 as the recipient, as described (30). The BL(DE3) rne131 and BL21(DE3) rne131ΔrhlB bacterial strains were used to study the consequent effects of ΔrhlB on activities of the PNPase complex (i.e., Fig. 5). The method used to construct Δeno was described previously (31), except that screening was performed on M9 agar supplemented with 0.2% glycerol/0.2% tryptone/40 mM succinate/1 mM l-arabinose plus antibiotics kanamycin (50 μg/ml) and ampicillin (100 μg/ml). BL21(DE3)Δeno was constructed also by P1 transduction by using BL21(DE3) (16) as the recipient, as described (30). When the isogenic strains BL21(DE3) and BL21(DE3)Δeno were used for the study of the effects of the Δeno mutation on the PNPase complex (i.e., Fig. 4), they were grown in M9 with above mentioned supplements plus 1% casamino acids. The DNA sequences of individual primers used in the construction of the Δeno mutant and their derivatives, as well as strain verification by Southern blotting analysis, are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
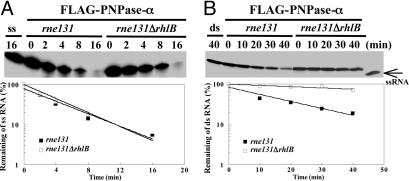
Fig. 5.
Determining the activity of PNPase-α affected by ΔrhlB mutation in the rne131 mutant. Strains contained pflag-PNP were incubated at 30°C in LB medium, and 0.5 mM isopropyl β-d-thiogalactoside was added for 2 h at OD600 = 0.6–0.1. Protein purification was as described (10). The FLAG–PNPase-α complexes were purified from the isogenic pair of E. coli strains BL21(DE3)rne131 and BL21(DE3)rne131ΔrhlB, as shown. The same amounts of FLAG–PNPase-α complexes with and without RhlB were used to determine the PNPase activity for single-stranded (A) and duplex RNA substrates (B), as described in Fig. 4.
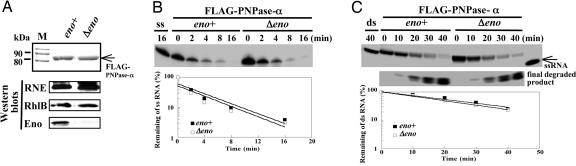
Fig. 4.
Determining the activity of PNPase-α affected by Δeno mutation. Strains containing pflag-PNP were incubated at 30°C in M9 medium plus 0.2% glycerol/40 mM succinate/1% casamino acids, then 0.05 mM isopropyl β-d-thiogalactoside was added for 2 h at OD540 = 0.6–0.1. Protein purification was as described (10). (A) Coomassie blue staining and Western blot analysis of purified FLAG–PNPase-α complexes from an isogenic pair of E. coli strains: eno wild-type [BL21(DE3), ref. 16] and eno mutant [BL21(DE3)Δeno], shown as eno+ and Δeno, respectively. The same amount of the FLAG–PNPase-α complex, with or without enolase, was used to study PNPase-α activity on single-stranded (ss) (B) and double-stranded (ds) RNA (C) substrates. “ss” indicates gel-purified 5′-32P labeled 22-mer RNA substrates incubated under the same conditions without enzyme (described in Supporting Materials and Methods); the ds-RNA unwinding reaction was carried out under the same conditions as in B, except 3 mM ATP was used. All reactions were performed at 30°C. Reaction time points are as shown. Individual reaction products in B were separated in 20% 7 M urea PAGE; the reaction products of duplex RNA in C were resolved by 16% native PAGE. Semilogarithmic plots show the remaining ss- or ds-RNA substrates revealed by the phosphorimager (FLA-5000, Fujifilm).
Immunoprecipitation. Cell cultures (100 ml) were grown at 37°C in LB medium to stationary phase. Cell pellets were resuspended in lysis buffer (50 mM Tris·HCl, pH 7.5/0.2 M NaCl/3 mM EDTA/5% glycerol/1 mM PMSF), lysed by French press twice, and centrifuged at 18,000 × g for 20 min at 4°C. The protein concentration of supernatant was estimated (32) by using BSA as the standard. Two milligrams of the supernatant was used for all immunoprecipitations with anti-PNPase, anti-RhlB, or anti-enolase antibodies at 4°C for 2 h and were precipitated by addition of Protein A-Sepharose (Amersham Pharmacia) overnight at 4°C. The protein-bead pellets were washed three times with buffer A (50 mM Tris·HCl, pH 7.5/0.2 M NaCl) and resuspended in SDS/PAGE loading buffer. The beads were removed by centrifugation, and the supernatants were heated at 100°C for 5 min and loaded onto a 8% SDS/PAGE gel. Western blot analyses were performed as described (10). To detect enolase primary antibodies, we used a mouse monoclonal anti-rabbit IgG light-chain specific secondary antibody (1:6,000, Jackson ImmunoResearch) to avoid detecting the IgG heavy chain, which has a molecular mass similar to that of enolase.
Plasmid Constructions and Complementation Assays. Plasmids pflag-PNP, -RhlB, and -ENO were previously described (27). Other plasmid constructions and complementation assays are described in Supporting Materials and Methods.
Protein Purification, in Vitro Reconstitution, and Gel Filtration. All FLAG-tagged individual proteins were purified from the BL(DE3) rne131 strain (16) by using an anti-M2 affinity column, as described (10). The purification of RhlBhis from BL21(DE3) rne131 followed the procedures outlined in the manual (Novagen). To purify the PNPase-α–RhlBhis complex, it was first concentrated by using a Centripep YM-3 (Amicon) with buffer A (50 mM Tris·HCl, pH 7.5/0.2 M NaCl), which permitted removal of an excess of the FLAG peptide used in the initial purification of the FLAG-tagged PNPase-α. In vitro reconstitution of PNPase-α–RhlBhis complex was carried out by incubating purified FLAG-tagged PNPase-α with anti-M2 gel resin (Sigma) at 4°C overnight. This mixture was then packed into a PolyPrep chromatography column (Bio-Rad), to which was added purified RhlBhis protein. The reconstituted FLAG-tagged PNPase-α–RhlB complex was washed with buffer A to remove weakly interacting proteins and then eluted by FLAG peptide. The FLAG-tagged PNPase-α–RhlB reconstituted complex was concentrated by using Amicon Ultra YM-5 (Amicon) and purified by gel filtration on a Superdex200 HR10/30 column (Amersham Pharmacia Biosciences), as described in Supporting Materials and Methods.
Denaturing and Native PAGE and Western Blot Analysis. The antibodies used for Western blotting and gel electrophoresis analysis are described in Supporting Materials and Methods.
RNA Degradation and Unwinding Assays. The RNA oligomer (22 mer; 5′-ACA GUA UUU-GGU ACU GCG CUC U) used as a single-stranded RNA substrate synthesized by Dharmacon Research, Lafayette, CO, is identical to the 5′-end sequences of RNAI, the antisense RNA of ColEI-type plasmids (33); the 22 oligomer was labeled with 32P at the 5′ end by using T4 polynucleotide kinase and purified as described (34, 35). A short 11-mer RNA oligo (5′-AGC GCA GUA CC) complementary to the 3′ end of the 22-mer (underlined region) was used to form duplex RNA for RNA unwinding assays (27). Single-stranded RNA cleavage and RNA unwinding assay procedures are described in Supporting Materials and Methods.
Results
E. coli Strains Producing RNase E Lacking the C-Terminal Region Show Direct Association of PNPase-α with RhlB Protein But Not Enolase. A polypeptide region near the C terminus of RNase E directly binds to all three degradosome component proteins, PN-Pase-α, enolase, and RhlB helicase (25, 36, 37). To eliminate possibly confounding effects of RNase E binding in the interpretation of interactions between other cellular proteins and PNPase-α, an E. coli strain [BL21(DE3) rne131, ref. 16] expressing a mutant RNase E lacking its C-terminal half was used to identify proteins associating directly with PNPase-α. PNPase was expressed in this strain at the physiological level from its single chromosomal locus. Immunoprecipitations using specific antibodies against PNPase-α, RhlB, or enolase reproducibly showed coprecipitation of PNPase-α with RhlB (Fig. 1A, lane 11), but not enolase (Fig. 1 A, lanes 5 and 17), whereas coprecipitation of PNPase-α with enolase was detected only from bacterial strains that produced full-length RNase E protein (Fig. 1 A, lanes 2, 8, and 14). In each lysate analyzed, the amounts of individual proteins detected were comparable (Fig. 1 A, lanes 1, 4, 7, 10, 13, and 16). Preimmunization sera from rabbits producing these antibodies detected no complexes of any of these proteins in lysates obtained from either the rne-wild-type or the rne131 mutant strain (Fig. 1 A, lanes 3, 6, 9, 12, 15, and 18). These results indicated that the PNPase-α and RhlB helicase, but not enolase, can directly form a protein complex when expressed at physiological levels from single-copy gene loci in the E. coli chromosome, and that interaction of PNPase-α and enolase depends on the RNase E C-terminal “scaffold” region.
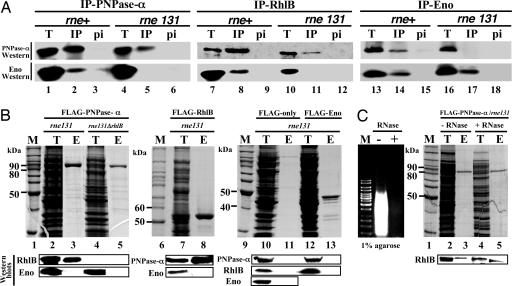
Fig. 1.
PNPase protein complexes in the rne131 mutant strain. (A) Western blotting analyses of the immunoprecipitation complexes. Immunoprecipitation assays using antibodies against PNPase (lanes 1–6), RhlB (lanes 7–12), or enolase (lanes 13–18) are shown. Immunoprecipitated complexes from rne wild type [BL21(DE3)] and the C-terminal truncated rne mutant [BL21(DE3)rne131] are labeled as rne+ and rne131, respectively. T, IP, and pi indicate total protein lysate (lanes 1, 4, 7, 10, 13, and 16), immunoprecipitation complex (lanes 2, 5, 8, 11, 14, and 17), and preimmune serum (lanes 3, 6, 9, 12, 15, and 18, as the negative controls), respectively. (B) Western blotting analyses of individual FLAG-tagged protein complexes purified by M2 affinity column. Strains containing pflag-PNP, -RhlB, -ENO, or -LRC (FLAG-only) were incubated at 30°C in LB medium, and 0.5 mM isopropyl β-d-thiogalactoside was added for 2 h at OD600 = 0.6–0.1. Nonspecific protein species associated with M2 column from FLAG-only vector expressed lysates are also shown (lanes 10 and 11). M is the standard protein marker; T is the total protein lysate; E is the elution fraction after FLAG-peptide application on to the M2 column. (C) The formation of the PNPase-α–RhlB complex is RNA-independent. FLAG–PNP-α complexes isolated from cell lysate with RNase (Benzonase Nuclease, Novagen) pretreatment (shown as +RNase) or no treatment (shown as –RNase) are shown. (Left) Agarose gel analysis of total RNAs extracted from aliquots of cell lysates with or without RNase treatment (shown as – and +, respectively) before affinity column purifications; M indicates DNA size markers. M2 affinity column purifications were performed as described (10). SDS/PAGE and Western blotting analyses are described in Supporting Materials and Methods.
During overexpression of individual proteins of FLAG-tagged PNPase-α, RhlB, or enolase in the rne131 mutant strain, association of PNPase-α with RhlB (Fig. 1B, lanes 3 and 8) but not enolase (Fig. 1B, lanes 3, 8, and 13) in a stable protein complex was again detected. In the rne131 or rne131ΔrhlB double mutation strain that overproduced FLAG-tagged PNPase-α, no association between PNPase-α and enolase was detected (Fig. 1B, lanes 3 and 5, respectively). Similarly, experiments that analyzed complexes of FLAG-tagged enolase showed no evidence of any association with PNPase-α (Fig. 1B, lane 13). In contrast, PNPase-α and RhlB were always observed in M2 affinity column purified protein complexes from rne131 mutant strains that overproduced either FLAG-tagged PNPase-α or RhlB (Fig. 1B, lanes 3 and 8, respectively). M2 affinity column protein purification from rne131 expressing only FLAG-tagged peptide did not detect any of PNPase-α, RhlB, or enolase, although these three proteins were present in the cell lysate (Fig. 1B, lane 10). To determine whether the interaction between PNPase-α and RhlB helicase was RNA-dependent, cell-extracted proteins were treated with RNase (Benzonase, Novagen) to digest RNA before M2 affinity purification (Fig. 1C Left). The results showed that complete digestion of RNA did not eliminate the binding between PNPase-α and RhlB helicase (Fig. 1C, lane 3 vs. lane 5). These results indicate that, even when overexpressed, enolase and PNPase do not interact in the absence of the RNase E C-terminal region and provide further evidence that the β-subunit observed in the B form PNPase complex is not enolase but is in fact RhlB helicase.
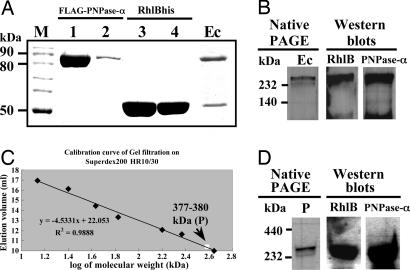
Interaction Between PNPase-α and RhlB Yields a α3β2-Form Enzymatically Active Protein Complex. To further characterize the stoichiometry of the complex formed by the interaction between PNPase-α and RhlB, we purified individual components by affinity column chromatography (Fig. 2A, indicated as FLAG-PNPase-α and RhlBhis, respectively) and reconstituted the PNPase-α–RhlB complex in vitro (Fig. 2 A, indicated as Ec). Analysis of the reconstituted complex by denaturing SDS/PAGE and Coomassie brilliant blue staining (Fig. 2 A, well no. 6, shown as Ec) showed that the complex (Ec) had molecular mass corresponding to three α polypeptides (≈88 kDa, i.e., FLAG-tagged PNPase-α) and two β polypeptides (≈52 kDa). The reconstituted complex analyzed by native gel electrophoresis showed a single band that was found by Western blot analysis to contain both the PNPase-α and RhlB proteins (Fig. 2B). The reconstituted PNPase-α–RhlB complex with the configuration of three α and two β polypeptides degraded double-stranded as well as single-stranded RNA (Fig. 6, which is published as supporting information on the PNAS web site). Gel filtration analysis showed that the reconstituted complex has a molecular mass calculated as 377–380 kDa (Fig. 2C, indicated as P). Moreover, using Western blot analysis, we confirmed that both PNPase-α and RhlB were present in the complex eluted from gels corresponding to a mass of 377–380 kDa (Fig. 2D). These results argue that PNPase-α and RhlB RNA helicase can be reconstituted into a B form α3β2 complex containing two types of chains in the proportions found in the native B form [i.e., three α-subunits (PNP) and two β-subunits (RhlB)], and that the RhlB β-subunit of the complex enables PNPase-α to degrade an RNA substrate containing 3′-ended double-stranded structure.
Fig. 2.
In vitro reconstitution of the PNPase-α–RhlB complex and molecular weight determination of the protein complex. (A) Reconstitution of the PNPase-α–RhlB complex. The individually purified protein fractions before (lanes 1 and 3) and after (lanes 2 and 4) M2 column elution and the reconstituted PNPase-α–RhlB protein complex eluted from the M2 affinity column (shown as Ec) were analyzed by SDS/PAGE. M, protein size marker. (B) Gradient native PAGE and Western blotting analyses of the in vitro reconstituted PNPase-α–RhlB complex in A.(C and D) Molecular weight and composition determination of the in vitro reconstituted PNPase-α–RhlB complex by Superdex G200 gel filtration, native PAGE, and Western blotting analyses. For native gel electrophoresis, the complex running as a higher molecular weight than the protein size markers might be due to the “doughnut” shape structure with the hole in the middle of PNPase-α trimers. P is the PNPase-α–RhlB complex after gel filtration. The details of the experiments are described in Supporting Materials and Methods.
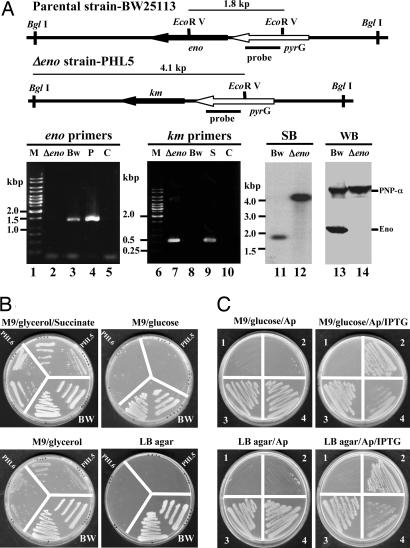
Deletion of the rhlB, But Not of the eno, Gene Affects 3′ to 5′ Exo-Ribonucleolytic Degradation of Duplex RNA by PNPase Protein Complex Isolated from the rne131 Strain. A minidegradosome reconstituted in vitro and containing the C-terminal region of RNase E, PNPase-α, and RhlB helicase has been shown to carry out RNA degradation mediated by the catalytic activity of PNPase together with the duplex RNA unwinding activity of RhlB helicase (38). This functional interaction between PN-Pase-α and RhlB helicase has been proposed to occur through the independent binding of these proteins to the RNase E C-terminal region (25). To learn whether RhlB can assist PNPase-α in degrading duplex RNA independently of the RNase E C-terminal region, we carried out in vitro RNA degradation assays using FLAG–PNPase-α complexes isolated directly from either the rne131 or the rne131ΔrhlB strain. To determine the role of enolase in assisting PNPase-α function, an identical experiment was performed with a Δeno strain containing a full-length rne gene, whose encoded C-terminal segment is required for complex formation of PNPase-α and enolase. In these experiments, the rne131ΔrhlB and Δeno strains were constructed by one-step PCR-based mutagenesis (ref. 31; see below) and P1 transduction, as described in Materials and Methods. Construction of the ΔrhlB strain has been described in an earlier publication (15). The procedure used for construction of the Δeno mutant is shown in Fig. 3A. The Δeno mutation was verified by using gene-specific primers, and PCR amplification that showed a chromosomal deletion of the eno gene with an inserted km gene in its place (Fig. 3A, lanes 2 and 7, respectively). The genotype and absence of enolase protein in Δeno mutants were confirmed by Southern and Western blotting analyses [Fig. 3A shown as SB (lane 12) and WB (lane 14), respectively]. Two independently isolated Δeno clones (PHL5 and -6) were tested for their ability to grow on different media. Glycolytic mutants require glycerol and a dicarboxylic acid (i.e., succinate) to grow normally and consequently cannot be cultured on M9 minimal agar plus glucose or LB agar (39, 40). PHL5 and -6 failed to grow on M9 minimal agar plus glucose or LB media in the absence of supplementation with either glycerol or succinate, which is consistent with the absence of enolase (Fig. 3B). Furthermore, bacteria containing Δeno led to a much slower growth rate compared with that of the parental strain (in this case, BW25113) at 37°C (75-vs. 48-min doubling time, respectively; Table 1, which is published as supporting information on the PNAS web site). A plasmid encoding a functional eno gene expressed under 1 mM isopropyl β-d-thiogalactoside induction was able to rescue both Δeno phenotypes [Fig. 3C and Table 1 (in this case, only PHL5 is shown)]. To determine the role of enolase in PNPase-α function, an identical amount of FLAG-tagged PNPase-α protein complex isolated from either the eno+ or Δeno (Fig. 4A) was used to perform kinetic RNA-degradation assays. Because association of PNPase-α and enolase depends of the RNase E C-terminal region, deletion of the eno gene had no effect on exonucleolytic degradation of either single-stranded or duplex RNAs, as shown in Fig. 4 B and C. In contrast, when compared with the catalytic activities for the FLAG–PNPase-α protein complexes of rne131 and rne131ΔrhlB, respectively, the PNPase-α protein complexes of rne131ΔrhlB showed decreased exonucleolytic degradation specifically of duplex RNA (Fig. 5B, rne131 vs. rne131ΔrhlB) but no loss of ability to degrade single-stranded RNA (Fig. 5A), consistent with evidence that the helicase is not required for PNPase transit through unpaired regions of RNA and for which, consequently, ATP is not required (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 3.
Δeno strain verification, phenotypic characterization, and complementation. (A) Δeno strain verification. The restriction enzyme maps show the eno locus in parental and Δeno strains. The DNA probe, containing the pyrG gene, the primers used for PCR amplifications, and the DNA size markers (lanes 1 and 6), are indicated. Bw, Δeno; S, P, and C are PCR products from parental strain-BW25113, Δeno, SU02 (ΔrhlB-kmr mutant), and plasmid pflag-ENO as positive control and negative control, without any DNA templates, respectively. Southern and Western blotting analyses are shown as SB and WB, respectively. Chromosomal DNAs were digested completely by EcoR V and BglI. The hybridized signals shown are 1.8 kbp for BW25113 and 4.1 kbp for Δeno, as predicted. (B) Phenotypes of Δeno mutants grown under different conditions, as shown. Photographs of individual plates were taken using a Hewlett–Packard P ScanJetIIC scanner. (C) Complementation study of Δeno mutant grown at 37°C, as described in Supporting Materials and Methods; 1 and 2 are Δeno (PHL5) containing pPW500 and pPW-eno, respectively; 3 and 4 are BW25113 containing pPW500 and pPW-eno, respectively. Plasmid selection in the presence of ampicillin (Ap) is shown.
Discussion
The discovery of PNPase by Grunberg-Manago et al. in 1955 (3) is generally viewed as a landmark in nucleic acid biochemistry and molecular biology, and multiple review articles have since summarized the extensive work carried out with this enzyme (5, 41, 42). The molecular weight of the PNPase holoenzyme purified from E. coli provided the first evidence of the existence of two PNPase protein complexes, which were termed A and B (23), as described above. Based on molecular weight comparisons, partial protease digestion, and N-terminal sequence analysis of PNPase-associated bands isolated from gels, the β-subunit of the B-form PNPase holoenzyme was thought to be the glycolytic enzyme enolase (11, 24). The results reported here indicate that the association of PNPase-α and enolase is indirect and occurs through their independent binding to the C-terminal region of the RNase E. Using genetic and biochemical approaches, we found that the β-subunit of the PNPase holoenzyme is not enolase but is in fact RhlB helicase and that the B form of PNPase protein complex is actually a complex consisting of the catalytic subunit of PNPase-α and RhlB helicase. Thus, the structure of the B-form of PNPase holoenzyme parallels the core structure of, and may be an evolutionary antecedent of, eukaryotic cell exosome complexes, which carry out 3′ to 5′ exonucleolytic RNA degradation (17, 18) and contain both PNPase-α type 3′ to 5′ exonucleases and an RNA helicase (17, 18, 43). The ability of RhlB helicase to assist exonucleolytic degradation by unwinding double-stranded RNA segments suggests a possibly analogous role for the eukaryotic helicase present in exosomes.
In early investigations of the quaternary structure of PNPase protein complex, Portier (23) proposed a molecular mass of 86,000 ± 5,000 and 48,000 ± 2,000 for the α- and β-subunits, respectively. An RNase E–PNPase complex purified by Carpousis et al. (24) contained 85-kDa PNPase-α, and a 48-kDa protein yielded a protease V8 partial digestion pattern that was similar to that observed for the Portier enzyme. Later, the RNase E-based degradosome complex found to contain the 50-kDa RhlB helicase protein (10, 11) as well as the PNPase catalytic unit and a 48-kDa protein; determination of the N-terminal amino acid sequences of the 48- and 50-kDa proteins indicated that these proteins corresponded to enolase and RhlB helicase, respectively (10, 11). On the basis of these results, enolase was determined to be the β-subunit of the PNPase protein complex (11). However, subsequent experiments using E. coli two-hybrid analysis and in vitro protein interaction assays showed that PNPase-α and RhlB can form a complex independently of RNase E (27), indicating that association of PNPase with this helicase does not result simply from binding of both proteins to the RNase E C-terminal scaffold region and opening the question of whether the designation of enolase as the β-subunit of the PNPase complex was correct. We used a combination of genetic and biochemical approaches to address this question and also examined the nature of the PNPase complex formed during the expression of the component proteins from single-copy chromosomal loci. Our demonstration that the PNPase β-subunit is not the 48-kDa enolase but is instead the 50-kDa RhlB protein not only rectifies a misconception existing since 1996 but also definitively resolves a question raised in 1973. The earlier conclusion of Py et al. (11) that enolase is the β-subunit of the PNPase holoenzyme may have resulted from association of these proteins through the scaffold region of RNase E in the complexes examined by these investigators.
The amino acid sequences of differently related PNPase-α encoded by bacteria and the nuclear genomes of plants, yeast, and mammals display a high degree of identity and feature similar motifs: two core domains related to the E. coli phosphorylase RNase PH, an α-helical domain between the two core domains followed by two adjacent RNA-binding domains KH and S1 (21, 44, 45). However, protein complexes containing homologous PNPase-α and RNA helicase from bacteria and eukaryotes have been found to have different constituents (21, 46); PNPase-α of the spinach chloroplast has been shown to form a homotrimeric complex and to lack any known interactions with other proteins (47). Domain analysis of chloroplast PNPase has revealed two core domains with distinct functions in RNA degradation and polyadenylation, respectively (47), and has led to the suggestion that RNA molecules in chloroplasts can be degraded only if poly(A) tails are added, presumably by the same enzyme (48). Interestingly, phylogenetic analysis of the two core domains (44, 47) has revealed that they separated very early during the evolution of PNPase-α, leading to distinct bacterial and organelle PNPase-α proteins on the one hand and eukaryotic exosome proteins on the other. Potentially, the polyadenylation required for degradation by the second core domain of chloroplast PNPase-α may be circumvented by the actions of an RNA helicase, because both RNA unwinding by RNA helicase (11) and polyadenylation (49) can enable PNPase-α to proceed through the RNA regions of secondary structure.
Earlier DNA microarray-based investigations of the steady-state abundance and decay of 4,289 E. coli mRNAs at single-gene resolutions (50) in bacteria carrying mutations in degradosome protein components indicate that the functions of all four components of the degradosome are necessary for normal mRNA turnover (15). Although the decay of some E. coli mRNAs in vivo depends on the action of assembled degradosomes, the formation of the α3β2 PNPase holoenzyme in vivo argues that different types of ribonuclease complexes exist, enabling bacterial cells to effectively process and degrade a wide range of complex RNA structures.
Supplementary Material
Acknowledgments
We thank Dr. S. J. Chang for helpful discussion throughout this study and Dr. Chwan-Deng Hsiao for the gel filtration analysis. The critical comments offered by the reviewers on the experiments, data presentation, and manuscript revision were highly appreciated. We thank English editing consultants Dr. K. Deen and G. M. Cohen. This work was supported by Grants NSC 91-2321-B-001-016 and 93-2311-B-001-023 from the National Science Council, Taiwan, and by an intramural fund grant from the Academia Sinica, Taiwan (to S.L.-C.).
Author contributions: P.-H.L. and S.L.-C. designed research; P.-H.L. performed research; P.-H.L. analyzed data; and P.-H.L. and S.L.-C. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PNPase, polynucleotide phosphorylase.
References
- 1.Mohanty, B. K. & Kushner, S. R. (2000) Proc. Natl. Acad. Sci. USA 97, 11966–11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yehudai-Resheff, S., Hirsh, M. & Schuster, G. (2001) Mol. Cell. Biol. 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grunberg-Manago, M., Ortiz, P. J. & Ochoa, S. (1955) Science 122, 907–910. [DOI] [PubMed] [Google Scholar]
- 4.Wood, J. N. & Hutchinson, D. W. (1976) Nucleic Acids Res. 3, 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godefroy-Colburn, T. & Grunberg-Manago, M. (1972) Enzymes 7, 533–574. [Google Scholar]
- 6.Peterkin, P. I. & Fitt, P. S. (1971) Biochem. J. 121, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estévez, A. M., Kempf, T. & Clayton, C. (2001) EMBO J. 20, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kessler, B. & Chen, D. (1964) Biochim. Biophys. Acta 80, 533–541. [DOI] [PubMed] [Google Scholar]
- 9.Fitt, P. S. & See, Y. P. (1970) Biochem. J. 116, 309–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miczak, A., Kaberdin, V. R., Wei, C.-L. & Lin-Chao, S. (1996) Proc. Natl. Acad. Sci. USA 93, 3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Py, B., Higgins, C. F., Krisch, H. M. & Carpousis, A. J. (1996) Nature 381, 169–172. [DOI] [PubMed] [Google Scholar]
- 12.Jäger, S., Fuhrmann, O., Heck, C., Hebermehl, M., Schiltz, E., Rauhut, R. & Klug, G. (2001) Nucleic Acids Res. 29, 4581–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, K. & Cohen, S. N. (2003) Mol. Microbiol. 48, 349–360. [DOI] [PubMed] [Google Scholar]
- 14.Liou G.-G., Jane, W.-N., Cohen, S. N., Lin, N.-S. & Lin-Chao, S. (2001) Proc. Natl. Acad. Sci. USA 98, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein, J. A., Lin, P.-H., Cohen, S. N. & Lin-Chao, S. (2004) Proc. Natl. Acad. Sci. USA 101, 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez, P. J., Marchand, I., Joyce, S. A. & Dreyfus, M. (1999) Mol. Microbiol. 33, 188–199. [DOI] [PubMed] [Google Scholar]
- 17.Anderson, J. S. & Parker, R. P. (1998) EMBO J. 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler, J. S. (2002) Trends Cell Biol. 12, 90–96. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M. & Tollervey, D. (1997) Cell 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 20.Carpousis, A. J. (2002) Biochem. Soc. Trans. 30, 150–155. [PubMed] [Google Scholar]
- 21.Symmons, M. F., Williams, M. J., Luisi, B. F., Jones G. H. & Carpousis, A. J. (2002) Trends Biochem. Sci. 27, 11–18. [DOI] [PubMed] [Google Scholar]
- 22.Portier, C., van Papenbusch, R., Thang, M. N. & Grunberg-Manago, M. (1973) Eur. J. Biochem. 40, 77–87. [DOI] [PubMed] [Google Scholar]
- 23.Portier, C. (1975) Eur. J. Biochem. 55, 573–582. [DOI] [PubMed] [Google Scholar]
- 24.Carpousis, A. J., Van Houwe, G., Ehretsmann, C. & Krisch, H. M. (1994) Cell 76, 889–900. [DOI] [PubMed] [Google Scholar]
- 25.Vanzo, N. F., Li, Y. S., Py, B., Blum, E., Higgins, C. F., Raynal, L. C., Krisch, H. M. & Carpousis, A. J. (1998) Genes Dev. 12, 2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauhut, R. & Klug, G. (1999) FEMS Microbiol. Rev. 23, 353–370. [DOI] [PubMed] [Google Scholar]
- 27.Liou, G.-G., Chang, H.-Y., Lin, C.-S. & Lin-Chao, S. (2002) J. Biol. Chem. 277, 41157–41162. [DOI] [PubMed] [Google Scholar]
- 28.Kuhnel, K. & Luisi, B. F. (2001) J. Mol. Biol. 313, 583–592. [DOI] [PubMed] [Google Scholar]
- 29.Roth, J. R. (1970) Methods Enzymol. 17a, 3–35. [Google Scholar]
- 30.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring. Harbor Lab. Press, Plainview, NY).
- 31.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 33.Lin-Chao, S. & Cohen, S. N. (1991) Cell 65, 1233–1242. [DOI] [PubMed] [Google Scholar]
- 34.Lin-Chao, S., Chen, W.-T. & Wong, T.-T. (1992) Mol. Microbiol. 6, 3385–3393. [DOI] [PubMed] [Google Scholar]
- 35.McDowall, K.J., Lin-Chao, S. & Cohen, S.N. (1994) J. Biol. Chem. 269, 10790–10796. [PubMed] [Google Scholar]
- 36.Kaberdin, V. R., Miczak, A., Jakobsen, J. S., Lin-Chao, S., McDowall, K. J. & von Gabain, A. (1998) Proc. Natl. Acad. Sci. USA 95, 11637–11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callaghan, A. J., Aurikko, J. P., Ilag, L. L., Gunter Grossmann, J., Chandran, V., Kuhnel, K., Poljak, L., Carpousis, A. J., Robinson, C. V., et al. (2004) J. Mol. Biol. 340, 965–979. [DOI] [PubMed] [Google Scholar]
- 38.Coburn, G. A., Miao, X., Briant, D. J. & Mackie, G. A. (1999) Genes Dev. 13, 2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zwaig, N., Kistler, W. S. & Lin, E. C. (1970) J. Bacteriol. 102, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irani, M. H. & Maitra, P. K. (1977) J. Bacteriol. 132, 398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grunberg-Manago, M. (1962) Annu. Rev. Biochem. 31, 301–332. [DOI] [PubMed] [Google Scholar]
- 42.Grunberg-Manago, M. (1963) Prog. Biophys. Molec. Biol. 13, 175–239. [DOI] [PubMed] [Google Scholar]
- 43.Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D. & Mitchell, P. (1999) Genes Dev. 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuo, Y. & Deutscher, M. P. (2001) Nucleic Acids Res. 29, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raijmakers, R., Egberts, W. V., van Venrooij, W. J. & Pruijn, G. J. (2002) J. Mol. Biol. 323, 653–663. [DOI] [PubMed] [Google Scholar]
- 46.Aloy, P., Ciccarelli, F. D., Leutwein, C., Gavin, A. C., Superti-Furga, G., Bork, P., Bottcher, B. & Russell, R. B. (2002) EMBO Rep. 3, 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yehudai-Resheff, S., Portnoy, V., Yogev, S., Adir, N. & Schuster, G. (2003) Plant Cell 15, 2003–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baginsky, S., Shteiman-Kotler, A., Liveanu, V., Yehudai-Resheff, S., Bellaoui, M., Settlage, R. E., Shabanowitz, J., Hunt, D. F., Schuster, G. & Gruissem, W. (2001) RNA 7, 1464–1475. [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, F. & Cohen, S. N. (1995) Nature 374, 180–183. [DOI] [PubMed] [Google Scholar]
- 50.Bernstein, J. A., Khodursky, A. B., Lin, P.-H., Lin-Chao, S. & Cohen, S. N. (2002) Proc. Natl. Acad. Sci. USA 99, 9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.