Abstract
Semaphorins are extracellular cell guidance cues that govern cytoskeletal dynamics during neuronal and vascular development. MICAL (molecule interacting with CasL) is a multidomain cytosolic protein with a putative flavoprotein monooxygenase (MO) region required for semaphorin-plexin repulsive axon guidance. Here, we report the 1.45-Å resolution crystal structure of the FAD-containing MO domain of mouse MICAL-1 (residues 1-489). The topology most closely resembles that of the NADPH-dependent flavoenzyme p-hydroxybenzoate hydroxylase (PHBH). Comparison of structures before and after reaction with NADPH reveals that, as in PHBH, the flavin ring can switch between two discrete positions. In contrast with other MOs, this conformational switch is coupled with the opening of a channel to the active site, suggestive of a protein substrate. In support of this hypothesis, distinctive structural features highlight putative protein-binding sites in suitable proximity to the active site entrance. The unusual juxtaposition of this N-terminal MO (hydroxylase) activity with the characteristics of a multiprotein-binding scaffold exhibited by the C-terminal portion of the MICALs represents a unique combination of functionality to mediate signaling.
Keywords: axon guidance, hydroxylase, monooxygenase, protein structure, signal transduction
To find their way through the developing nervous system, axonal growth cones must sense and respond to guidance cues in their environment. Plexins act as the signal transducing receptors for semaphorins, a family of secreted and cell surface-attached proteins best characterized by their chemorepulsive role in axon guidance (1). The extracellular portions of semaphorins and plexins share a distinctive β-propeller fold termed the sema domain (2, 3); the plexin cytosolic regions are of unknown structure. Molecules of the MICAL [molecule interacting with CasL (4)] family link signaling from the cytosolic regions of class A plexins to the cytoskeleton (5). MICALs are conserved from flies to mammals, with one MICAL gene identified in Drosophila and three (MICAL-1, MICAL-2, and MICAL-3) found in mammals, each with several isoforms (6). MICALs are large (>1,000 aa), multidomain, cytosolic proteins expressed in specific neuronal and nonneuronal (thymus, lung, spleen, and testis) tissues both during development and in adulthood (4).
From sequence analysis, it has been shown that MICALs contain two protein-protein interaction domains implicated in signal transduction and cytoskeletal organization, a calponin homology (CH) domain (7) and a LIM domain (8), plus a proline-rich region for Src homology 3 (SH3) domain recognition that mediates interaction with CasL, a multidomain docking protein localized at focal adhesions and stress fibers (4). Human MICAL-1 associates with the small GTPase Rab1 (6, 9) and with vimentin (4), a major component of intermediate filaments. In addition to the SH3 domain-binding motif, the C-terminal region (of ≈250 residues) contains coiled-coil motifs and binds the cytosolic domain of class A plexins (5). Thus, the MICALs are protein-binding scaffolds, but, uniquely, they combine this property with a highly conserved N-terminal region of some 500 residues, characterized by sequence analyses and functional studies as a putative flavoprotein monooxygenase (MO) required for semaphorin-plexin-mediated axon guidance (5).
Flavoenzymes bind the cofactor FAD as an integral part of their structure. Despite <20% sequence identity between disparate members of this family, they share a similar fold and essentially identical FAD-binding sites (10). In contrast, the catalytic reactions carried out by the flavoenzymes are varied, and their active-site architectures differ accordingly. The structure of p-hydroxybenzoate hydroxylase (PHBH) provides the paradigm for the flavoprotein MO (hydroxylase) subset of flavoenzymes (11). Flavoprotein MOs act on a broad range of small molecules (e.g., p-hydroxybenzoate, steroids, and amino acids). The substrate(s), mode of action, and, indeed, function of the putative MO region in the MICALs are unknown.
Our structural and biophysical analyses on the N-terminal portion of murine MICAL-1 confirm that this region has the architecture and characteristics of a flavoenzyme of the MO family, demonstrate the enzymatic activity to be NADPH-dependent, and reveal a mechanism for controlled substrate access to the active site, which is strongly indicative of large (potentially protein) substrates.
Methods
Protein Expression and Purification. The mMICAL489 expression construct (amino acids 1-489 of the mouse MICAL-1 gene plus C-terminal His-tag) was generated by ligation-independent cloning (Gateway Technology, Invitrogen), overexpressed in Escherichia coli (DE3)pLysS (Novagen), and purified with Ni affinity and size-exclusion chromatography; all stages used the high-throughput pipeline of the Oxford Protein Production Facility (see Supporting Text, which is published as supporting information on the PNAS web site). Before crystallization, the protein solution was concentrated to 10 mg/ml in 10 mM Tris·HCl, pH 7.5/200 mM NaCl.
Crystallization and Data Collection. Crystallization trials by sitting drop-vapor diffusion (drop size of 200 nl) used previously reported robotic technologies and protocols (12). mMICAL489 crystallized at 20°C in 0.1 M Na acetate, pH 4.6/30% (wt/vol) polyethylene glycol 2000 monomethyl ether/0.2 M ammonium sulfate. A native crystal frozen in reservoir solution plus 20% glycerol diffracted to 1.45 Å at the European Synchrotron Radiation Facility (ESRF)-ID29 (88.3% complete with an Rmerge of 0.058). A single anomalous dispersion (SAD) data set was collected at ESRF-ID23 from a native crystal soaked in p-chloromercurybenzoate-saturated crystallization solution for 1 h. Crystals of the reduced form were obtained by soaking a native crystal in crystallization solution containing 15 mM NADPH for 1 min. Data to the diffraction limit (2.9 Å) were collected on a MAR345 imaging plate detector (MAR Research, Hamburg) mounted on a microfocus Micromax 007 generator with a confocal multilayer (Rigaku, Tokyo/MSC, The Woodlands, TX). X-ray data were processed and scaled with hkl (13) (see also Table 1, which is published as supporting information on the PNAS web site).
Structure Determination and Analysis. The structure was determined by SAD analysis. The positions of 20 mercury atoms were determined by using shelxd (14) with a correlation coefficient of 49.3% (correlation coefficient, weak = 27.1%). This solution was input into autosharp (15) for phase calculation and improvement (figure of merit = 0.37-2.3 Å). An initial model was built automatically by using resolve (16) and completed by hand using o (17). After a few cycles of refinement with refmac5 (18), the structure was used as a molecular replacement model in epmr (19) against the native data to 3 Å. This solution was input into arp/warp (20) for automated model building and manually adjusted and refined by using o and refmac5. The final model of mMICAL489 (residues 7-489, one FAD molecule, a sulfate, and a chloride ion) has an R factor of 0.179 [Rfree = 0.219; rms deviation (rmsd) bond lengths of 0.012 Å] using all data between 30 and 1.45 Å. The 2.9-Å structure of the reduced form was determined by molecular replacement using epmr and was refined with refmac and o. The structures have no residues in disallowed regions in the Ramachandran plot. Additional crystallographic statistics are provided in Table 1, and sample electron density is provided in Fig. 7, which is published as supporting information on the PNAS web site.
Results
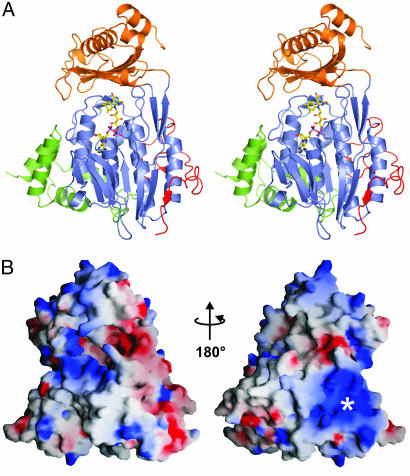
Crystal Structure and Topology. The crystal structure of mMICAL489 contains two molecules (each with residues 7-489 and the cofactor FAD) per crystallographic asymmetric unit, which are essentially identical (rmsd on 476 Cα pairs = 0.42 Å). mMICAL489 folds into a compact structure of three distinct domains and a C-terminal linker region (Fig. 1A). The N-terminal 85 residues form a four-helix bundle that rigidly abuts the core structure. Solvent-exposed arginine, lysine, and histidine residues from this domain (Lys-52, -61, -66, -69, and -86; Arg-35 and -70; His-11, -13, and -49) make an extensive patch of basic charge on the surface (Fig. 1B). This area of positive electrostatic potential is a very striking feature and gives mMICAL489 a basic charge. The spatial arrangement of the four helices does not correspond closely to any other known structures, although, intriguingly, the nearest equivalent found by the Dali server (www.ebi.ac.uk/dali) is the tetratricopeptide repeat of collagen prolyl 4-hydroxylase (rmsd of 3.9 Å for 57 Cα atoms with 9% sequence identity, PDB ID code 1TJC), a motif implicated in protein-protein interactions. The C-terminal region (residues 445-489; Fig. 1A) is also tightly interfaced to the core structure and is well ordered to Glu-489, the last mMICAL-1 residue in the expression construct.
Fig. 1.
Crystal structure of mMICAL489. (A) Stereoview of mMICAL489 with four-helix bundle domain (1-85, green), FAD-binding domain (86-234 and 367-444, slate), MO domain (235-366, orange) and linker region (445-489, red) shown. The FAD molecule is depicted as sticks. (B) Solvent-accessible surface of mMICAL489 colored by electrostatic potential contoured at ±15 kT using grasp (36) (red, acidic; blue, basic). Left-hand view is shown, as in A. The asterisk marks a patch of basic potential.
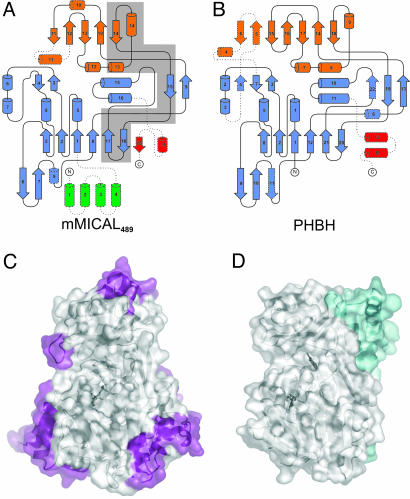
The core of the mMICAL489 structure comprises 358 residues and is formed jointly by the FAD-binding domain (residues 86-234 and 367-444; Fig. 1 A) and the MO domain (residues 235-366; Fig. 1 A). The closest structural matches are with PHBH (rmsd of 2.9 Å for 291 Cα atoms with 10% sequence identity, PDB ID code 1PBE) and phenol hydroxylase (rmsd of 3.7 Å for 310 Cα atoms with 14% sequence identity, PDB ID code 1PN0). The FAD-binding and MO domains are connected by a long, two-stranded β-sheet with the MO domain inserted between β-strands 9 and 15 of the FAD-binding domain. The normally rather complicated topology of homologous structures such as PHBH, where the polypeptide chain passes several times between the FAD-binding domain and the MO domain, is absent (Fig. 2 A and B). This simplified topology arises because a substantial portion of the MO domain of PHBH (and other MOs) is missing from the mMICAL489 structure (Fig. 2 C and D), a structural difference that may significantly affect function.
Fig. 2.
Structural comparison of mMICAL489 and PHBH. (A) Topology of mMICAL489 (β-strands, arrows; α-helices, cylinders). Domains are colored as in Fig. 1 A. Dotted lines denote unique structural elements. The gray-shaded area is deleted in the human splice isoform MICAL-1B (6); this deletion appears to be incompatible with formation of a stable molecule. (B) Equivalent diagram for PHBH. (C) Solvent-accessible surface of mMICAL489 with parts unique to mMICAL489 (compared with PHBH) highlighted in violet (orientation is as in Fig. 1 A). (D) Solvent-accessible surface of PHBH (oriented to superpose on mMICAL489) with parts unique to PHBH (compared with mMICAL489) highlighted in cyan.
The central part of the FAD-binding domain consists of a predominantly parallel β-sheet (β-strands 5, 2, 1, 8, 17, 16, and 18; Figs. 1A and 2A) flanked on one side by several α-helices (helices 5, 8, 15, and 16) and on the other by a β-hairpin (β-strands 6 and 7), followed by the short helix α9. The main feature of the MO domain is a large, five-stranded, antiparallel β-sheet (β-strands 11, 12, 13, 10, and 14). An edge strand in this sheet (β11) is only well ordered in the heavy-atom soaked crystal structure (where it is involved in lattice contacts). In the high-resolution crystal structure of the native molecule, the electron density and crystallographic B factors indicate that this secondary structure element is very flexible in both copies of the molecule. The upper surface (Fig. 1A orientation) of the antiparallel β-sheet is capped by three helices (α10, α12, and α13); the bottom surface forms hydrogen bonds and hydrophobic interactions with the FAD-binding domain and interacts with the isoalloxazine ring of the FAD. The total surface area buried in the interface between the MO and FAD-binding domains is 1,950 Å2.
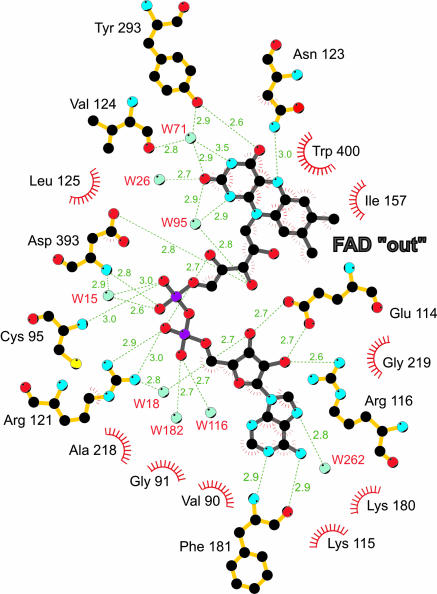
The FAD-Binding Site. The FAD cofactor is well ordered for all copies of mMICAL489 in the heavy-atom soaked and high-resolution crystal structures. As observed in other flavoproteins (10), it is bound in an extended conformation with the isoalloxazine of the flavin located at the interface between the FAD-binding domain and the MO domain (Fig. 1A). The adenine dinucleotide portion of the FAD is deeply embedded within the FAD-binding domain. The adenosine moiety abuts the parallel β-sheet of the domain, in the pocket formed between the end of strand β1 and the start of β2. As predicted from sequence analysis (5), this part of the MICAL fold (β1α5β2) is an example of the dinucleotide-binding Rossmann fold. The central part of this domain requires the consensus motif GXGXXG (21), which, in mMICAL489, corresponds to Gly-91, Gly-93, and Gly-96 (Fig. 8, which is published as supporting information on the PNAS web site). The N terminus of helix α5 points toward the FAD pyrophosphate moiety, providing charge compensation. The main-chain nitrogen atoms of Cys-95 and Asp-393, the side chain of Arg-121, and four water molecules (Fig. 3) form a network of hydrogen bonds to the two phosphate groups. The extended conformation of the adenine dinucleotide portion of the cofactor is further stabilized by one of the phosphate oxygen atoms forming a hydrogen bond to the second ribityl hydrogen group. The side chain of Glu-114 interacts by means of hydrogen bonds with the two OH groups of the AMP ribosyl moiety, and, finally, the position of the adenine moiety is stabilized by hydrogen bonds to the main chain of Phe-181 and a water molecule (Fig. 3).
Fig. 3.
Schematic representation of the FAD-apoprotein interactions in mMICAL489. View on the si face of the flavin with the FAD and interacting residues depicted as sticks [N, blue; O, red; P, violet; S, yellow; C (protein), orange; C (FAD), gray] and water molecules shown as cyan spheres. H bonds are shown in green with lengths in Å. Red “eyelashes” show hydrophobic interactions.
The three rings of the isoalloxazine form an almost perfect plane, and the flavin adopts a conformation that partially exposes the ring system to bulk solvent. This position is stabilized by ring stacking between the re side of the isoalloxazine and the side chain of Trp-400 so that the indole system forms a coplanar π-complex with the isoalloxazine. The si face of the isoalloxazine makes van der Waals interactions to Ile-157. The flavin O(4) hydrogen-bonds to the side-chain OH of Tyr-293, while the side-chain nitrogen of Asn-123 forms a hydrogen bond to the flavin N(5). The Asn-123 side-chain oxygen is coordinated by a network of hydrogen bonds (residues Asn-243, Thr-291, and Asp-360) whose proton donor and acceptor contributions lock the orientation of the Asn-123 side chain so that the nitrogen acts as a hydrogen-bond donor to the flavin N(5), implying that the isoalloxazine is in the oxidized state. A network of hydrogen bonds involving the side-chain OH of Tyr-293, the main-chain oxygen of Val-124, three water molecules (Fig. 3), and the N(1), O(2), and N(3) atoms of the isoalloxazine satisfy the remaining hydrogen-bonding potential of the ring system. Hydrogen bonds from the side chain of Asp-393 and a water molecule to one of the ribityl oxygen atoms provide the final contributions to the stability of this flavin conformation.
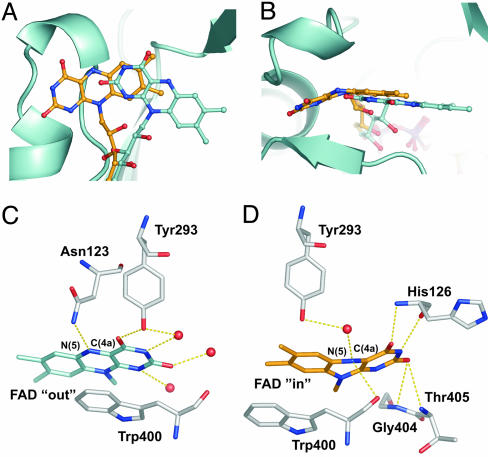
In PHBH, the flavin ring can adopt two very different positions, corresponding to “out” and “in” conformations (11, 22, 23), and the ability to switch between these two conformations is essential for the catalytic activity. Comparison with PHBH shows that the flavinring in the high-resolution mMICAL489 structure is in the out position (24). In contrast, the position of the flavin ring in most MO structures corresponds to the in conformation of PHBH (10), which places the reactive isoalloxazine in position to contribute to catalysis. Some MOs are permanently locked into the in conformation, but, for the PHBH family of hydroxylases, the ability to switch between in and out conformations is essential to allow access to the active site for substrate binding and product release. The catalytic cycle of the PHBH family also depends on NADPH (to reduce the flavin, which is then returned to an oxidized state during catalysis), and a comparison of the PHBH and mMICAL489 structures indicates that PHBH residues implicated in NADPH binding [by biochemical analyses of PHBH mutants (25, 26)] are conserved in mMICAL489 (Fig. 8). We therefore investigated whether mMICAL489 had NADPH-binding properties.
NADPH Triggered Changes in FAD Conformation. We found that addition of NADPH to mMICAL489 in solution results in an instantaneous loss of the yellow color that characterizes samples containing this flavoprotein. Mass spectrometry indicates that the NADPH is oxidized to NADP+ (data not shown). Soaking a mMICAL489 crystal in 15 mM NADPH resulted in a loss of color and a rapid deterioration in crystal quality; however, x-ray diffraction data were successfully collected (albeit at a reduced resolution of 2.9 Å; see Supporting Text). The resultant electron density maps showed no evidence for a bound NADPH [the transient nature of this interaction has precluded direct visualization of the complex with any native PHBH-type flavoenzyme, although a complex has been reported for a mutant PHBH (24)]. However, the flavin ring had clearly switched position (presumably as a result of an interaction having taken place between NADPH and mMICAL489; Figs. 4 A and B and 7C). The change in FAD position is at full occupancy for one of the two copies of mMICAL489 in the crystallographic asymmetric unit, whereas for the second copy, both conformations are observed (and refined as such). All further analysis of the NADPH soaked crystal structure of mMICAL489 (mMICAL489*) presented here is based on the single conformation copy.
Fig. 4.
Comparison of the reduced and oxidized forms of mMICAL489.(A and B) Superposition of the two forms. The FAD molecules are drawn as balls and sticks (carbons of oxidized mMICAL489, cyan; carbons of reduced mMICAL489, orange). The main chain of the oxidized form is depicted as a ribbon. B is rotated by 90° about the x axis relative to A. (C and D) Coordination of the isoalloxazine ring in the oxidized (C) and reduced (D) forms viewed from a common orientation. The isoalloxazine ring and selected residues are depicted as sticks (orange, carbon of reduced isoalloxazine; gray, protein carbon), waters are shown as spheres, and H bonds are shown as yellow dashes.
The isoalloxazine ring, positioned in the out conformation in the native (high resolution) crystal structure, occupies an in conformation (corresponding to that observed for PHBH) in mMICAL489* (Fig. 4). The position of the adenine dinucleotide portion of the FAD remains unchanged, clamped within the FAD-binding domain. The pivot point for the two FAD conformations is provided by the ribityl, which has the properties of a flexible hinge within the cofactor, allowing the orientation of the isoalloxazine ring to switch by some 20° between conformations (Fig. 4 A and B). In the mMICAL489* structure, the isoalloxazine is buried at the interface of the MO and FAD-binding domains, in part occupying a cavity filled by three water molecules in the native crystal structure. The interactions of the flavin for the in conformation are detailed in Fig. 4 C and D and also in Fig. 9, which is published as supporting information on the PNAS web site. New hydrogen bonds are formed from the main-chain oxygen and nitrogen of His-126 to the N(3) and O(4) atoms of the isoalloxazine, respectively. The O(2) atom is coordinated by hydrogen bonds to the main-chain nitrogens of Gly-404 and Thr-405. N(5) is involved in a network of hydrogen bonds with the main-chain oxygen of Trp-400, a water molecule, and the hydroxyl group of Tyr-293. The isoalloxazine ring adopts a “butterfly” conformation with an angle between the two wings of 155° (Fig. 4 B and D), indicative of a switch to the reduced state. Furthermore, the changes in environment and hydrogen-bond network are consistent with stabilization of a reduced flavin, with the hydrogen bond between the side-chain nitrogen of Asn-123 and the isoalloxazine N(5) replaced by a hydrogen-bonding acceptor, the main-chain oxygen of Trp-400 (Fig. 4 C and D).
How might the interaction of mMICAL489 with NADPH trigger the repositioning of the cofactor? In the oxidized state, the out conformation of the flavin is stabilized by ring stacking between the isoalloxazine and Trp-400. The inability to form this coplanar π-complex on reduction of the flavin, combined with the change in the hydrogen-bonding properties of the isoalloxazine N(5), provides a plausible mechanism to trigger the switch to the in conformation. The observed (i.e., reduced) flavin ring conformation fits snugly with the MO and FAD-binding domain interface for the in conformation, whereas a modeled planar (i.e., oxidized) conformation generates steric clashes. Trp-400 appears to play a role comparable to that of the flavin-shielding aromatic residue that regulates electron transfer in the flavoenzyme-NADP+ reductase family (27). In this case, Trp-400 would stabilize the flavin ring in the out conformation before being displaced during attack on the reface of the isoalloxazine ring by NADPH.
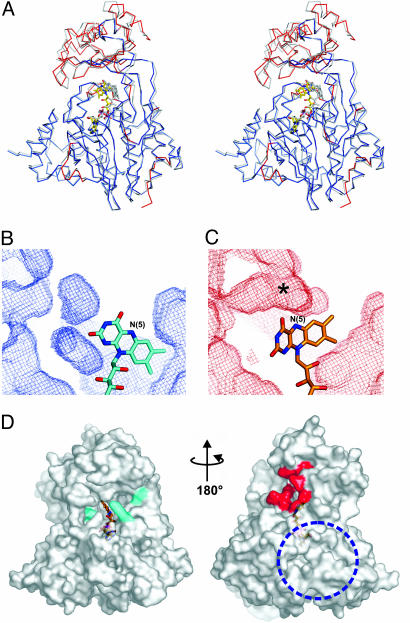
Domain Reorientation in Response to the Flavin Redox State. Several secondary structure elements that make major contributions to the active site in PHBH are missing in mMICAL489 (Fig. 2). Conversely, the PHBH substrate-binding cavity, when mapped to the equivalent position in native mMICAL489, is filled by the side chains of residues Tyr-287 and Tyr-293, which remains the case in the mMICAL489* structure. However, there are significant changes in the overall structure of the protein in response to the switch in flavin conformation between in (primed for catalytic activity) and out (positioned for NADPH interaction). The relative orientation of the MO and FAD-binding domains differs by 6.5° between the native mMICAL489 and mMICAL489* structures (Fig. 5A). No such rigid body shift is observed in PHBH or related hydroxylases.
Fig. 5.
NADPH-induced conformational changes of mMICAL489. (A) Stereo-view of the superposition of oxidized and reduced (gray) mMICAL489. FAD molecules are drawn as sticks. The oxidized form is colored red where there are significant differences between the structures, whereas blue parts are super-imposable [significance level is defined by default criteria of the program escet (37)]. (B and C) Cavities in the oxidized (B) and reduced (C) forms drawn as chicken wire (red, oxidized; cyan, reduced). Both have identical depth slab and orientation, centered on the MO/FAD-binding domain interface (protein atoms are omitted for clarity). The asterisk marks the increase of cavity volume in the reduced form (channel). (D) The solvent-accessible surface of mMICAL489* with the channel entrance highlighted in red (orientation is as in Fig. 1B). Potential NADPH-binding residues are colored cyan. A blue dotted circle marks the patch of basic electrostatic potential. The orientation is as in Fig. 1B.
Two unique structural features of mMICAL permit the domain reorientation. First, in other MOs, the interactions to the isoalloxazine are made exclusively by residues of the FAD-binding domain, but, in native mMICAL489, Tyr-293 hydrogen-bonds to the flavin O4. The loss of this hydrogen bond on reduction of the flavin may be a major factor driving the rearrangement of the interface between the MO and FAD-binding domains. Secondly, the interface between the MO and FAD-binding domains is inherently less rigid in mMICAL because of the less complex fold topology (Fig. 2A). The MO domain is formed by a single insert in the FAD-binding domain, allowing β-strands 9 and 15 to act as a simple hinge. The ability of the MO domain to reorientate in response to the oxidation state of the flavin has important implications for substrate access to the active site.
A Gated Mechanism for Substrate Access to the Active Site. The catalytic activity of flavoenzymes depends on substrate being brought into close proximity with the N(5) and C(4a) atoms of the reduced isoalloxazine ring (10) (Fig. 4D). For the in (active) conformation of the PHBH family, the reduced ring is embedded in the interface between the MO and FAD-binding domains, with the small molecule substrate-binding pocket abutting the N(5) and C(4a) atoms. The active site is thus shielded from bulk solvent (as required for a redox reaction), but a mechanism is needed to load substrate into the binding pocket.
In PHBH, the out conformation allows substrate access to the active site (22, 23). The equivalent route in mMICAL-1 is closed off by Asn-123, which hydrogen-bonds N(5) in the out conformation (Fig. 5B). However, the 6.5° domain reorientation triggered by the change in oxidation state of the flavin (and the associated switch from out to in conformation) opens a channel that leads directly from the molecular surface to the heart of the active site (Fig. 5C). The residues lining this channel are highly conserved across all members of the MICAL family (Fig. 10, which is published as supporting information on the PNAS web site), suggesting that it is of functional importance. The length and width of this channel are sufficient to allow insertion of a substrate amino acid side chain. The channel opens out on the opposite side of the mMICAL surface to that occupied by the putative NADPH-binding site (Fig. 5D). This direction is not one typically used for substrate access in the hydroxylases; however, a channel running from this surface to the active site is used for substrate access in polyamine oxidase (28).
Discussion
Mutagenesis of the fly MICAL dinucleotide-binding motif, essential for FAD-binding and catalytic activity in other MOs (29), indicated that the MICAL N-terminal region is essential for semaphorin signaling in vivo (5). The results reported here establish that this region has a structure consistent with the flavoprotein MO (hydroxylase) subset of flavoenzymes and imply that it is enzymatically active with a catalytic cycle dependent on binding NADPH. This 500-residue region shows a high level of sequence identity between all MICALs both within and across species (e.g., 62% between fly and human MICAL-1; Fig. 10); thus, all of these molecules are likely to show similar enzymatic activity. This prediction is consistent with the finding that the green tea-derived compound epigallocatechin gallate, a specific inhibitor for NADPH-dependent hydroxylases such as PHBH, abrogates mammalian Sema3A-mediated axon repulsion and growth cone collapse in vitro (5). The addition of the MICALs to the flavoprotein MO family introduces the hydroxylase functionality to a completely different type of molecule: one that is multidomain, bears a plethora of protein-binding sites, and is implicated in signal transduction. Classically, the substrates of the hydroxylases (for example, PHBH) have been small molecules (30), but several lines of argument suggest that the MICALs may show a unique functionality, that of targeting protein substrate(s).
Unlike most wild-type hydroxylase crystal structures, the structure of mMICAL489 shows the isoalloxazine ring in the out position. For mMICAL, this position appears to be a highly stable conformation for the isoalloxazine ring when in the oxidized state and is maintained by interactions with residues unique to the MICALs [in particular, ring stacking interactions with Trp-400 and a hydrogen-bonding network involving N(5) and Asn-123]. NADPH binding and consequent reduction of the isoalloxazine ring [by hydrogenation at the N(5) position to produce N(5)H] triggers a switch to the in conformation of the mMICAL489* crystal structure. Structural and fluorescence data (see Supporting Text) indicate that for mMICAL489, in the absence of substrate, the in conformation is inherently less stable, implying that docking of a macromolecular substrate is tightly synchronized with the switch to the catalytically active state. In flavoenzymes, the addition of oxygen to a reduced isoalloxazine ring results in production of C(4a)-hydroperoxide (30). At this point, unless the C(4a)-hydroperoxide and N(5)H groups are sequestered from bulk solvent, there is rapid decay to hydrogen peroxide and oxidized flavin. In the prototypic hydroxylases, such as PHBH, this nonproductive reaction is avoided by the complete burial of the isoalloxazine ring between the MO and FAD-binding domains for the in conformation, but in the MICALs, binding of a macromolecular substrate appears necessary to block solvent access to the flavin C(4a)-hydroperoxide group. Phenylalanine and tyrosine are obvious candidate targets for a hydroxylation reaction, because they are directly analogous to the PHBH small molecule substrate p-hydroxybenzoate; however, biologically relevant oxidative modification of other amino acid side chains can occur (e.g., hydroxylation of lysine and proline, formation of methionine sulfoxide, and modification of cysteine) (31). An extensive patch of basic potential (Fig. 1B), conserved in all MICALs, is suggestive of a binding surface for a positively charged protein substrate, and it is noteworthy not only that actin and many actin-related proteins are highly acidic (32) but also that oxidation of actin leads to disassembly of actin filaments and collapse of actin networks (33). In addition to actin (which could be presented by first binding to the CH domain of MICAL), candidate substrates currently include many other components of the semaphorin-mediated signaling cascade (34).
Regardless of the substrate identity, the MICALs must function in a more complex fashion than the prototypic hydroxylases; their multidomain architecture (Fig. 6) implies that enzymatic activity is controlled by a hierarchy of protein-protein interactions. These interactions could modulate the enzymatic activity indirectly, serving to control access to substrate by localization, to capture substrate before active-site binding, or to juxtapose product with the next element in the signaling cascade. However, intra- and inter-molecular interactions appear likely to directly control the nature and rate of enzymatic activity, because either NADPH binding must be stringently regulated to occur only when substrate is available or the active site must be sealed in the absence of substrate (otherwise, MICALs would function as hydrogen peroxide pumps, generating reactive oxygen species) (35). The protein-binding domains of MICAL could play a role in this regulation. For example, in the absence of substrate, the CH domain could switch off enzymatic activity by stopping NADPH binding, but on interaction with a cytoskeleton protein, the CH domain could reorientate to allow NADPH binding and present the substrate for catalysis. The structure of this hybrid family of flavoenzyme-signaling molecules links plexin binding with hydroxylation of substrate. An elucidation of the mechanism by which these two functionalities can modulate each other must be the aim of further structural and functional studies.
Fig. 6.
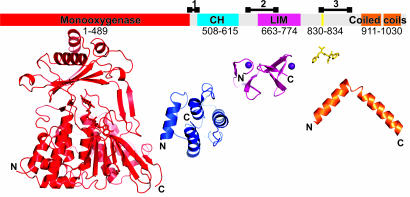
Domain organization of MICAL. In Upper, the domain structure of murine MICAL-1 is mapped onto a bar representing the linear sequence. The boundaries for the enzymatic (MO) domain (red) are taken from the mMICAL489 crystal structure. Boundaries for the CH (blue) and LIM (magenta) domains are defined from sequence alignments with homologous structures. A Src homology 3 (SH3) domain-binding motif is marked in yellow. Two coiled-coil regions are highlighted in orange, and potentially flexible linker regions are indicated by black lines above the bar (region 1, 486-512; region 2, 635-679; region 3, 830-883). Lower shows, to scale, the available structural information: CH domain, LIM domain, SH3 domain-binding motif, and examples of coiled coil (PDB ID codes 1BKR, 1IML, 1N5Z, and 1I84, respectively). Structures are orientated sequentially, with N and C termini marked.
Supplementary Material
Acknowledgments
We thank the staff of the European Synchrotron Radiation Facility and European Molecular Biology Laboratory outstation (Grenoble, France) for assistance with data collection, J. Brown for thermal shift assays, and R. Aplin for mass spectrometry. This work was supported by the European Community Integrated Program (Structural Proteomics in Europe) (QLG2-CT-2002-00988), the U.K. Medical Research Council (MRC), and Cancer Research UK (CR-UK). The Oxford Protein Production Facility is funded by the MRC. D.I.S. is a MRC Research Professor, and E.Y.J. is a CR-UK Principal Research Fellow.
Author contributions: C.S. and E.Y.J. designed research; C.S., N.B., T.S.W., and K.H. performed research; R.J.O., D.I.S., J.R.T., A.L.K., and R.J.P. contributed new reagents/analytic tools; C.S., D.I.S., and E.Y.J. analyzed data; and C.S., D.I.S., A.L.K., R.J.P., and E.Y.J. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MICAL, molecule interacting with CasL; PHBH, p-hydroxybenzoate hydroxylase; MO, monooxygenase; CH, calponin homology; rmsd, rms deviation.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2BRY (mMICAL489) and 2C4C (mMICAL489*)].
References
- 1.Huber, A. B., Kolodkin, A. L., Ginty, D. D. & Cloutier, J. F. (2003) Annu. Rev. Neurosci. 26, 509-563. [DOI] [PubMed] [Google Scholar]
- 2.Antipenko, A., Himanen, J. P., van Leyen, K., Nardi-Dei, V., Lesniak, J., Barton, W. A., Rajashankar, K. R., Lu, M., Hoemme, C., Puschel, A. W. & Nikolov, D. B. (2003) Neuron 39, 589-598. [DOI] [PubMed] [Google Scholar]
- 3.Love, C. A., Harlos, K., Mavaddat, N., Davis, S. J., Stuart, D. I., Jones, E. Y. & Esnouf, R. M. (2003) Nat. Struct. Biol. 10, 843-848. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki, T., Nakamoto, T., Ogawa, S., Seo, S., Matsumura, T., Tachibana, K., Morimoto, C. & Hirai, H. (2002) J. Biol. Chem. 277, 14933-14941. [DOI] [PubMed] [Google Scholar]
- 5.Terman, J. R., Mao, T., Pasterkamp, R. J., Yu, H. H. & Kolodkin, A. L. (2002) Cell 109, 887-900. [DOI] [PubMed] [Google Scholar]
- 6.Weide, T., Teuber, J., Bayer, M. & Barnekow, A. (2003) Biochem. Biophys. Res. Commun. 306, 79-86. [DOI] [PubMed] [Google Scholar]
- 7.Gimona, M., Djinovic-Carugo, K., Kranewitter, W. J. & Winder, S. J. (2002) FEBS Lett. 513, 98-106. [DOI] [PubMed] [Google Scholar]
- 8.Kadrmas, J. L. & Beckerle, M. C. (2004) Nat. Rev. Mol. Cell. Biol. 5, 920-931. [DOI] [PubMed] [Google Scholar]
- 9.Fischer, J., Weide, T. & Barnekow, A. (2005) Biochem. Biophys. Res. Commun. 328, 415-423. [DOI] [PubMed] [Google Scholar]
- 10.Fraaije, M. W. & Mattevi, A. (2000) Trends Biochem. Sci. 25, 126-132. [DOI] [PubMed] [Google Scholar]
- 11.Wierenga, R. K., de Jong, R. J., Kalk, K. H., Hol, W. G. & Drenth, J. (1979) J. Mol. Biol. 131, 55-73. [DOI] [PubMed] [Google Scholar]
- 12.Walter, T. S., Diprose, J. M., Mayo, C. J., Siebold, C., Pickford, M. G., Carter, L., Sutton, G. C., Berrow, N. S., Brown, J., Berry, I. M., et al. (2005) Acta Crystallogr. D 61, 651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 14.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 58, 1772-1779. [DOI] [PubMed] [Google Scholar]
- 15.Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003) Acta Crystallogr. D 59, 2023-2030. [DOI] [PubMed] [Google Scholar]
- 16.Terwilliger, T. C. (2003) Acta Crystallogr. D 59, 1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 18.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-355. [DOI] [PubMed] [Google Scholar]
- 19.Kissinger, C. R., Gehlhaar, O. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484-491. [DOI] [PubMed] [Google Scholar]
- 20.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol 6, 458-463. [DOI] [PubMed] [Google Scholar]
- 21.Eppink, M. H., Schreuder, H. A. & Van Berkel, W. J. (1997) Protein Sci. 6, 2454-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatti, D. L., Palfey, B. A., Lah, M. S., Entsch, B., Massey, V., Ballou, D. P. & Ludwig, M. L. (1994) Science 266, 110-114. [DOI] [PubMed] [Google Scholar]
- 23.Schreuder, H. A., Mattevi, A., Obmolova, G., Kalk, K. H., Hol, W. G., van der Bolt, F. J. & van Berkel, W. J. (1994) Biochemistry 33, 10161-10170. [DOI] [PubMed] [Google Scholar]
- 24.Wang, J., Ortiz-Maldonado, M., Entsch, B., Massey, V., Ballou, D. & Gatti, D. L. (2002) Proc. Natl. Acad. Sci. USA 99, 608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eppink, M. H., Schreuder, H. A. & van Berkel, W. J. (1998) J. Biol. Chem. 273, 21031-21039. [DOI] [PubMed] [Google Scholar]
- 26.Eppink, M. H., Schreuder, H. A. & van Berkel, W. J. (1998) Eur. J. Biochem. 253, 194-201. [DOI] [PubMed] [Google Scholar]
- 27.Deng, Z., Aliverti, A., Zanetti, G., Arakaki, A. K., Ottado, J., Orellano, E. G., Calcaterra, N. B., Ceccarelli, E. A., Carrillo, N. & Karplus, P. A. (1999) Nat. Struct. Biol. 6, 847-853. [DOI] [PubMed] [Google Scholar]
- 28.Binda, C., Coda, A., Angelini, R., Federico, R., Ascenzi, P. & Mattevi, A. (1999) Structure Fold. Des. 7, 265-276. [DOI] [PubMed] [Google Scholar]
- 29.Dym, O. & Eisenberg, D. (2001) Protein Sci. 10, 1712-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghisla, S. & Massey, V. (1989) Eur. J. Biochem. 181, 1-17. [DOI] [PubMed] [Google Scholar]
- 31.Shacter, E. (2000) Drug Metab Rev 32, 307-326. [DOI] [PubMed] [Google Scholar]
- 32.Otterbein, L. R., Graceffa, P. & Dominguez, R. (2001) Science 293, 708-711. [DOI] [PubMed] [Google Scholar]
- 33.Pollard, T. D. & Borisy, G. G. (2003) Cell 112, 453-465. [DOI] [PubMed] [Google Scholar]
- 34.Pasterkamp, R. J. & Kolodkin, A. L. (2003) Curr. Opin. Neurobiol. 13, 79-89. [DOI] [PubMed] [Google Scholar]
- 35.Finkel, T. (1998) Curr. Opin. Cell Biol. 10, 248-253. [DOI] [PubMed] [Google Scholar]
- 36.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281-296. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, T. R. (2004) Acta Crystallogr. D 60, 2269-2275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.