Abstract
Atherosclerosis is a chronic inflammatory process and progresses through characteristic morphologic stages. We have shown previously that chronically injecting bone-marrow-derived vascular progenitor cells can effect arterial repair. This repair capacity depends on the age of the injected marrow cells, suggesting a progressive decline in progenitor cell function. We hypothesized that the progression of atherosclerosis coincides with the deteriorating repair capacity of the bone marrow. Here, we ascribe patterns of gene expression that accurately and reproducibly identify specific disease states in murine atherosclerosis. We then use these expression patterns to determine the point in the disease process at which the repair of arteries by competent bone marrow cells ceases to be efficient. We show that the loss of the molecular signature for competent repair is concurrent with the initiation of atherosclerotic lesions. This work provides a previously unreported comprehensive molecular data set using broad-based analysis that links the loss of successful repair with the progression of a chronic illness.
Keywords: genomic, vascular progenitor cell, aging, inflammation, bone marrow obsolescence
Atherosclerosis is a pathological inflammation of the arteries preceded by endothelial dysfunction. After injury, the arterial wall becomes lipid-laden and undergoes characteristic morphologic changes through a wound-healing-like process (1-3). Ultimately, as atherosclerosis progresses, aberrant vascular remodeling occurs, leading to arterial lumen narrowing, aneurysm formation, and atheroma plaque instability. Although therapies targeting inflammation and cholesterol homeostasis have proven effective in preventing thromboembolic sequelae of atherosclerosis, atherosclerosis remains a leading health problem worldwide, and a better understanding of the disease process is needed.
The regenerative potential of bone-marrow-derived progenitor cells has stirred great excitement for disease processes previously thought to be irreversible (4). Recently, our group was the first to demonstrate that chronic i.v. injection of bone-marrow-derived vascular progenitor cells (BM-VPCs) can attenuate the progression of atherosclerosis in mice that spontaneously develop lesions in their aorta (5). When the recipient aortas are examined for where the injected bone marrow cells localize, many of the injected bone marrow cells are found to reconstitute the endothelium at sites prone to atherosclerosis. This reparative effect is only found in mice with markedly elevated cholesterol (1,200-1,400 mg/dl) but does not affect the cholesterol level of the recipient mice. However, the reparative effect is associated with a decrease in IL-6 levels proportional to the therapeutic effect of the bone marrow cells. Remarkably, the repair capacity of the BM-VPCs seems to be progressively lost with aging in the presence of sustained arterial injury. Our findings have led us to postulate that BM-VPCs may play a physiologic role in preventing atherosclerosis and that a reduced capacity for arterial repair coincides with disease advancement (5, 6).
We also recently developed an approach using large-scale microarray analysis to identify patterns of expression that are highly correlated with atherosclerotic burden in human aortas (7). Because microarrays functionally assess loci by measuring the expression of thousands of genes simultaneously, our approach represents a relatively unbiased strategy for prioritizing genes related to atherosclerosis. A great majority of the genes identified have not yet been implicated in atherosclerosis and remain important targets for future investigations. Our initial human study focused on aortas that were either minimally or severely diseased and identified likely genes chronically associated with atherosclerosis. However, the complement of genes affecting each morphologic transition in atherosclerosis is likely to be distinct. For example, variants of genes such as ABCA1 and LDLr may predispose to early onset of disease, but variants of genes such as MEF2A may predispose to thromboembolic sequelae in the presence of established disease (8-12). Therefore, a better understanding of the genes and a definition of the biological processes at the molecular level that contribute to the initiation and progression of atherosclerosis are needed.
We hypothesize (i) that discrete expression programs contribute to different stages of atherosclerosis underscoring distinct genomic phenotypes at each stage and (ii) that a major event in the atherosclerotic process corresponds to the progressive loss of successful repair provided by specialized bone marrow progenitors and, perhaps, additional marrow lineages. Here, we define the natural molecular history of atherosclerosis using gene expression and use this database to assess the timing of the changing phenotype of vascular progenitor cells and its association with the progression of atherosclerosis. We link the advancement of a chronic disease with a change in the bone-marrow-mediated repair phenotype.
Materials and Methods
Mouse Aorta Samples. Wild-type (WT) and apoE-/- C57BL/6J mice purchased from The Jackson Laboratory were used to breed the mice used in the study. Pups were weaned at 3 weeks (wk) of age and fed either a chow or a Western diet ad libitum (Teklad, Madison, WI). Mice were euthanized at 6, 12, or 16 wk. At the time of euthanization, aortas were rinsed in PBS and either fixed in RNALater (Ambion, Austin, TX) or 10% formalin. Samples fixed in RNALater were homogenized, and total RNA was extracted by using an RNA Minikit (Qiagen, Valencia, CA). All mouse work was approved by the Duke University Institutional Animal Care and Use Committee.
Assessment of Lesion Burden. Formalin-fixed aortas were stained with the Oil Red O and photographed. Each aorta image was projected to a reference aorta template, derived from the average size and shape of all aortas surveyed. Extent of disease was quantified by counting pixels stained with Oil Red O (13). For each segment of the aorta, the fraction of the total area that stained positively with Oil Red O was determined. These values were logit transformed to enhance normality of the data, as was done in the Pathobiological Determinants of Atherosclerosis in Youth study (13). The Student-Newman-Keuls procedure was used to control for multiple testing.
Microarray Hybridization. After verifying the 28S:18S rRNA ratio to ensure quality, equal fractions of RNA from two or three aortas was pooled and sent to the Duke Microarray Core Facility for cDNA synthesis and hybridization to a MG-U74Av2 oligonucleotide microarray (Affymetrix, Santa Clara, CA). Hybridization was done by using the standard Affymetrix GeneChip protocol. Quality of microarray experiments was verified by inspection of the array image and assessment of scaling factors and of housekeeping genes.
Expression Phenotyping. Expression data were analyzed by using the method of West et al. (14). After background correction with the Affymetrix MAS5 algorithm, data were log2 transformed and quantile normalized. Genes then were filtered to eliminate those that do not vary across experimental conditions. Genes were selected for correlation with experimental outcome. Metagenes were created from the principal components of these top genes and then were used as predictors in probit regression models. Models were evaluated by “hold-one out” cross-validation. Candidate genes were identified by their correlation with outcome and their relative weight in the metagenes. For prospective data analysis, data from the “test set” were first quantile normalized to the medians of the “training data” and then assigned classification by the probit regression model created using the test set.
Gene Annotation. Candidate genes were annotated by using the Duke Integrated Genomics Database (https://dig.cgt.duke.edu) and fatigo (15). Gene Ontology (GO) terms for Biological Process that are overrepresented for each predictor were identified by using the GOHyperG function in the gostats package for bioconductor (16). Comparison of the distribution of genes in GO categories for different gene sets was done by first mapping each probe ID to GO terms using bioconductor, then counting the number of genes in each GO category, and finally calculating Pearson's correlation coefficient for these GO distributions. Significance of the correlation was determined by comparing the observed correlation coefficient with the correlation coefficients for 10,000 randomly selected predictors.
Comparison of Mouse Predictors. To assess the statistical significance of the number of overlapping genes in our predictors, we generated experimental predictors A and B to assess the probability of gene overlap by random chance. Predictor A was created by randomly picking x genes and predictor B was created by randomly picking a set of y genes. This process was repeated 10,000 times. For each set of A and B, the number of overlapping genes was determined, thus creating an empiric distribution and allowing for an estimate of the chance of overlap between two predictors of the same size as A and B. P values indicate the fraction of random predictors with more genes in common than we observed experimentally.
Comparison of Murine and Human Expression Data. Probe sets on the HG-U95Av2 array were mapped to homologous probe sets on the MG-u74Av2 using resourcerer (The Institute for Genomic Research, Rockville, MD) (17). Significance of the overlap between mouse and human predictors was assessed as described above for the mouse predictors. For classification of human samples with the mouse predictors, only genes present on both murine and human arrays were considered. By using this trimmed gene set, the mouse data then were used as the training data to rederive the gene expression predictors. These mouse predictors then were used to classify the human data (test set). Details on disease severity indices for human aortas, Sudan IV and raised lesion scores, are described in ref. 7.
Bone-Marrow-Treated Aortas. Mice treated with bone marrow were i.v. administered 1 × 106 whole bone marrow cells from either young (3-5 wk) or old (1 year) apoE-/- donor mice every 2 wk (n = 10) from 4 wk of age and maintained on a high-fat diet. All recipient mice were killed at 16 wk of age, after a total of six bone marrow injections. RNA was extracted from these aortas as described above.
Results
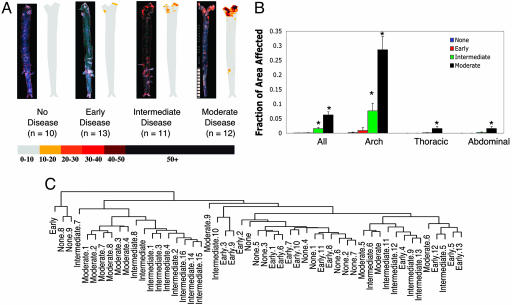
Gene Expression Phenotypes of Atherosclerosis. With our first set of experiments, we surveyed the dynamic nature of gene expression as a function of time in an established model of atherosclerosis. We created a large database of gene expression data at discrete, consecutive stages of the disease process using apolipoprotein E-null (apoE-/-) C57BL6/J mice, because they have been shown to closely mimic human atherosclerosis both in the spontaneous appearance of lesions and the distribution of lesions within the vasculature (18-20). Because WT mice have intrinsic resistance to atherosclerosis, aortas from 6-wk-old WT C57BL6/J mice fed a chow diet served as the baseline for absence of disease (21). We took 6-wk-old apoE-/- mice fed a chow diet to represent “early” disease because these mice do not have grossly detectable disease but are known to have monocytes adherent to their aortic endothelium (22). For advanced disease, increasing age and a high-fat diet is needed in the apoE-/- model (22). Therefore, we assigned 12-wk-old apoE-/- mice and 16-wk-old apoE-/- C57BL6/J mice fed a high-fat diet as having “intermediate” and “moderate” disease, respectively. We confirmed the difference in disease extent of these four groups with Oil Red O staining, a measure of atherosclerotic burden that has been extensively validated in human and murine atherosclerosis (Fig. 1 A and B) (13, 20). Samples with intermediate and moderate disease have significantly increased disease burden compared with the other groups (P < 0.05 using the Student-Newman-Keuls correction for multiple comparisons). Of note, no significant difference in lesion burden between the groups with no disease and early disease was observed.
Fig. 1.
Lesion burden of groups. (A) For each group, a representative aorta stained with Oil Red O and an averaged image for all aortas in the group is given. Regions of the aorta that stain deeply with Oil Red O have high lipid content. For composite images, color indicates proportion of aortas in the group with positive staining for that pixel (see color bar). (B) Quantitative comparison of disease burden across groups. The y axis shows percent of aortic segment that stained positive with Oil Red O. * denotes groups with significantly different disease burdens from the other groups (P < 0.05) after logit transformation. Error bars reflect SEM. (C) Dendrogram showing results of hierarchical clustering of samples based on all gene expression data. Distances were computed by using centered Pearson's correlation, and centroid clustering was performed. Samples with similar disease state are grouped together.
Once we were able to define groups of mice with reproducible levels of disease, we extracted RNA from representative aortas for hybridization to Affymetrix MG-U74Av2 microarrays (20). In total, we ran 10, 14, 17, and 10 array experiments for the groups with no disease, early disease, intermediate disease, and moderate disease, respectively. Each array experiment used RNA from a unique pool of two or three aortas. We initially used hierarchical clustering to determine whether patterns of gene expression correlate with disease extent. Fig, 1C shows a trend where samples with the same degree of disease tend to cluster together based on gene expression profiling. This result confirms our prior finding in human aortas that gene expression patterns correlate with disease severity and also suggests that distinct sets of genes may mark the transition from one disease stage to the next (7).
To identify which genes are associated with the morphologic progression of atherosclerosis, we compared expression profiles of mice with (i) no disease vs. early disease, (ii) early vs. intermediate disease, (iii) intermediate vs. moderate disease, and (iv) no disease vs. moderate disease. We elected to use predictive modeling to associate genes with each disease interval, because predictive modeling not only identifies genes that are associated with a phenotype but also allows for testing of the reproducibility of these patterns through cross-validation and with independent data sets. To define our predictors, we used the methodology of West et al. (14), which we successfully used in our prior study of human atherosclerosis (20). This approach groups genes into “metagenes” that share common gene expression patterns and then uses the metagenes to create a standard binary regression model capable of classifying samples. Candidate genes make up the most predictive metagenes (14). This approach is unique in that sets of related genes are assessed for their predictive ability based on their collective expression and is particularly relevant for atherosclerosis, because the disease likely results from the additive effects of numerous genes and their variable expression. To guard against overfitting and to provide an unbiased estimate of the true predictive accuracy of the model, standard hold-one-out of sample cross-validation was used.
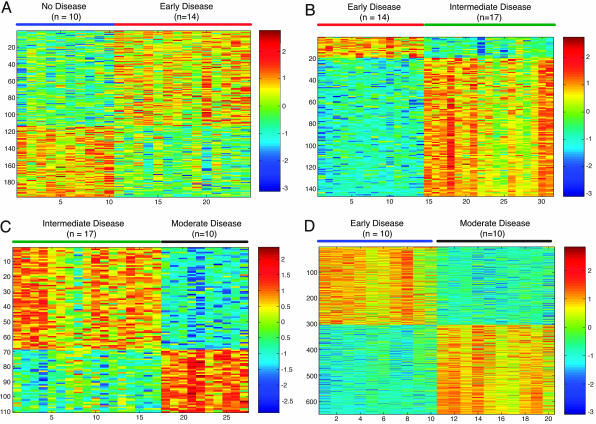
We identified 197 genes (see Table 1, which is published as supporting information on the PNAS web site) associated with the transition from no disease to early disease (24/24 = 100% of samples correctly classified on cross-validation), 146 genes (see Table 2, which is published as supporting information on the PNAS web site) associated with the transition from early to intermediate disease (31/31 = 100% of samples correctly classified on cross-validation), 110 genes (see Table 3, which is published as supporting information on the PNAS web site) associated with the transition from intermediate to moderate disease (26/27 = 96% of samples correctly classified on cross-validation), and 650 genes (see Table 4, which is published as supporting information on the PNAS web site) associated with the transition from no disease to moderate disease (20/20 = 100% of samples correctly classified on cross-validation). Differential expression of these genes across each disease interval is depicted in Fig. 2.
Fig. 2.
Gene expression predictors of disease intervals. (A) Color plot showing expression patterns of genes used to build predictor of undiseased aortas and aortas with early disease. Rows depict standardized expression of genes across samples (columns). Blue represents relatively lower expression compared with red (higher expression). Undiseased and early disease samples are indicated by blue and red lines, respectively. (B) Expression pattern of genes that predict aortas with early disease from aortas with intermediate disease. Samples with early and intermediate disease are indicated by red and green lines, respectively. (C) Expression pattern of genes that predict aortas with intermediate disease from aortas with moderate disease. Samples with intermediate and moderate disease are indicated by green and black lines, respectively. (D) Expression pattern of genes that predict aortas with no disease from aortas with moderate disease. Samples without disease and with moderate disease are indicated by blue and black lines, respectively.
Our initial approach to identify the relevance of these genes was to determine whether they participate in processes already known to relate to atherosclerosis. Therefore, we looked for GO terms more commonly associated with these genes compared with randomly chosen genes (20, 23). For disease initiation, we found genes relating to metabolic processes, such as “lipid metabolism” (P = 0.024) and “lipoprotein metabolism,” to be significantly overrepresented (P = 0.051). Genes known to modify the metabolic state of the arterial wall by affecting cholesterol efflux, such as apoE and ABCA1, are in this predictor. apoE and ABCA1 are well known to modify susceptibility to atherosclerosis (10, 11, 24). For the progression of early disease, genes associated with “defense response” (or inflammation, P < 7 × 10-6) are significantly overrepresented. Genes in this predictor include SPP1/osteopontin, MMP12, SAA3, CTSS, CCL8, and IL1-RN, all of which are known to modify the inflammatory response. The predictor that distinguishes intermediate and moderate disease is enriched for genes that contribute to “nuclear organization and biogenesis” (P = 0.002) and “morphoogenesis” (P = 0.016) and may contribute to the changing cellular phenotypes that occur during vascular remodeling in advanced lesions. The gene set for this predictor has numerous transcription factors known to have roles in cell differentiation and tissue patterning, such as MAFF, EPAS-1, and RXR-α. The expression of individual genes that are annotated to the significantly overrepresented GO categories are given in Fig. 5A, which is published as supporting information on the PNAS web site.
Our experimental design is such that we identified genes associated with subtle changes in disease phenotype and a group of genes associated with a larger change in phenotype. Therefore, one would expect that the genes corresponding to smaller disease intervals should have significant overlap with the gene set that spans the larger disease process. Such an analysis represents an assessment of the reproducibility of our results. Indeed, there is significant overlap. The predictor comparing undiseased and moderately diseased aortas shares 21 genes with the predictor for disease initiation (P = 0.0010), 122 genes with the predictor for early disease progression (P < 0.0001), and 61 genes with the predictor for intermediate disease progression (P < 0.0001) (20). Such a high degree of concordance between these separate analyses helps to validate our approach and results. Interestingly, there is much less overlap between the predictors of the more subtle disease changes, suggesting that we have identified ordered programs of gene expression that relate to atherosclerosis.
As part of our prior large-scale survey of gene expression in human aortas, we have defined an expression pattern that differentiates minimally from severely diseased human aortas (7). Therefore, as a stringent assessment of both the validity and reproducibility of our results, we compared the results of our parallel human and mouse studies and found significant concordance (20). We identified 40 genes (of 650 unique genes, P < 0.0001) in the mouse predictor comparing undiseased with moderately diseased aortas that are also an integral part of our human predictor. The overlapping genes for the mouse and human studies are indicated in Tables 1-4 by asterisks. Interestingly, the concordance of our mouse and human studies extends beyond individual genes to the GO biological processes. The distribution of genes within GO categories for the mouse predictor and our human predictor are nearly identical (r2 = 0.97, P < 0.0001) (20).
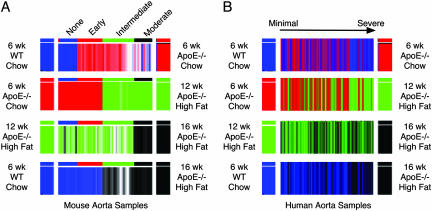
Recently, others have shown that expression signatures of pathways are conserved across species (25). Thus, a third and extreme verification of our results would be to verify that the different sets of genes we have identified in murine disease vary similarly in human and mouse aorta samples with advancing disease severity (20). Genes on the mouse microarray were mapped to the human microarray by using resourcerer (The Institute for Genomic Research) as described in ref. 26. Next, each of the predictors differentiating murine aorta samples was asked to assign classifications to the human samples from our previous work (7). Fig. 3 shows how the predictions vary as a function of disease severity in the mouse and human samples. Comparison of Fig. 3 A and B shows that classifications progress across mouse and human disease stages with similar patterns, providing additional support to the associations we have identified in the mouse samples and suggesting that gene expression phenotypes of atherosclerosis are conserved across species.
Fig. 3.
Conserved expression phenotypes for atherosclerosis. (A) Classification of mouse samples ordered from no disease (samples on left) to moderate disease (samples on right) by mouse predictors. For each predictor, mouse samples are assigned a classification as indicated by the color on either end. For example, the top row shows classification of mouse samples by the predictor for disease initiation. The collective expression of genes in this predictor is more similar to the undiseased samples in the samples colored blue and more similar to samples with early disease in the samples colored red. Shade of coloring is graded by strength of the prediction. (B) Classification of human samples ordered from minimal (samples on left) to advanced (samples on right) disease by mouse predictors. For each predictor, human samples are assigned a classification as indicated by the color on either end, as in A. Coloring is graded by strength of the prediction.
Gene Expression Signature of Arterial Repair. Having defined patterns of genes associated with specific atherosclerotic disease states, we tested our second hypothesis that lesion inception coincides with the loss of successful arterial repair, repair that is based, at least to a significant extent, on the availability of circulating bone marrow progenitor cells. We have previously shown chronic injection of BM-VPCs to slow the progression of atherosclerosis in recipient apoE-/- mice (5). The reparative effect can be accomplished by cells from young (3-5 wk), but not old (>6 months), apoE-/- mice. We noted that the injected cells engrafted onto the aortic surface at sites prone to atherosclerosis, with the majority of injected cells becoming endothelial cells and a smaller population of cells expressing markers of other arterial cell types. Instructively, the fraction of BM-VPCs we identified to become endothelial cells markedly decreased with aging, suggesting that exhaustion of BM-VPCs may be related to the advancement of atherosclerosis. Here, we surmised that BM-VPCs may exert their antiatherosclerotic effect by repairing damaged endothelium and that such an activity might have an identifiable gene expression signature. If such an activity would be detectable by gene expression survey, we hoped to define, using our “natural history” of gene expression in the course of atherosclerosis, the specific time point in the disease process when the reparative effect of BM-VPCs ceased.
We first added to our database expression profiles obtained from aortas of moderately diseased mice (16-wk-old apoE-/- mice fed a high-fat diet) treated with weekly i.v. injections of whole bone marrow from old apoE-/- mice (>1 year old) or young apoE-/- mice (3-5 wk old) (20). Ten unique array experiments were run for each group. The rationale for the selection of the two data sets is based on the premise that moderately diseased aortas rescued by young apoE-/- BM-VPCs would provide an expression profile that reflects successful ongoing arterial repair, whereas the expression profile resulting from old (>1 year old) apoE-/- BM-VPC treatment, if any different from the intrinsic atherosclerotic aorta profile, would display accentuated inflammation markers. We next developed a gene expression model predicting aortas treated with young vs. old BM-VPCs. This model is made up of 181 genes (see Table 5, which is published as supporting information on the PNAS web site; see also Fig. 3A) and has ≈75% (15/20 samples) predictive accuracy by cross-validation. For this predictor of repair phenotype, genes associated with “angiogenesis” (P = 0.018), “blood vessel development” (P = 0.032), “cell growth regulation” (P = 0.005), and “regulation of cell size” (P = 0.013) are significantly overrepresented. Of interest, this predictor has numerous genes involved in cardiac myocyte morphogenesis such as MDFI, GATA4, XIN, MEF2C, and FOXH1. Additionally, several genes associated with cell recruitment such as ICAM1, MCAM, and CCR2 are in this predictor. Thus, the genes of this predictor seem to be involved in processes likely to affect cell recruitment and arterial repair. The expression of selected individual genes across an atherosclerotic gradient is given in Fig. 5B.
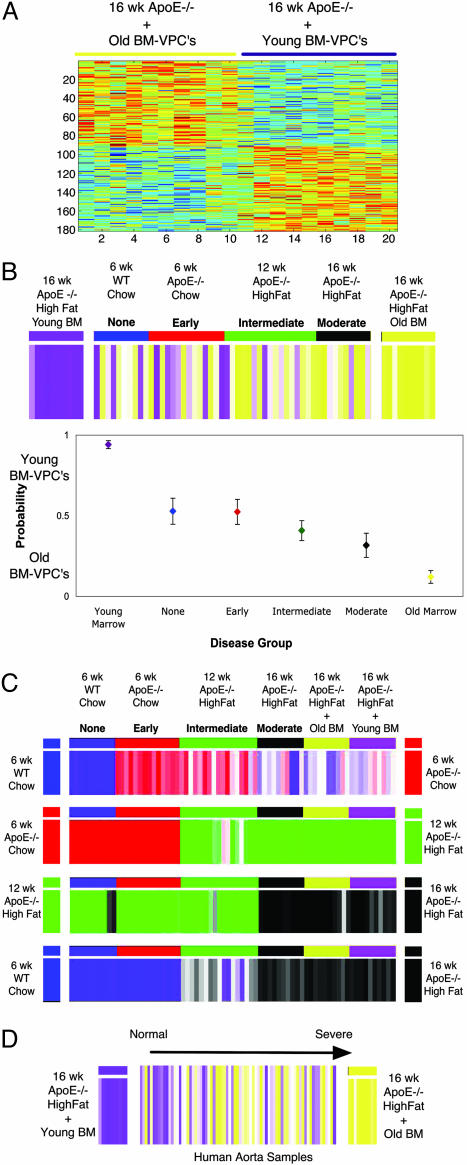
To determine whether a change in the repair phenotype is associated with the progression of atherosclerosis, we next asked how this model would classify samples along a disease gradient. Fig. 4B shows that mice with no disease are generally assigned an indeterminate classification but those with early disease have an expression profile more similar to that of the aortas treated with young BM-VPCs. By contrast, aorta samples with intermediate and moderate disease have an expression pattern more similar to that of the aortas treated with the old BM-VPCs. To understand how the changing phenotype of the BM-VPC repair process coincides with the other processes that we associated with atherosclerosis, we used our stage-specific predictive models to assign classifications to samples with different degrees of atherosclerosis. Interestingly, the point in the natural history of the atherosclerotic disease process where the expression patterns of aorta begins to switch from young to old BM-VPCs treatments coincides with the point at which the inflammatory response become most marked and gross lesions of atherosclerosis become detectable.
Fig. 4.
Predictor of BM-VPC repair phenotype. Each predictor was used to classify additional samples of differing disease severity. Blue samples, no disease (n = 10); red, early disease (n = 14); green, intermediate disease (n = 17); black, moderate disease (n = 10). (A) Expression pattern of genes that predict aortas treated with young BM-VPCs from aortas treated with old BM-VPCs. Samples treated with young and old BM-VPCs are indicated by purple and yellow lines, respectively. (B) Classification of samples by model differentiating aortas treated with young (purple) and old (yellow) BM-VPCs. For this set of genes, samples in yellow and purple have an expression pattern more similar to that of the aortas treated with young and old BM-VPCs, respectively. Shade of coloring is proportional to the strength of prediction. Adjacent plot shows mean ± SEM probability of being classified as an aorta treated with young (probability of 1) or old (probability of 0) BM-VPCs. Classification of samples treated with BM-VPCs are included in the plot as controls for the model. (C) Classification of mouse samples by disease stage specific predictors. For each predictor, samples of additional disease stages are assigned a classification as indicated by the color on either end. Coloring is graded by strength of the prediction. (D) Classification of human samples ordered from minimal (samples on left) to advanced (samples on right) disease by mouse model differentiating aortas treated with young (purple) and old (yellow) BM-VPCs. Coloring is graded by strength of the prediction.
To illustrate the relevance of our results away from murine models, we sought to verify that the signature for arterial repair varies similarly with advancing disease severity in human and mouse aorta samples. Fig. 4D shows a trend where human aorta samples with minimal disease have an expression pattern similar to the mice treated with young bone marrow cells, and human samples with more advanced disease have an expression pattern similar to mouse aortas treated with old bone marrow cells. These results suggest not only that the bone marrow repair signature is conserved in human samples but also that a changing BM-VPC repair phenotype is associated with the advancement of human atherosclerosis.
Discussion
Atherosclerosis is a complex genetic disease that remains the most significant health problem worldwide. Considerable advances have been made in treating atherosclerotic disease, but with the increasing age of the population, atherosclerosis will remain a major health issue in the future (27, 28). One strategy for identifying better diagnostic and treatment modalities is a better understating of the basic molecular processes that contribute to atherosclerotic lesion initiation and progression.
This work presents a previously unreported systematic and comprehensive survey of gene expression throughout the natural history of atherosclerosis. By using a previously undescribed microarray-based approach, we have identified specific patterns of genes whose expression is associated with the initiation and progression of atherosclerosis. Our results provide considerable insight into the kinetics of the disease process and therefore the basic pathogenesis of atherosclerosis. The genes we have associated with the progression of atherosclerosis may represent important modifiers of atherosclerotic susceptibility and resistance. Experimentally, several of the genes identified by our analysis have been shown to retard or hasten lesion formation. Manipulation of VCAM1, CTSS, IL-18BP, and ABCA1 slows the onset of atherosclerosis, whereas IL-1RA null mice and SPP1 transgenic mice have enhanced atherosclerosis (29-34). Our work also has potentially important therapeutic implications because the genes identified in our analysis may represent previously undescribed targets for intervention, and the expression patterns we have identified may be used to stage disease.
Our group has previously noted that BM-VPCs may have therapeutic benefit in attenuating the progression of atherosclerosis. However, the present work associates a loss of the BM-VPC repair capacity with the stepwise advancement of disease, therefore providing a physiologic role for the bone marrow in repairing a damaged end organ. Our results are consistent with the theory, although do not prove it, that in the presence of a chronic insult (hypercholesterolemia in our model) the breakpoint in the tissue response corresponds to the onset of failing of the arterial repair provided by competent bone marrow cells. Although much work in the past two decades has focused on the metabolic and inflammatory basis of atherosclerosis, our work suggests that bone marrow may have an important physiologic role in repairing arterial tissues after injury and is potentially a major advance in our understanding of atherosclerosis. Our findings are also consistent with others' findings, namely that endothelial progenitor cells lose their ability to differentiate into mature endothelial cells when exposed to traditional risk factors for atherosclerosis (35). Additionally, our previous work demonstrated a decline in competent BM-VPCs within the bone marrow with aging. Such findings call for additional studies on the mechanisms for the progressing obsolescence of the bone marrow as it relates to capacity for end organ repair. Furthermore, although we have studied atherosclerosis here, the changing phenotype of bone-marrow-mediated repair is likely to be associated with the progression of other chronic diseases as well.
Supplementary Material
Acknowledgments
The work was supported by National Institute of Aging Pilot Grants AG 11268 (to D.S.) and AG023073 (to P.J.G.-C.) and John A Hartford Foundation Grant 990339 through the Duke University Pepper Older Americans Independence Center (to D.S.); National Institutes of Health Program Project Grant HL073042 (to P.J.G.-C.) through the National Heart, Lung, and Blood Institute; National Institutes of Health Research Project Grant HL71536 (to P.J.G.-C.); a Duke University Medical School Eugene A. Stead Scholarship (to R.K.); a Howard Hughes Medical Student Fellowship (to S.V.); and a gift from Bill and Peggy Britt.
Author contributions: R.K., S.V., C.D., J.R.N., P.J.G.-C., and D.S. designed research; R.K., S.V., E.E.H., X.S., K.S., and D.S. performed research; R.K., J.R.N., M.W., and D.S. contributed new reagents/analytic tools; R.K., C.D., E.E.H., P.J.G.-C., and D.S. analyzed data; and R.K., P.J.G.-C., and D.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: BM-VPC, bone marrow-derived vascular progenitor cell; GO, Gene Ontology.
References
- 1.Ross, R. (1999) N. Engl. J. Med. 340, 115-126. [DOI] [PubMed] [Google Scholar]
- 2.Libby, P. (2002) Nature 420, 868-874. [DOI] [PubMed] [Google Scholar]
- 3.Lusis, A. J. (2000) Nature 407, 233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korbling, M. & Estrov, Z. (2003) N. Engl. J. Med. 349, 570-582. [DOI] [PubMed] [Google Scholar]
- 5.Rauscher, F. M., Goldschmidt-Clermont, P. J., Davis, B. H., Wang, T., Gregg, D., Ramaswami, P., Pippen, A. M., Annex, B. H., Dong, C. & Taylor, D. A. (2003) Circulation 108, 457-463. [DOI] [PubMed] [Google Scholar]
- 6.Goldschmidt-Clermont, P. J. (2003) Am. Heart J. 146, S5-12. [DOI] [PubMed] [Google Scholar]
- 7.Seo, D. M., Wang, T., Dressman, H., Herderick, E. E., Iversen, E. S., Dong, C., Vata, K., Schulteis, R., Milano, C. A., Rigat, F., et al. (2004) ATVB 24, 1922-1927. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. S. & Goldstein, J. L. (1976) Cell 9, 663-674. [DOI] [PubMed] [Google Scholar]
- 9.Davis, C. G., Lehrman, M. A., Russell, D. W., Anderson, R. G., Brown, M. S. & Goldstein, J. L. (1986) Cell 45, 15-24. [DOI] [PubMed] [Google Scholar]
- 10.Bodzioch, M., Orso, E., Klucken, J., Langmann, T., Bottcher, A., Diederich, W., Drobnik, W., Barlage, S., Buchler, C., Porsch-Ozcurumez, M., et al. (1999) Nat. Genet. 22, 347-351. [DOI] [PubMed] [Google Scholar]
- 11.Rust, S., Rosier, M., Funke, H., Real, J., Amoura, Z., Piette, J. C., Deleuze, J. F., Brewer, H. B., Duverger, N., Denefle, P. & Assmann, G. (1999) Nat. Genet. 22, 352-355. [DOI] [PubMed] [Google Scholar]
- 12.Wang, L., Fan, C., Topol, S. E., Topol, E. J. & Wang, Q. (2003) Science 302, 1578-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathobiological Determinants of Atherosclerosis in Youth Research Group (1993) Arterioscler. Thromb. 13, 1291-1298. [DOI] [PubMed] [Google Scholar]
- 14.West, M., Blanchette, C., Dressman, H., Huang, E., Ishida, S., Spang, R., Zuzan, H., Olson, J. A., Jr., Marks, J. R. & Nevins, J. R. (2001) Proc. Natl. Acad. Sci. USA 98, 11462-11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Shahrour, F., Diaz-Uriarte, R. & Dopazo, J. (2004) Bioinformatics 20, 578-580. [DOI] [PubMed] [Google Scholar]
- 16.Ihaka, R. & Gentleman, R. (1996) J. Comput. Graph. Stat. 5, 299-314. [Google Scholar]
- 17.Tsai, J., Sultana, R., Lee, Y., Pertea, G., Karamycheva, S., Antonescu, V., Cho, J., Parvizi, B., Cheung, F. & Quackenbush, J. (2001) Genome Biol. 2, software0002.1-software0002.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, S. H., Reddick, R. L., Piedrahita, J. A. & Maeda, N. (1992) Science 258, 468-471. [DOI] [PubMed] [Google Scholar]
- 19.Plump, A. S., Smith, J. D., Hayek, T., Aalto-Setala, K., Walsh, A., Verstuyft, J. G., Rubin, E. M. & Breslow, J. L. (1992) Cell 71, 343-353. [DOI] [PubMed] [Google Scholar]
- 20.Daugherty, A. (2002) Am. J. Med. Sci. 323, 3-10. [DOI] [PubMed] [Google Scholar]
- 21.Breslow, J. L. (1996) Science 272, 685-688. [DOI] [PubMed] [Google Scholar]
- 22.Nakashima, Y., Plump, A. S., Raines, E. W., Breslow, J. L. & Ross, R. (1994) Arterioscler. Thromb. 14, 133-140. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson, P. W., Schaefer, E. J., Larson, M. G. & Ordovas, J. M. (1996) Arterioscler. Thromb. Vasc. Biol. 16, 1250-1255. [DOI] [PubMed] [Google Scholar]
- 25.Sweet-Cordero, A., Mukherjee, S., Subramanian, A., You, H., Roix, J. J., Ladd-Acosta, C., Mesirov, J., Golub, T. R. & Jacks, T. (2005) Nat. Genet. 37, 48-55. [DOI] [PubMed] [Google Scholar]
- 26.Grigoryev, D. N., Ma, S. F., Irizarry, R. A., Ye, S. Q., Quackenbush, J. & Garcia, J. G. (2004) Genome Biol. 5, R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonow, R. O., Smaha, L. A., Smith, S. C., Jr., Mensah, G. A. & Lenfant, C. (2002) Circulation 106, 1602-1605. [DOI] [PubMed] [Google Scholar]
- 28.Bonow, R. O. & Smith, S. C., Jr. (2004) Circulation 109, 817-820. [DOI] [PubMed] [Google Scholar]
- 29.Cybulsky, M. I., Iiyama, K., Li, H., Zhu, S., Chen, M., Iiyama, M., Davis, V., GutierrezRamos, J. C., Connelly, P. W. & Milstone, D. S. (2001) J. Clin. Invest. 107, 1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukhova, G. K., Zhang, Y., Pan, J. H., Wada, Y., Yamamoto, T., Naito, M., Kodama, T., Tsimikas, S., Witztum, J. L., Lu, M. L., et al. (2003) J. Clin. Invest. 111, 897-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singaraja, R. R., Fievet, C., Castro, G., James, E. R., Hennuyer, N., Clee, S. M., Bissada, N., Choy, J. C., Fruchart, J. C., McManus, B. M., et al. (2002) J. Clin. Invest. 110, 35-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallat, Z., Corbaz, A., Scoazec, A., Graber, P., Alouani, S., Esposito, B., Humbert, Y., Chvatchko, Y. & Tedgui, A. (2001) Circ. Res. 89, E41-E45. [DOI] [PubMed] [Google Scholar]
- 33.Isoda, K., Sawada, S., Ishigami, N., Matsuki, T., Miyazaki, K., Kusuhara, M., Iwakura, Y. & Ohsuzu, F. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 1068-1073. [DOI] [PubMed] [Google Scholar]
- 34.Isoda, K., Kamezawa, Y., Ayaori, M., Kusuhara, M., Tada, N. & Ohsuzu, F. (2003) Circulation 107, 679-681. [DOI] [PubMed] [Google Scholar]
- 35.Hill, J. M., Zalos, G., Halcox, J. P., Schenke, W. H., Waclawiw, M. A., Quyyumi, A. A. & Finkel, T. (2003) N. Engl. J. Med. 348, 593-600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.