Abstract
Streptococcus pyogenes is an important bacterial pathogen afflicting humans. A striking feature is its extraordinary biological diversity, evident in the wide range of diseases it can cause and the antigenic heterogeneity present on its surface. The T antigens form the basis of a major serological typing scheme that is often used as an alternative or supplement to M typing. Unlike M typing, the genetic basis for T typing is poorly understood. In this report, the tee6 gene is localized to a position ≈3.3 kb downstream from prtF1 (or sfbI), which encodes the Fn-binding protein, protein F, a key virulence factor. Comparison of this portion of the genome with those of four additional strains reveals the presence of genes encoding a collagen-binding protein (Cpa) and a second Fn-binding protein (PrtF2 or PfbpI). This chromosomal region—here designated the FCT region—is ≈11 to 16 kb in length and is flanked at both ends by long stretches of highly conserved sequence. For each of the five strains, the FCT region contains a unique combination of semiconserved loci, indicative of extensive intergenomic recombination. The data provide evidence that the highly recombinatorial FCT region of the S. pyogenes genome is under strong selection for change in response to the host environment.
Group A streptococci (GAS; Streptococcus pyogenes) are among the most prevalent bacterial pathogens, and humans are their only known biological host. A striking feature of GAS is the large variety of diseases that they can cause (6). Decades of field epidemiology, combined with serological typing of GAS, has been central to our understanding of the natural history of streptococcal diseases and the biological diversity among strains. The primary typing scheme for GAS is based on M protein, a key virulence determinant giving rise to surface fibrils. Determinants of type specificity lie at the N-terminal fibril tips, which also contain targets of protective immunity. More than 80 M types are defined. Serological typing is gradually being replaced by emm sequence typing, and >150 emm types are now recognized (4; http://www.cdc.gov/ncidod/biotech/infotech_hp.html).
T typing is a serologically based scheme that is often used as an alternative or supplement to M typing. T antigens are trypsin-resistant surface antigens that exhibit extensive antigenic diversity, although there are fewer known T types than M types. Isolates of a given M type frequently share the same T agglutination pattern (1, 14). A specific role for T antigens in virulence remains unknown. The gene encoding one T antigen—T type 6 (tee6)—was cloned and sequenced (26). Southern blots of DNA restriction digests derived from strains representing 25 T types show that tee genes have tremendous genetic heterogeneity (15). Despite completion of the genome sequence for an M type 1 strain (6), and the partial genome sequence for an M type 5 strain (http://www.sanger.ac.uk), the genomic location of the tee locus has remained elusive.
In this report, the genomic location of tee6 is mapped to a region adjacent to the locus encoding protein F, a surface protein that mediates binding of the organism to fibronectin (Fn), a principal component of the extracellular matrix of the human host. Protein F is present in some, but not all, GAS strains (9, 21). It mediates adherence of GAS to epithelial cells (11, 28) and intracellular invasion of the bacterium (12, 20) and thereby plays a key role in virulence. Comparative analysis of this portion of the genome for five GAS strains reveals a highly recombinatorial zone, containing loci encoding Fn- and collagen-binding proteins and T antigen, referred to as the FCT region.
MATERIALS AND METHODS
Bacterial strains.
The clinical and epidemiological features of the GAS strains in this study are listed in Table 1.
TABLE 1.
GAS strains undergoing nucleotide sequence determination and/or analysis
| Strain | M or emm type | T type | Tissue source | Disease | Yr | Location | Comments |
|---|---|---|---|---|---|---|---|
| SF370 | 1 | NDa | Wound | Invasive | ND | USA | Genome sequence at http://www.ou.genome.edu (accession no. AE004092); ATCC strain 700294 |
| Manfredo | 5 | ND | Throat | Pharyngitis; rheumatic fever | 1958 | Chicago | Partial genome sequence at http://www.sanger.ac.uk |
| D471 | 6 | 6 | ND | ND | 1971 | Egypt | Sequence determined for this study; source for rofA and prtF1.6 (accession no. U01312 and L10919) |
| A735 | 12 | ND | Throat | ND | 1964 | ND | Sequence determined for this study; source for pfbpI sequence (accession no. AF071083) |
| A374 | 12 | ND | Throat | Acute glomerulonephritis | 1960 | Trinidad | Source for pfbpI sequence (accession no. AF071083) |
| B737 | 49 | ND | Skin | Impetigo | 1957 | Minnesota | Sequence determined for this study |
| CS101 | 49 | ND | Skin | Impetigo | 1957 | Minnesota | Derivative of B737; source for partial FCT region sequence (accession no. U49397) |
| 100076 | 49 | ND | ND | ND | ND | ND | Source for prtF2 sequence (accession no. U31980) |
ND, not determined.
Nucleotide sequence determination and computational analysis.
Chromosomal DNA used as a template for PCR was purified from bacteria following mutanolysin treatment, as previously described (2). For PCR amplifications, the annealing temperature was 55°C. For generating PCR products <6 kb in length, a standard Taq DNA polymerase (Qiagen, Inc., Valencia, Calif.) was used. For larger PCR products, the Expand Long Template PCR system (Roche Diagnostics GmbH, Mannheim, Germany) was used according to the manufacturer's instructions. Amplicons were purified by standard methods and subjected to nucleotide sequence determination of both strands by primer walking. The nucleotide sequence was determined for overlapping PCR products in order to construct chromosomal maps. DNAStar software was used for contig assembly and identifying open reading frames (ORFs).
The extent of nucleotide identity between the sequences of two GAS strains was determined by pairwise BlastN 2 analysis using default settings (http://www.ncbi.nlm.nih.gov). Pairwise comparisons were made for all possible combinations of the five strains under study. The National Center for Biotechnology Information server was used for BlastP analysis (protein query-protein database), and COGnitor was used for functional assignment of clusters of orthologous groups (COGs); both were performed using default settings. G+C content was calculated using DNAStar.
Nucleotide sequence accession numbers.
New sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY049087 to AY049089 and AF447492.
RESULTS
Sequence homology between the SF370 genome and tee6.
The recent publication of the complete genome sequence of strain SF370 (M type 1 [Table 1]) failed to identify the location of a putative tee gene (6). However, a BlastN search using tee6 plus flanking sequences (GenBank accession number M32978; derived from M type 6 strain D471) as the query revealed a short stretch of high nucleotide sequence identity (91% over 70 bp) between sequences immediately downstream of the tee6 ORF and the intergenic region of the SPy0133 and SPy0135 loci on the SF370 genome. In strain SF370, this site of high homology lies 8.05 kb upstream of rofA, a global regulator of transcription.
The BlastN search was also significant for 98% nucleotide identity (over 80 bp) between the transcriptional-terminator region of tee6 and a downstream region of the prtF1 (or prtF or sfbI) gene, also derived from strain D471; prtF1 encodes the Fn-binding protein, protein F. Although strain SF370 lacks a prtF1 locus, prtF1 in D471 lies immediately upstream of rofA, whose product (RofA) up-regulates prtF1 expression (8). Overall, the data point to a possible relationship between tee6 and loci positioned upstream of rofA.
Comparative genome structure of the rofA region.
Previous studies have indicated that there are extensive regions of both high and low sequence homology between the chromosomal region surrounding rofA in strain SF370 and the nra region of M type 49 strain CS101 (24). Figure 1 depicts the structural relationships between 40-kb sections of the SF370 genome (6) and the M type 5 Manfredo strain partial genome sequence (http://www.sanger.ac.uk). A striking feature is the two long stretches (>15 kb) of very high nucleotide identity (>95%) between SF370 and Manfredo, which is disrupted by an ≈11-kb region in which nucleotide sequence identity drops to <70%, except for a few short segments. The left and right boundaries of the central region of lower homology are marked by the SPy0123 and SPy0136 loci, respectively. This patchwork arrangement of nucleotide sequence identity and divergence is highly suggestive of genetic reassortment within the ≈11-kb zone of lower homology.
FIG. 1.
Partial genome maps of strains SF370, Manfredo, and CS101. ORFs are indicated by open boxes placed above the line (transcribed left to right) or below the line (oppositely transcribed). The percent nucleotide identity (nt id) between pairs of sequences was established by pairwise BlastN 2 and is depicted for regions showing ≥70% identity. The percent nucleotide identity is presented for SF370 versus Manfredo and for Manfredo versus CS101. Strain CS101 displays 98% nucleotide identity with SF370 at bp 112089 through 114912 (data not shown). The vertical dotted lines depict the boundaries for high nucleotide sequence identity. The distance scale (in kilobases) is shown for strain SF370; distances for Manfredo and CS101 are presented for easy visual alignment but are not drawn precisely to scale. In SF370, the emm locus lies ≈283 kb from SPy0123 (hsp33).
Comparison of strain SF370 to a 10.84-kb region of strain CS101 (24) confirms the position of the left boundary of high nucleotide sequence homology at SPy0123 (Fig. 1). SF370 has several short segments of 70 to 90% nucleotide identity with CS101 between portions of rofA and nra, cpa.1 and cpa.49, and SPy0129 and the 3′ portion of the so-called etfLSL locus (data not shown). Like rofA, nra is a global regulator of transcription; cpa.49 encodes a collagen-binding protein (24). As observed for CS101, Manfredo lacks significant overall homology with SF370 in the SPy0124-to-SPy0135 region (Fig. 1). However, strains Manfredo and CS101 have extensive regions of high nucleotide sequence identity across the entire region of interest, unlike their comparisons to SF370.
Chromosomal mapping of tee6 in strain D471.
As stated above, there is a short stretch of high nucleotide sequence identity between the DNA sequence downstream of tee6 and the intergenic SPy0133-SPy0135 region of SF370 (Fig. 1). Thus, we sought to determine whether tee6 is located near the SPy0136 locus in the parent strain, D471. Using oligonucleotide primers specific for tee6 and SPy0136, PCR amplification yielded products whose size placed the 3′ ends of the two oppositely transcribed loci ≈1.0 kb apart. In a second PCR amplification, primers corresponding to a region upstream of tee6 and to the 3′-end region of prtF1 were paired to yield a product of ≈4.0 kb. The nucleotide sequence was determined for these and other overlapping PCR amplicons, and a chromosomal map for D471 was constructed (Fig. 2). A single large ORF of 3.1 kb lies between the genes encoding protein F and the T6 antigen of strain D471.
FIG. 2.
Complete FCT region of strain D471 and comparison to SF370. The percent nucleotide identity (nt id) between D471 and SF370 was established by pairwise BlastN 2 and is depicted for regions having ≥70% identity. The vertical dotted lines mark the FCT region boundaries for high nucleotide sequence identity. The distance scale (in kilobases) is shown for strain SF370; distances for D471 are presented for easy visual alignment but are not drawn precisely to scale. The sources for strain D471 sequences are GenBank (Table 1) and this study. The nomenclature for the protein F gene (prtF1) indicates its source as being an M type 6 strain (prtF1.6).
Based on the presence of genes encoding Fn- and collagen-binding proteins and the T antigen, this portion of the genome is designated the FCT region.
Genome localization of prtF2/pfbpI to the FCT region.
Analysis of the FCT region of the Manfredo strain (Fig. 1) revealed the presence of an ORF displaying several regions of high nucleotide sequence identity to genes, known as prtF2 (13) and pfbpI (25), encoding a second Fn-binding protein. PrtF2 was originally derived from an M type 49 strain (100076 [Table 1]), and displays ≈90% amino acid sequence identity to PfbpI, whose sequence is a composite derived from two M type 12 strains (A374 and A735). The high amino acid sequence homology extends through their Fn-binding domains. However, PrtF2 and PfbpI are structurally distinct from protein F.
The prtF2/pfbpI-like ORF of Manfredo lies adjacent to the highly conserved right boundary locus, SPy0136, of the FCT region (Fig. 1). It was of interest to determine whether the pfbpI locus from an M type 12 strain was also contained within the FCT region. DNA from strain A735 (Table 1) was used as a template for PCR amplification with msmRL- and SPy0136-specific primers, yielding an ≈5.2-kb amplicon, which subsequently underwent nucleotide sequence determination.
Sequence alignment of the msmRL-SPy0136 product from strain A735 with the FCT region of Manfredo shows 99% nucleotide identity over the left-end 1.05 kb and 96% nucleotide identity over the right-end 1.3 kb (Fig. 3). At the left end, there is a small gap followed by an additional region of 99% nucleotide identity over 0.3 kb. The regions of high homology correspond to the msmRL region through the 5′ end of the prtF2/pfbpI-like locus and to SPy0136 through the 3′ end of the prtF2/pfbpI-like locus, including the portion which encodes the Fn-binding region of PfbpI. Furthermore, the pfbpI gene derived from strain A735 exhibits 99% nucleotide identity with the composite pfbpI sequence from M type 12 strains A374 and A735 (data not shown). The data indicate that the location of prtF2/pfbpI is within the FCT region of the genome.
FIG. 3.
Alignments of the prtF2/pfbpI gene and flanking regions in three strains. The percent nucleotide identity (nt id) between pairs of sequences was established by pairwise BlastN 2 and is depicted for regions having ≥70% identity. The percent nucleotide identity is presented for Manfredo versus A735 and for A735 versus B737. Since A735 and B737 are highly related in sequence, the percent nucleotide identity between Manfredo and B737 closely parallels that between A735 and B737. The prtF2/pfbpI.5 allele of Manfredo is ≈1.4 kb smaller than the prtF2/pfbpI.12 and prtF2/pfbpI.49 alleles.
It was also of interest to determine whether the partial FCT region of the M type 49 strain CS101 has a prtF2/pfbpI-like locus adjacent to its msmRL locus (Fig. 1). DNA from B737, the strain from which CS101 was derived (Table 1), was used as a template for PCR amplification with msmRL- and SPy0136-specific primers. Compared to strain A735, 99% nucleotide identity over the left-end 1.08 kb, 94% nucleotide identity over the next 1.2 kb, and 98% nucleotide identity over the right-end 2.5 kb were evident (Fig. 3). Only a 0.36-kb segment, corresponding to the central portion of the prtF2/pfbpI locus, failed to display significant similarity by pairwise BlastN 2 analysis. Furthermore, the complete prtF2/pfbpI gene from strain B737 exhibited 99% nucleotide identity with the prtF2 gene derived from another M type 49 strain, 100076 (Table 1 and data not shown). The data indicate that all three prtF2/pfbpI genes under study, derived from strains of three different M types (5, 12, and 49), lie within the FCT region.
Variation in the size of the FCT region.
For the strains of M types 1, 5, 6, and 49, the FCT region is ≈10.5, 10.7, 11.0, and 12.7 kb in length, respectively, extending from the 5′ end of hsp33 to the 3′ end of SPy0136 (Table 2). The remaining portion of the FCT region of M type 12 strain A735 was also characterized and was found to be considerably larger than the other FCT regions, measuring ≈16 kb in length (Fig. 4). Therefore, it appears that the structure of the FCT region may be larger and more complex for some strains.
TABLE 2.
Size and G+C content of FCT region and flanking regions
| Strain | emm type | Region | Length (bp) | % G+C |
|---|---|---|---|---|
| SF370 | 1 | FCT | 10,525 | 35.64 |
| Manfredo | 5 | FCT | 10,690 | 34.25 |
| D471 | 6 | FCT | 10,981 | 36.19 |
| A735 | 12 | FCT | 16,207 | 36.40 |
| B737/CS101 | 49 | FCT | 12,730 | 35.28 |
| SF370 | 1 | Left flank | 14,891 | 38.95 |
| Manfredo | 5 | Left flank | 14,890 | 38.97 |
| SF370 | 1 | Right flank | 14,584 | 38.25 |
| Manfredo | 5 | Right flank | 14,587 | 38.24 |
FIG. 4.
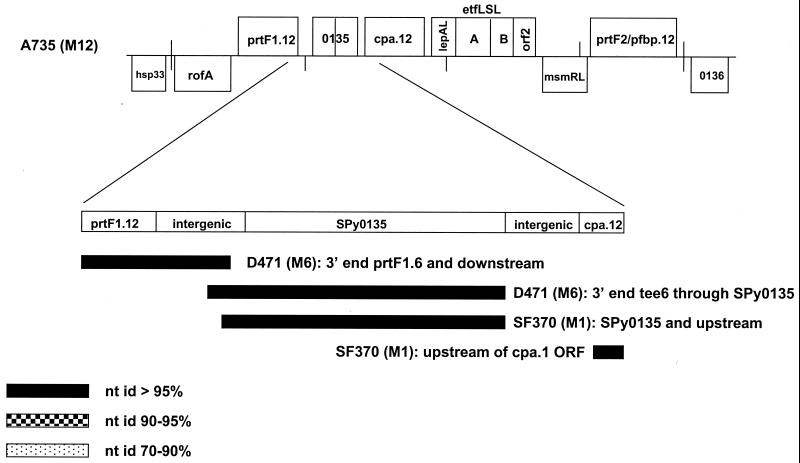
Complete FCT region of strain A735 and a possible hot spot for recombination. The complete FCT region for strain A735 is shown. The categories for each locus, and homology to other FCT region loci, are summarized in Table 3. The vertical lines indicate ≈4-kb intervals. The percent nucleotide sequence identity (nt id) to FCT region genes from other strains is shown for a 1.8-kb region spanning from the 3′-end region of prtF1.12 through the 5′-end region of cpa.12. The relative positions of the aligned regions are shown in Fig. 1 (for strain SF370) and 2 (for strain D471) and indicate genomic rearrangements.
Structural properties of FCT region loci and predicted proteins.
The loci of the FCT region can be classified according to four main structural groups, based on nucleotide sequence (Table 3). Category 1 loci are defined as highly conserved, having >95% nucleotide sequence identity for all alleles, and are present in all strains. The boundary loci, hsp33 (SPy0123) and SPy0136, are category 1, displaying nearly complete nucleotide identity for all five strains. Category 2 loci are also highly conserved (>95% nucleotide identity for all alleles), but they are not present in all strains and thus are regarded as semiconserved.
TABLE 3.
Classification and strain distribution of loci of the FCT region
| Locus | Category | No. of aaa residues | (Putative) function | COGnitor search results | BlastP search resultsb
|
Distribution of loci in FCT region ofc:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Best alignment | BlastE value | % aa identity (total no. of residues) | SF370 (M1) | Manfredo (M5) | D471 (M6) | A735(M12) | CS101/B737 (M49) | |||||
| hsp33 (SPy0123) | 1 | 264 | Heat shock protein | COG1281 | Chaperonin SP2188 (S. pneumoniae) | e-110 | 75 (264) | + | + | + | + | + |
| SPy0136 | 1 | 221 | Unknown | None | NA | Above cutoff | NA | + | + | + | + | + |
| rofA (SPy0124) | 2 | 439 | Transcriptional regulator | None | Nra (S. pyogenes) | e-148 | 64 (441) | + | − | + | + | − |
| nra | 2 | 511 | Transcriptional regulator | None | RofA (S. pyogenes) | e-160 | 61 (499) | − | + | − | − | + |
| lepAL | 2 | 173 | Signal peptidase I | COG0681 | SPy0127 (S. pyogenes) | 2e-23 | 44 (132) | − | + | − | + | + |
| etfLSL (B) (3′ portion) | 2 | 241 | Unknown | None | SPy0129 (S. pyogenes) | 5e-64 | 46 (240) | − | + | − | + | + |
| orf2 | 2 | 195 | Unknown | None | SPy0130 (S. pyogenes) | 3e-14 | 29 (123) | − | + | − | + | + |
| msmRLd | 2 | 401 | AraC-type DNA-binding domain-containing protein | COG2027 | AraC-like activator (S. agalactiae) | 3e-12 | 31 (164) | − | + | − | + | + |
| SPy0135 | 2 | 227 | Fimbria-associated protein (putative sortase) | None | Putative sortase SP0467 (S. pneumoniae) | 2e-44 | 50 (178) | + | − | + | + | − |
| prtF/prtF1/sfbI | 3 | Varies | Fn-binding protein | None | Cpa (S. pyogenes) | 1e-65 | NA | − | − | + | + | − |
| cpa (SPy0125) | 3 | Varies | Collagen-binding protein | None | Protein F (S. pyogenes) | 2e-20 | NA | + | + (partial) | − | + | + |
| prtF2/pfbpI | 3 | Varies | Fn-binding protein | None | Protein F; opacity factor (S. pyogenes) | 1e-21; 1e-16 | NA | − | + | − | + | + |
| etfLSL (A) (5′ portion) | 3 | Varies | Unknown | None | SPy0128 (S. pyogenes) | 5e-57 | 43 (240) | − | + | − | + | + |
| SPy0127 (lepAL.1) | 4 | 185 | Signal peptidase I | COG0681 | LepAL (S. pyogenes) | 1e-30 | 43 (171) | + | − | − | − | − |
| SPy0128 | 4 | 340 | Unknown | None | EtfLSL (S. pyogenes) | 4e-41 | 35 (355) | + | − | − | − | − |
| SPy0129 | 4 | 237 | Unknown | None | EtfLSL (S. pyogenes) | 1e-60 | 46 (240) | + | − | − | − | − |
| SPy0130 | 4 | 215 | Unknown | None | Orf2 (S. pyogenes) | 1e-14 | 29 (173) | + | − | − | − | − |
| SPy0131 | 4 | 450 | Unknown | None | Unknown (Pseudomonas) | 7e-50 | 32 (426) | + | − | − | − | − |
| SPy0133 | 4 | 116 | Transposase | COG3436 | IS 66 family element (S. pneumoniae) | 1e-46 | 77 (116) | + | − | − | − | − |
| fctX | 4 | 1,036 | Unknown | None | Cell wall surface anchor family SP0462 (S. pneumoniae) | 3e-6 | 24 (392) | − | − | + | − | − |
| tee6 | 4 | 537 | Unknown | None | NA | Above cutoff | NA | − | − | + | − | − |
aa, amino acid.
Reported using cutoff E values at e-4. For category 3 loci only, values are reported for best alignment to another locus within S. pyogenes. All other values are based on highest score with entire National Center for Biotechnology Information database (all species). NA, not applicable.
SPy0135 in A735 contains a disrupted ORF (stop codon; frameshift). The eftLSL 5′ portion of CS101/B737 is a separate ORF in Manfredo and A735; all share stretches of high nucleotide identity at their 5′ and 3′ ends. +, present; −, absent.
Composite of strains CS101 and B737.
Structural analysis of predicted proteins can provide additional insights into relationships between different loci. COGnitor analysis, which identifies COGs, was performed for each predicted gene product of the FCT region loci (Table 3). Products of several category 1 and 2 loci can be assigned a COG, indicating that they share significant structural similarity with biologically characterized gene products that are distributed among numerous species. However, for most category 2 loci, the best alignment by BlastP is with a locus present within the FCT region of another GAS strain: RofA and Nra, LepAL and SPy0127, EtfLSL-3′ (EtfLSL.B) and SPy0129, and Orf2 and SPy0130. The data suggest that these protein pairs may be paralogs (derived by gene duplication; evolved new functions) or orthologs (evolved in separate species) that underwent recent interspecies gene transfer.
Category 3 loci are also semiconserved, present in some, but not all, strains (Table 3). However, they differ from category 2 loci in that they share extended regions of high nucleotide sequence identity (>95%) interrupted by regions of lower homology. The genes giving rise to the extracellular matrix-binding proteins—protein F (prtF/prtF1/sfbI), PrtF2/PfbpI (prtF2/pfbpI), and Cpa (cpa)—are classified as category 3. When analyzed for amino acid alignment by BlastP, weak structural similarities among the products of these three loci become apparent.
The loci classified as category 4 (Table 3) lack high nucleotide sequence identity with any known FCT region locus. However, as our knowledge of the FCT region loci in other GAS strains increases, it is likely that several category 4 loci will be reclassified as category 2 or 3.
The FCT region of M type 6 strain D471 has several striking parallels to a portion of the Streptococcus pneumoniae genome. Together, GAS and pneumococci are the most important streptococcal species causing human disease. Although the putative function of the fctX locus of D471 is unknown, its highest BlastP score is with SP0462 from S. pneumoniae strain TIGR4 (Table 3); SP0462 has both an LPXTG cell wall anchor motif and a signal peptide (29). Additional BlastP analysis was performed for predicted proteins of flanking pneumococcal genes. Adjacent to SP0462 is SP0461, which encodes a protein showing the highest amino acid sequence homology (28% identity) with RofA of GAS. On the far side of SP0461 lies a transposase gene (SP0460; COG3464). The FCT region of GAS strain SF370 also has a putative transposase (SPy0133; COG3436 [Table 3]). On the opposite side of SP0462 lies SP0463, which has 22% amino acid sequence identity with the T6 antigen of strain D471. Proteins encoded by SP0466, SP0467, and SP0468 each have relatively high BlastP scores with SPy0135. SP0469 is disrupted by a putative transposase. Using DNA microarrays based on the TIGR4 strain for comparative genome analysis, two other pneumococcal strains tested both lack SP0463 through SP0468 (29). The data suggest that there is a common origin for the FCT region of GAS and SP0461 through SP0468 of pneumococci and that these loci might be subject to horizontal transfer by a mechanism involving mobile genetic elements.
Genetic recombination involving the FCT region.
Intergenomic recombination can give rise to unique combinations of loci. The FCT regions of Manfredo (M5) and CS101/B737 (M49) contain nra, cpa, and prtF2/pfbpI (Table 3). However, the FCT regions of the other strains are quite different: SF370 (M1) has rofA and cpa, D471 has rofA and prtF1, and A735 has rofA, prtF1, cpa, and prtF2/pfbpI. Thus, for these five strains at three discrete chromosomal map positions, four different combinations of loci are observed.
It stands to reason that crossover sites within the FCT region, leading to intergenomic recombination via a homologous mechanism, can lie within any of the numerous stretches of nucleotides that display high sequence identity between strains (Fig. 1 through 3 and Table 3). The presence of a putative transposase within the FCT region of strain SF370 (SPy0133; COG 3436 [Table 3]) raises the possibility that site-specific recombination can also contribute to allelic rearrangements within the FCT region. Strain A735 (M12) reveals some unusual gene rearrangements that might provide further clues to the nature of recombination within the FCT region (Fig. 4). In A735, both a prtF1 and a cpa allele are present, separated by a region of high homology to SPy0135 and upstream sequences. As stated previously, there is a short stretch of high nucleotide sequence identity between sequences immediately downstream of the tee6 ORF and the intergenic region of the SPy0133 and SPy0135 loci on the SF370 genome. The 3′ end of tee6 and its downstream region (in strain D471) also have high nucleotide identity to a downstream region of prtF1 in both D471 and A735 (Fig. 4). This intergenic region appears to be a hot spot for recombination, bringing the SPy0135 locus adjacent to prtF1.12 in strain A735. Whether a SPy0133 putative transposase, similar to that observed in SF370 (M1), has a role in site-specific recombination within the FCT region of A735 is not known, but it seems a reasonable possibility.
The percent G+C content of the five complete FCT regions was ascertained (Table 2). For the FCT regions, bounded by the 5′ end of hsp33 and the 3′ end of SPy0136, the G+C content ranged from 34.25 to 36.40%. The values for the FCT region are ≈2 to 5% lower than the G+C content of the entire SF370 M1 genome, which is 38.5%, and 39.1% for ORFs (6). The ≈15 kb of nucleotide sequence flanking the left side of the FCT region is nearly 39.0% G+C in both the SF370 and Manfredo strains, more closely reflecting the G+C content for the entire SF370 genome. The G+C content at the right flank of the FCT region is also higher, at 38.25%. The lower G+C content of the FCT region is consistent with the possibility that it was acquired by S. pyogenes following horizontal transfer from another donor species.
DISCUSSION
In this report, we characterize the structure of a portion of the GAS genome that plays a central role in virulence. By virtue of its highly recombinatorial nature, it appears that the FCT region plays a critical role in the adaptation of GAS to different host environments.
Several products of the FCT region are surface proteins that interact with the human host during infection. Protein F and PrtF2/PfbpI are structurally distinct proteins which bind host Fn in order to mediate GAS adherence to the epithelium and/or intracellular invasion (11, 12, 18, 20, 28). The notion that intracellular invasion leads to persistent throat infection is supported by an epidemiological study in which organisms recovered in cases of antibiotic treatment failure harbored the prtF1 gene (22, 27). A role in pathogenesis for T antigen, or for the collagen-binding protein Cpa, remains to be established. However, T antigens are under strong selection from the host immune response. Furthermore, the putative products of the four cpa alleles (cpa.1, cpa.5, cpa.12, and cpa.49) are structurally heterogenous, suggesting that they, too, are under strong selection for change.
A striking feature of the FCT region is that it is flanked by long stretches of highly conserved sequences. The SPy0140 locus (yqiL), a well-studied housekeeping locus located ≈4.5 kb from the right boundary of the FCT region (SPy0136), is present in every GAS isolate tested (n > 200) (3). The maximal nucleotide divergence among 22 yqiL alleles is 1.4%, suggesting that the FCT region is flanked by a highly conserved region in all GAS isolates.
The lower G+C content of the FCT region, compared to both the immediate flanking nucleotide sequences and the genomic average, suggests that an ancestral GAS strain may have acquired the FCT region en bloc from another species following a horizontal transfer event. A skewed G+C content is a hallmark feature of the pathogenicity islands found in many gram-negative bacterial species.
The FCT region shows extensive heterogeneity in gene content, indicative of intergenomic recombination. The genetic mosaicism extends across the whole FCT region. Numerous regions of high nucleotide sequence identity between strains provide potential crossover sites and opportunities for homologous recombination. The alleles of yqiL and other neutral housekeeping genes are randomly associated among GAS strains, indicating that GAS have high intrinsic rates of recombination, leading to disruption of genetic linkages between loci (5, 16a). The finding of a putative transposase (SPy0133) within the FCT region of one strain raises the possibility that site-specific recombination has also contributed to the generation of diversity. Conceivably, GAS can also acquire divergent genes from other bacterial species (16). Another possible explanation for some of the observed genetic diversity within the FCT region is that a progenitor cell had the full complement of loci (Table 3) but its descendants underwent deletion of different subsets of genes.
All five FCT regions under study contain either an rofA or nra locus at the left-end boundary. These genes encode regulators of gene transcription—RofA and Nra—that display ≈60% amino acid sequence identity (8, 10, 24). Transcriptional control of prtF1 in strain D471 is regulated by RofA in response to aerobic and anaerobic growth (7). Nra negatively regulates cpa and prtF2 in an M type 49 strain but is not influenced by atmospheric conditions (24). In another M type 49 strain, prtF2 expression is up-regulated in an O2-enriched atmosphere (13). In addition, Nra negatively regulates mga, which encodes a positive regulator of emm gene transcription; Nra may play an important role in facilitating GAS persistence in an intracellular environment (19). Expression of prtF1, cpa, and prtF2/pfbpI is complex and may involve signal transduction through multiple regulatory pathways. Among the five strains under study, both cpa and prtF2/pfbpI are present within FCT regions which also contain either rofA or nra. Perhaps the generation of unique combinations of semiconserved loci within the FCT region provides an avenue for the fine tuning of virulence.
The presence of both nra and rofA within the same strain was reported by others using Southern blots and includes an unnamed M type 5 strain (24). Whether experimental conditions were sufficiently stringent to block annealing to putative nra and/or rofA homologs is not known. In our analysis, the incomplete Manfredo (M type 5) genome has a single nra match by BlastN analysis but no matches with rofA. Furthermore, both Manfredo and a second M type 5 strain (1RP144) were negative for PCR using hsp33- and rofA-specific primers and failed to yield products with internal rofA primers (data not shown). Neither the SF370 nor the Manfredo genome has multiple copies of hsp33 (via BlastN). While it is possible that some strains have multiple FCT regions, our limited study provides no such evidence.
T antigens are trypsin-resistant surface antigens to which specific antisera have been raised. However, this definition provides no clue to whether T antigens share a common genetic origin. Southern blots of DNA restriction digests derived from strains representing 25 T types show that only 40% of the T types hybridize with tee6 probes; several distinct combinations of partial tee6 gene probes hybridize to different-size fragments (15). Yet, all 25 strains hybridize with a probe corresponding to 1.3 kb downstream of tee6, indicating there is a common genetic feature tightly linked to tee6. In strain D471, the 1.3 kb downstream of tee6 include both the semiconserved SPy0135 locus and the widely conserved SPy0136 right-boundary locus. It is possible that some T antigens are unrelated to T6 in sequence and their loci lie outside of the FCT region.
The GAS proteins which bind the extracellular matrix of the human host (Fn and collagen) are encoded by category 3 loci, meaning that they are present in some, but not all, strains and have some regions of high nucleotide identity which define them as a locus. Presumably, the tee locus will also fall into this category (15). The generation of diversity within category 3 loci could be the result of accumulated mutations, coupled with strong diversifying selection by the host immune response. Alternatively, the category 3 gene products might be multifunctional, having discrete structural domains that are arranged in several different combinations as a result of intragenic recombination. For example, protein F recognizes two functionally distinct sites within Fn (23), and in addition, it can directly bind human fibrinogen, as well as Fn (17). Functional-domain swapping via intragenic recombination is another possible mechanism for the fine tuning of virulence.
The individual strains that compose the global GAS population exhibit a wide array of clinical and epidemiological phenotypes which in turn are reflected in both the types of diseases they cause and their relative incidence. The ability of many GAS strains to become highly adapted to just one of the two principal ecological niches of their human hosts—the upper respiratory tract and the epidermal layer of the skin--is one driving force behind their extensive biological diversity. The FCT region, by virtue of its high capacity for intergenomic recombination leading to substantive genetic change, seems a likely candidate for providing the raw material upon which natural selection can act.
Acknowledgments
We thank David Chu for expert technical assistance. We are grateful for the release of partial genome sequence data for the Manfredo strain by the Sanger Centre.
This work was supported by grants from the National Institutes of Health (GM-60793 and AI-28944) to D.E.B.
Editor: E. I. Tuomanen
REFERENCES
- 1.Beall, B., R. Facklam, J. Elliott, A. Franklin, T. Hoenes, D. Jackson, L. Laclaire, T. Thompson, and R. Viswanathan. 1998. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J. Med. Microbiol. 47:1-6. [DOI] [PubMed] [Google Scholar]
- 2.Bessen, D. E., M. W. Izzo, T. R. Fiorentino, R. M. Caringal, S. K. Hollingshead, and B. Beall. 1999. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J. Infect. Dis. 179:627-636. [DOI] [PubMed] [Google Scholar]
- 3.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationship between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Facklam, R., B. Beall, A. Efstratiou, V. Fischetti, E. Kaplan, P. Kriz, M. Lovgren, D. Martin, B. Schwartz, A. Totolian, D. Bessen, S. Hollingshead, F. Rubin, J. Scott, and G. Tyrrell. 1999. Report on an international workshop: demonstration of emm typing and validation of provisional M-types of group A streptococci. Emerg. Infect. Dis. 5:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feil, E. J., E. C. Holmes, D. E. Bessen, M.-S. Chan, N. P. J. Day, M. C. Enright, R. Goldstein, D. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogg, G., and M. Caparon. 1997. Constitutive expression of fibronectin binding in Streptococcus pyogenes as a result of anaerobic activation of rofA. J. Bacteriol. 179:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fogg, G. C., C. M. Gibson, and M. G. Caparon. 1994. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an mu-gamma-delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol. Microbiol. 11:671-684. [DOI] [PubMed] [Google Scholar]
- 9.Goodfellow, A. M., M. Hibble, S. R. Talay, B. Kreikemeyer, B. J. Currie, K. S. Sriprakash, and G. S. Chhatwal. 2000. Distribution and antigenicity of fibronectin binding proteins (SfbI and SfbII) of Streptococcus pyogenes clinical isolates from the Northern Territory, Australia. J. Clin. Microbiol. 38:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granok, A., D. Parsonage, R. Ross, and M. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanski, E., and M. Caparon. 1992. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89:6172-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jadoun, J., V. Ozeri, E. Burstein, E. Skutelsky, E. Hanski, and S. Sela. 1998. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J. Infect. Dis. 178:147-158. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe, J., S. Natanson-Yaron, M. G. Caparon, and E. Hanski. 1996. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Mol. Microbiol. 21:373-384. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, D. R., and E. L. Kaplan. 1993. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J. Med. Microbiol. 38:311-315. [DOI] [PubMed] [Google Scholar]
- 15.Jones, K. F., O. Schneewind, J. M. Koomey, and V. A. Fischetti. 1991. Genetic diversity among the T-protein genes of group A streptococci. Mol. Microbiol. 5:2947-2952. [DOI] [PubMed] [Google Scholar]
- 16.Kalia, A., M. C. Enright, B. G. Spratt, and D. E. Bessen. 2001. Directional gene movement from human-pathogenic to commensal-like streptococci. Infect. Immun. 69:4858-4869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16a.Kalia, A., B. G. Spratt, M. C. Enright, and D. E. Bessen. Influence of recombination and niche separation on the population genetic structure of the pathogen Streptococcus pyogenes. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 17.Katerov, V., A. Andreev, C. Schalen, and A. Totolian. 1998. Protein F, a fibronectin-binding protein of Streptococcus pyogenes, also binds human fibrinogen: isolation of the protein and mapping of the binding region. Microbiology 144:119-126. [DOI] [PubMed] [Google Scholar]
- 18.Molinari, G., M. Rohde, C. A. Guzman, and G. S. Chhatwal. 2000. Two distinct pathways for the invasion of Streptococcus pyogenes in non-phagocytic cells. Cell Microbiol. 2:145-154. [DOI] [PubMed] [Google Scholar]
- 19.Molinari, G., M. Rohde, S. R. Talay, G. S. Chhatwal, S. Beckert, and A. Podbielski. 2001. The role played by the group A streptococcal negative regulator Nra on bacterial interactions with epithelial cells. Mol. Microbiol. 40:99-114. [DOI] [PubMed] [Google Scholar]
- 20.Molinari, G., S. R. Talay, P. Valentin-Weigand, M. Rohde, and G. S. Chhatwal. 1997. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun 65:1357-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natanson, S., S. Sela, A. E. Moses, J. M. Musser, M. G. Caparon, and E. Hanski. 1995. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J. Infect. Dis. 171:871-878. [DOI] [PubMed] [Google Scholar]
- 22.Neeman, R., N. Keller, A. Barzilai, Z. Korenman, and S. Sela. 1998. Prevalence of internalisation-associated gene, prtF1, among persisting group-A streptococcus strains isolated from asymptomatic carriers. Lancet 352:1974-1977. [DOI] [PubMed] [Google Scholar]
- 23.Ozeri, V., A. Tovi, I. Burstein, S. Natanson-Yaron, M. G. Caparon, K. M. Yamada, S. K. Akiyama, I. Vlodavsky, and E. Hanski. 1996. A two-domain mechanism for group A streptococcal adherence through protein F to the extracellular matrix. EMBO J. 15:989-998. [PMC free article] [PubMed] [Google Scholar]
- 24.Podbielski, A., M. Woischnik, B. A. B. Leonard, and K.-H. Schmidt. 1999. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31:1051-1064. [DOI] [PubMed] [Google Scholar]
- 25.Rocha, C. L., and V. A. Fischetti. 1999. Identification and characterization of a novel fibronectin-binding protein of the surface of group A streptococci. Infect. Immun. 67:2720-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characterization of the trypsin-resistant T6 surface protein of group A streptococci. J. Bacteriol. 172:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sela, S., R. Neeman, N. Keller, and A. Barzilai. 2000. Relationship between asymptomatic carriage of Streptococcus pyogenes and the ability of the strains to adhere to and be internalised by cultured epithelial cells. J. Med. Microbiol. 49:499-502. [DOI] [PubMed] [Google Scholar]
- 28.Talay, S. R., P. Valentin-Weigand, P. G. Jerlstrom, K. N. Timmis, and G. S. Chhatwal. 1992. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect. Immun. 60:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]