Abstract
Studies of mouse models of endotoxemia and sepsis with gram-negative bacteria have shown that the host response is genetically controlled. Mice infected with the gram-negative bacterium Salmonella enterica serovar Typhimurium exhibit marked genetic differences in disease manifestation, and the wild-derived strain Mus musculus molossinus MOLF/Ei is extremely susceptible to S. enterica serovar Typhimurium. The kinetics of bacterial proliferation within the liver and the spleen and histological examination of tissue sections have suggested that MOLF/Ei mice do not succumb to infection because of overwhelming bacterial growth in the reticuloendothelial organs or massive tissue necrosis, as observed in other Salmonella-susceptible strains. MOLF/Ei mice respond normally to lipopolysaccharide (LPS) in vivo and in vitro, as determined by the production of tumor necrosis factor alpha and spleen cell mitogenesis. However, they have a unique cytokine profile in response to infection compared to that observed for other Salmonella-susceptible mice. There was increased expression of mRNA of the interleukin-1α (IL-1α) and IL-1β genes as the infection in the spleens and livers of MOLF/Ei mice progressed. Despite the fact that MOLF/Ei mice have the ability to respond to LPS and the fact that there are significant increases in IL-1α and IL-1β mRNA, Nos2 in the spleen is not upregulated and nitrite production by spleen cells is reduced. At the central level, the inflammatory response is characterized by strong upregulation of the inhibitory factor kappa B alpha and Toll-like receptor 2 genes, two genes known to be regulated by LPS and IL-1 in the brain. The high levels of IL-1 expression in the spleens and livers of MOLF/Ei mice may have important implications for the activation of peripheral and central innate immune mechanisms.
Salmonella enterica serovar Typhimurium is a gram-negative facultative intracellular bacterium that causes a systemic infection in mice reminiscent of human typhoid fever (4). According to the World Health Organization, approximately 16 million cases of typhoid fever are reported annually, and close to 600,000 deaths occur in areas of endemicity in Africa and Asia (28). Mouse models of infection have allowed elucidation of some of the host defense mechanisms involved in both innate and adaptive immune responses to S. enterica serovar Typhimurium. During natural infection with S. enterica serovar Typhimurium, only a small fraction of the ingested bacteria cross the intestinal epithelium, reach the bloodstream through the mesenteric lymph nodes, and disseminate further into the reticuloendothelial system (RES) (spleen and liver) of the host to cause disease (2). After systemic dissemination of Salmonella, the courses of natural and experimental parenteral infections are equivalent, as shown by similar histopathological changes in mouse reticuloendothelial organs (5).
Mouse models of experimental infection with S. enterica serovar Typhimurium have been used to define four phases in the progression of infection (reviewed in reference 19). The initial phase involves rapid clearance of the majority of the inoculum from the circulation by macrophages and neutrophils (6, 7). This is followed, in the second phase, by exponential bacterial growth within the spleen and the liver, which is characterized by the production of proinflammatory cytokines (tumor necrosis factor alpha [TNF-α] and gamma interferon [IFN-γ]), which in the third phase activate host cells nonspecifically to suppress the invading pathogen. Mice eventually acquire specific immunity during the late phase of infection through B- and T-lymphocyte activation.
Classical inbred strains of mice show genetic variation in susceptibility to infection with S. enterica serovar Typhimurium; C57BL/6J, BALB/cJ, and C3H/HeJ mice are highly susceptible to infection, while 129/Sv mice are extremely resistant. Other strains, such as DBA/2J and A/J, show intermediate susceptibility (9, 29, 34). Critical genes known to influence the course of experimental infection and to be responsible for the observed differences among inbred stains of mice have been identified. Two of these genes, Nramp1 (Nramp1G169) and Tlr4 (Tlr4Pro712), have been shown to play an important role in innate immunity (10, 30, 33, 42). Mutated alleles of Nramp1 in C57BL/6J and BALB/cJ mice and of Tlr4 in C3H/HeJ mice are responsible for the extreme susceptibility of these hosts to Salmonella (26, 31, 41). These two genes are known to be involved in the early control of bacterial replication within the macrophages and polymorphonuclear cells of mice.
We have identified a wild-derived strain of mice, Mus musculus molossinus MOLF/Ei, that is extremely susceptible to infection with S. enterica serovar Typhimurium despite the presence of normal Nramp1 and Tlr4 alleles (33, 39). MOLF/Ei mice succumb to infection with S. enterica serovar Typhimurium within 1 week of intravenous challenge regardless of the dose used (G. Sebastiani and D. Malo, unpublished data). To identify the genetic components of S. enterica serovar Typhimurium susceptibility in MOLF/Ei mice, we previously performed a genome-wide study using an F2 intercross between C56BL/6J and MOLF/Ei inbred mice and described the mapping of two quantitative trait loci (QTL) which significantly affect survival time following lethal infection with S. enterica serovar Typhimurium in MOLF/Ei mice (39). A Salmonella-resistant phenotype (Ity2, immunity to typhimurium 2) was linked to a region on mouse chromosome 11 (maximum LOD score, 7.0 at D11Mit5), and a second QTL (Ity3), conferring recessive susceptibility, was located on mouse chromosome 1 approximately 25 cM distal to Nramp1 (maximum LOD score, 4.8 at D1Mit100) in a chromosomal region harboring the gene encoding another member of the Toll-like receptor family, Tlr5 (38). Together with Nramp1, these QTL accounted for approximately 55% of the variance in survival time following infection with S. enterica serovar Typhimurium in our mouse model. These studies clearly demonstrated that the genetic components underlying this phenotype in our wild-derived murine model of infection are complex. Although these linkage analyses provided important information about the inheritance of resistance to Salmonella infection in MOLF/Ei mice, the role of each of the underlying genes in the resistance to infection is still unknown. In the present study, we examined the immune response of MOLF/Ei mice infected with S. enterica serovar Typhimurium in order to provide a rational approach for eliminating or incriminating attractive candidate genes.
MATERIALS AND METHODS
Preparation of S. enterica serovar Typhimurium inoculum.
One milliliter of a frozen culture of S. enterica serovar Typhimurium strain Keller (originally obtained from Hugh Robson, Royal Victoria Hospital, Montreal, Canada) was used to seed 100 ml of Trypticase soy broth (TSB) at an optical density at 600 nm of 0.1 to 0.2. The amount of bacteria in the resulting culture was determined by plating serial 10-fold dilutions onto petri dishes containing solid TSB agar. Inocula were prepared by diluting the S. enterica serovar Typhimurium culture to concentrations of 0.5 × 104 to 1.0 × 104 CFU per 200 μl of phosphate-buffered saline (PBS). The exact dose of an inoculum used to infect mice was verified by plating serial dilutions.
Infection with S. enterica serovar Typhimurium.
All procedures involving animals were performed in accordance with regulations of the Canadian Council on Animal Care. Each mouse in groups of C57BL/6J (n = 5), MOLF/Ei (n = 4), and 129/Sv (n = 5) mice was infected with 0.2 ml of PBS containing 0.5 × 104 to 1.0 × 104 CFU of S. enterica serovar Typhimurium in the caudal vein. To determine the growth rate of the bacteria within the reticuloendothelial organs of each mouse, mice were sacrificed by CO2 asphyxiation at zero time and on days 1, 3, 4, 5, 7, and 8, and their spleens and livers were recovered. Half of each organ was homogenized in 2 ml (spleen) or 5 ml (liver) of isotonic saline with a Polytron homogenizer (Brinkmann Instruments). Serial dilutions of each homogenate were plated on TSB agar to enumerate the CFU within each organ. The remaining half of each organ was snap frozen in liquid nitrogen and used for RNA extraction. The spleen and liver from one additional mouse of each strain for all time points were fixed in Bouin's solution and used for histopathological analysis.
Histopathology.
Two C57BL/6J mice and two MOLF/Ei mice were sacrificed by cervical dislocation in the terminal stages of infection (days 4 and 5, respectively), and the spleens, livers, kidneys, lungs, brains, hearts, small intestines, and colons were removed, fixed in 10% buffered formaldehyde, and embedded in paraffin. Sections (4 μm thick) were cut and stained with hematoxylin-phloxin-saffron. All sections were examined by the same pathologist. Salmonella-resistant 129/Sv mice were sacrificed on the same days for comparison purposes.
In vivo LPS stimulation.
C57BL/6J (n = 2), C3H/HeJ (n = 2), MOLF/Ei (n = 3), and 129/Sv (n = 2) mice were each injected with 0.5 ml of PBS containing 25 μg of protein-free (<0.008%) Escherichia coli K235 lipopolysaccharide (LPS) intraperitoneally. After 90 min, the mice were sacrificed, and serum was collected by cardiac puncture. Control mice were injected intraperitoneally with PBS. TNF-α concentrations in serum samples were measured with a mouse TNF-α DuoSet kit (Genzyme) used according to the manufacturer's instructions. The lower limit of detection for this TNF-α enzyme-linked immunosorbent assay is 31 pg/ml.
Spleen cell mitogenic response.
Cell suspensions were prepared as previously described from spleens harvested from MOLF/Ei mice (n = 3) (27). Cells, resuspended in RPMI medium supplemented with 20% fetal bovine serum, were plated at a density of 2 × 105 cells/well and were stimulated with 1 to 5 μg of LPS (E. coli K235; Sigma) per ml or 1.5 μg of concanavalin A (ConA) per ml. After 48 h of incubation at 37°C in the presence of 5% CO2, [3H]thymidine was added, and incubation was continued for an additional 16 h. Cells were harvested, and the incorporation of radioactivity was measured by scintillation counting. The amounts of proliferation in response to LPS and ConA are expressed below as stimulation ratios obtained by comparing stimulated cells to unstimulated cells. C3H/HeJ and C57BL/6J mice were used as negative and positive controls, respectively.
Flow cytometry.
The phenotypes of spleen cells were characterized by single-color staining of surface proteins. Fluorescein isothiocyanate-conjugated monoclonal antibodies (MAb) against mouse CD3 (clone 145-2C11, hamster immunoglobulin G [IgG]), CD4 (clone RM4-5, rat IgG2A), CD8 (clone 53-6.7, rat IgG2a), B220 (clone RA3-6B2, rat IgG2b), and Gr-1 (clone RB6-8C5, rat IgG2b) were used. MAb raised against MAC-1 and F4/80 were purchased from Serotec (Oxford, United Kingdom). All other MAb-fluorescein isothiocyanate conjugates were obtained from PharMingen (San Diego, Calif.). Isotype-matched MAbs were used as negative controls. Freshly isolated spleen cells (1 × 106 cells/preparation) were stained with 2 μg of MAb for 30 min in buffer consisting of PBS supplemented with 1% bovine serum albumin and 0.1% sodium azide. After three washes, the cells were resuspended in PBS containing 1% fetal calf serum and 0.1% sodium azide. All staining steps were performed at 4°C. Acquisition of the cells was carried out immediately after staining using FACScan (Becton Dickinson, Mountain View, Calif.). Calibrite beads (Becton Dickinson) were used for calibration, and the data were analyzed with CellQuest software (Becton Dickinson). Approximately 10,000 cells were sampled and gated for each staining analysis.
Northern blot analysis.
RNA was extracted from the spleens of at least two mice for each time point (except on day 5, when only one of the B6 mice survived) by using TRIZOL (Gibco BRL). Ten micrograms of total RNA was used to prepare Northern blots (Hybond-N; Amersham). All membranes were prehybridized for at least 2 h in a solution containing 50% formamide, 10% dextran sulfate, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2× Denhardt's reagent, 1% sodium dodecyl sulfate, and 200 μg of herring sperm DNA per ml. Then 1 × 106 cpm of [α-32P]dATP-labeled cDNA probe for NOS2 (nucleotides 231 to 1993) per ml was hybridized for 20 h at 42°C in the same hybridization solution. The blots were washed in 0.1× SSC-0.1% sodium dodecyl sulfate (final stringency) at 60°C for 20 min. The signal was visualized by autoradiography after overnight exposure. A human β-actin cDNA probe (Clontech) was hybridized to the same blots to control for loading and integrity of the RNA.
Mouse peritoneal macrophages.
Mouse peritoneal macrophages were isolated by peritoneal lavage with 10 ml of RPMI medium 72 h after mice were injected with 1 ml of 3% Brewer's thioglycolate (Difco Laboratories, Detroit, Mich.). Following lysis of red blood cells, the peritoneal cells were washed with PBS. The cells were subsequently plated at a density of 1 × 106 cells/well and allowed to adhere for 2 h. After thorough washing with RPMI medium, the adherent population was stimulated with various concentrations of IFN-γ (100 to 500 U/ml) and/or LPS (10 to 1,000 ng/ml) for 48 h. Supernatants were collected and assayed for NO2− production using Greiss reagent (11).
RNase protection assay.
The RiboQuant multiprobe RNase protection assay system (Pharmingen Canada, Mississauga, Canada) was used to determine levels of cytokine expression following infection with S. enterica serovar Typhimurium. The mCK-2b multiprobe set (containing templates encoding interleukin-12p35 [IL-12p35], IL-12p40, IL-10, IL-1α, IL-1β, IL-1Ra, IL-18, IL-6, IFN-γ, migration inhibition factor [MIF], L32, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) was labeled with [α-32P]UTP and subsequently hybridized for 16 h to 3 μg of total liver or spleen RNA isolated from infected mice. After RNase treatment and purification of the protected fragments, the samples were electrophoresed on a 5% denaturing polyacrylamide gel at a constant power of 30 W. The protected fragments were visualized and quantified by using a STORM 860 PhosphorImager and ImageQuant software (Molecular Dynamics).
In situ hybridization histochemistry.
Two 129/Sv mice, two MOLF/Ei mice, and two C57BL/6J mice were infected with S. enterica serovar Typhimurium, and mice of each strain that were not infected were used as controls. Each animal was deeply anesthetized in the terminal stage of infection (day 5) with an intraperitoneal dose of a mixture containing ketamine (50 mg/kg), xylazine (5 mg/kg), and acepromazine (1 mg/kg), and then it was rapidly perfused transcardially with 0.9% saline and then with 4% paraformaldehyde in buffer containing 0.1 mol of borax per liter (pH 9.5 at 4°C). The brains were removed from the skulls, postfixed for 1 to 2 days, and then placed in 10% sucrose in 4% paraformaldehyde-borax buffer overnight. The brains were mounted on a microtome (Reichert-Jung, Cambridge Instrument Co., Deerfield, Ill.), frozen with dry ice, and cut into 20-μm-thick coronal sections from the olfactory bulb to the end of the medulla. The slices were collected in a cold cryoprotectant solution (0.05 mol of sodium phosphate buffer [pH 7.3] per liter, 30% ethylene glycol, 20% glycerol) and stored at −20°C. Hybridization histochemical localization of inhibitory factor kappa B alpha (IκBα) and Toll-like receptor 2 (Tlr2) mRNA was performed for every sixth section of the whole rostrocaudal extend of each brain. All solutions were treated with diethyl pyrocarbonate (DEPC) and sterilized to prevent degradation of the RNA. The slices were mounted on gelatin- and poly-l-lysine-coated slides, vacuum dried, fixed in 4% paraformaldehyde for 20 min, and treated with proteinase K (10 μg/ml in 100 mM Tris HCl [pH 8.0]-50 mM EDTA [pH 8.0]) at 37°C for 25 min. Then the brain sections were rinsed in sterile DEPC-treated water and then in a 100 mM triethanolamine (pH 8) solution, acetylated with 0.25% acetic anhydride in 100 mM triethanolamine, and dehydrated with graded concentrations of alcohol (50, 70, 95, and 100%). After vacuum drying, 90 μl of hybridization mixture (106 cpm) was spotted on each slide, sealed under a coverslip, and incubated at 60°C for 15 to 20 h on a slide warmer. The coverslips were then removed, and the slices were rinsed in 4× SSC at room temperature. The sections were treated with RNase A (20 μg/ml, 37°C, 30 min), rinsed in descending concentrations of SSC (2×, 1×, and 0.5×), washed in 0.1× SSC for 30 min at 60°C, and dehydrated with graded concentrations of alcohol. After they were dried under a vacuum, the sections were exposed at 4°C to X-ray film (Kodak) for 18 h (IκBα) or for 45 h (Tlr2), defatted in xylene, and dipped in NTB2 nuclear emulsion (Kodak) that was diluted 1:1 with distilled water. The slides were exposed for 7 days (IκBα) or for 11 days (Tlr2), developed in D19 developer (Kodak) for 3.5 min at 14 to 16°C, washed for 15 s in water, and fixed in rapid fixer (Kodak) for 5 min. Then the tissues were rinsed under running water for 1 to 2 h, counterstained with thionin (0.25%), dehydrated with graded concentrations of alcohol, cleared in xylene, and covered with coverslips by using distrene plasticizer xylene.
cRNA probe synthesis.
The pBluescript SK plasmid containing the 1.114-kb full-length coding sequence of the mouse IκBα cDNA (provided by Alain Israel, Institut Pasteur, Paris, France) was linearized with BamHI and HindIII to obtain antisense and sense riboprobes, respectively. The PCR-blunt II-topo plasmid containing the mouse TLR2 cDNA fragment (2.278 kb) was linearized with EcoRV and SpeI to obtain antisense and sense riboprobes, respectively (unpublished data). Radioactive cRNA copies were synthesized by incubating 250 ng of linearized plasmid in a solution containing 6 mM MgCl2, 40 mM Tris (pH 7.9), 2 mM spermidine, 10 mM NaCl, 10 mM dithiothreitol, 0.2 mM ATP, 0.2 mM GTP, 0.2 mM CTP, 200 μCi of α-35S-UTP (NEG 039H; Dupont NEN), 40 U of RNasin (Promega, Madison, Wis.), and 20 U of the appropriate RNA polymerase (T7 for antisense IκBα and sense TLR2 probes, SP6 for antisense TLR2 probe, and T3 for sense IκBα probe) for 60 min at 37°C. The unincorporated nucleotides were removed by adding 100 μl of a DNase solution (1 μl of DNase, 5 μl of a 5-mg/ml tRNA solution, 94 μl of 10 mM Tris-10 mM MgCl2) for 10 min, followed by phenol-chloroform extraction. The cRNA was precipitated with 80 μl of 5 M ammonium acetate and 500 μl of 100% ethanol for 20 min on dry ice. After centrifugation, the pellets were dried and resuspended in 100 μl of 10 mM Tris-1 mM EDTA. The probes (107 cpm/ml) were mixed into 1 ml of hybridization solution containing 822 μl of solution I (500 μl of formamide, 60 μl of 5 M NaCl, 10 μl of 1 M Tris [pH 8.0], 2 μl of 0.5 M EDTA [pH 8.0], 20 μl of 50× Denhardt's solution, 200 μl of 50% dextran sulfate, and 30 μl of DEPC-treated water), 50 μl of a 10-mg/ml tRNA solution, and 10 μl of a 1 M dithiothreitol solution (118 μl of DEPC-treated water − volume of probe used) and heated for 5 min at 65°C before they were spotted onto slides.
Semiquantitative analysis.
Semiquantitative analysis of the hybridization signals for the IκBα and TLR2 transcripts was carried out with X-ray film over the choroid plexus for each mouse. Transmittance values (referred to here as optical density) for the hybridization signals were determined with a Northern Light desktop illuminator (Imaging Research) by using a Sony camera video system attached to a Micro-Nikkor 55-mm Vivitar extension tube set for a Nikon lens and coupled to a Macintosh computer (Power PC 7100/66) with Image software (version 1.61, non-FPU; W. Rasband, National Institutes of Health). All choroid plexus preparations displaying a clear positive signal were evaluated within the linear optical density curve to avoid pixel saturation and underestimation. Sections from control and infected animals were digitized and subjected to densitometric analysis, which yielded mean densities. Each optical density value was then corrected for the average background signal by subtracting the optical densities of areas without positive signals located immediately outside the digitized choroid plexus.
Statistical analysis.
Differences between groups of mice were analyzed by Student's t test using StatView 4.5 software, and P values less than 0.05 were considered significant. Data shown in Fig. 7 were analyzed by a 3 × 2 analysis of variance, followed by a Bonferroni-Dunn test for each postinjection time (Statview 4.01). Factors were identified as follows: strain of mice, which was composed of three levels (129/Sv, MOLF/Ei, and C57BL/6J); and treatment, which combined two levels (control and infected with Salmonella).
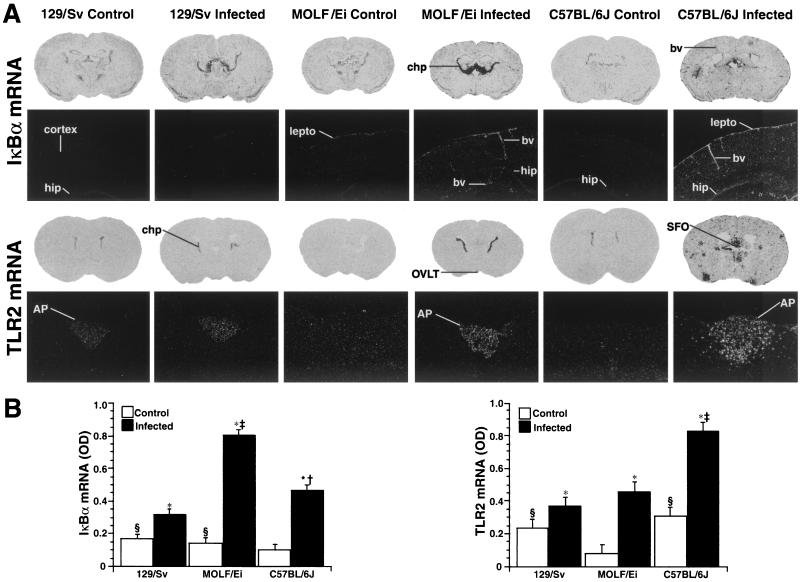
FIG.7.
Effect of S. enterica serovar Typhimurium infection on transcription of the IκBα and Tlr2 genes in the brains of 129/Sv, MOLF/Ei, and C57BL/6J mice. (A) Photomicrographs of coronal sections (first and third rows), showing the hybridization signals for IκBα and Tlr2 mRNA in the brains of control and infected mice. The brain sections (thickness, 25 μm; X-ray film; Biomax; Kodak) show that there were robust hybridization signals in the choroid plexus of MOLF/Ei and C57BL/6J infected mice. Numerous inflammatory clusters were also found in the brains of C57BL/6J infected mice. Dark-field photomicrographs (second and fourth rows; magnifications, ×9 and ×22.5, respectively) of nuclear emulsion-dipped sections showing that IκBα mRNA was expressed mostly in the leptomeninges and blood vessels of MOLF/Ei and C57BL/6J infected mice and Tlr2 mRNA was expressed in the area postrema. Note the spreading of hybridization signal from the area postrema to adjacent parenchymal regions in C57BL/6J infected mice. Abbreviations: AP, area postrema; bv, blood vessels; chp, choroid plexus; hip, hippocampus; lepto, leptomeninges; OVLT, organum vasculosum of the lamina terminalis; SFO, subfornical organ. (B) Average optical densities (OD) for the IκBα (left panel) and Tlr2 (right panel) mRNA signals in the choroid plexus of control and infected animals. The values are means ± standard errors of the means, and statistical analysis was performed by using a 3 × 2 analysis of variance followed by a Bonferroni-Dunn post hoc test (Statview 4.01). An asterisk indicates that the value is significantly different (P < 0.0001) from the value for the control mice; a section sign indicates that the value is significantly different (P < 0.05) from the value for the C57BL/6J control group (IκBα mRNA) or the MOLF/Ei control group (Tlr2 mRNA); a dagger indicates that the value is significantly different (P < 0.0001) from the value for the 129/Sv infected group (IκBα mRNA); and a double dagger indicates that the value is significantly different (P < 0.0001) from the values for the 129/Sv and C57BL/6J infected groups (IκBα mRNA) and from the values for the 129/Sv and MOLF/Ei infected groups (Tlr2 mRNA). For more information on image analysis, see Materials and Methods.
RESULTS
Kinetics of S. enterica serovar Typhimurium infection.
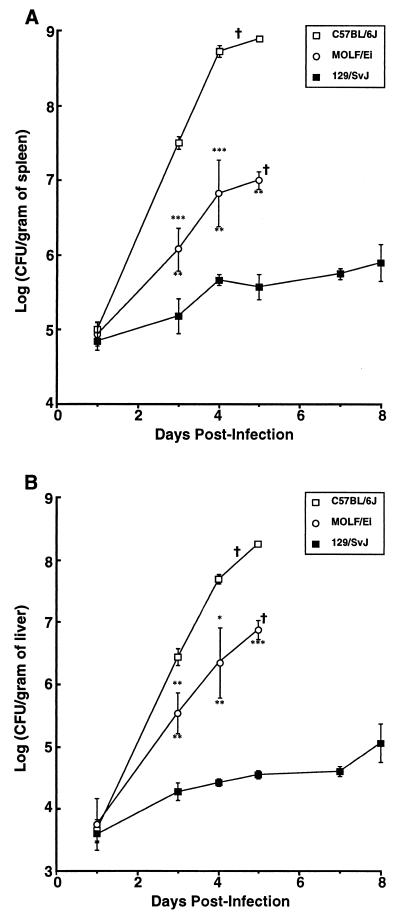
To further investigate the extreme susceptibility of MOLF/Ei mice to Salmonella, MOLF/Ei mice, as well as Salmonella-susceptible C57BL/6J mice and Salmonella-resistant 129/Sv mice, were infected intravenously with 0.5 × 104 to 1 × 104 CFU of S. enterica serovar Typhimurium. At specific times following infection (zero time and days 1, 3, 4, 5, 7, and 8), three to five mice from each group were sacrificed, and their spleens and livers were harvested. The numbers of CFU in each organ were determined by plating serial dilutions. The resistant and susceptible controls behaved as expected. C57BL/6J mice, which have an Nramp1s allele, could not limit multiplication of Salmonella within the spleen and the liver (Fig. 1). Soon after infection, there were drastic increases in the numbers of Salmonella CFU in the spleens and livers, with the levels reaching 1 × 108 to 1 × 109 CFU/g of organ on days 4 and 5. Consequently, C57BL/6J mice died within 5 days, before a specific immune response was established. Resistant 129/Sv mice (Nramp1r) inoculated with the same dose of S. enterica serovar Typhimurium survived for more than 4 weeks (unpublished data). This correlates with the ability of these mice to prevent the bacteria from dividing uncontrollably in both the liver and the spleen during the first week of infection and the subsequent establishment of a plateau phase (Fig. 1). In fact, even after day 8, the number of CFU in the spleen and liver remained low (1 × 104 to 1 × 106 CFU/g of organ) compared to the number observed in C57BL/6J mice. In MOLF/Ei mice, the Salmonella replication profile was intermediate between the profile in the resistant 129/Sv control and the profile in the susceptible C57BL/6J control. As Fig. 1 shows, MOLF/Ei mice infected with S. enterica serovar Typhimurium had 40- to 100-fold-lower levels of bacteria in their spleens and approximately 35-fold-lower levels of bacteria in their livers than C57BL/6J mice. MOLF/Ei mice died from infection on day 5 or 6 with less than 107 CFU per g of organ. These results suggest that MOLF/Ei mice allow replication of the bacteria within the spleen and liver, fail to eliminate Salmonella, and ultimately succumb to infection but with a lower bacterial load than C57BL/6J mice.
FIG. 1.
Bacterial counts in RES organs of Salmonella-infected mice. C57BL/6J, MOLF/Ei, and 129/Sv mice were infected with 0.5 × 104 to 1.0 × 104 CFU of S. enterica serovar Typhimurium intravenously. The numbers of CFU per gram of spleen (A) and liver (B) were determined by plating serial dilutions of homogenized organ on TSB agar. Each data point represents the data for groups of three to five mice. Results are expressed as means ± standard errors of the means. All of the C57BL/6J mice succumbed by day 5 postinfection, all of the MOLF/Ei mice succumbed by day 6 postinfection, and no 129/Sv mice succumbed by day 8 postinfection. Asterisks above the error bars indicate the levels of significance for comparisons of C57BL/6J and MOLF/Ei mice, whereas asterisks below the error bars indicate the levels of significance for comparisons of MOLF/Ei and 129/Sv mice (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001). †, time at which animals start to succumb from infection.
Histopathology of Salmonella-induced lesions.
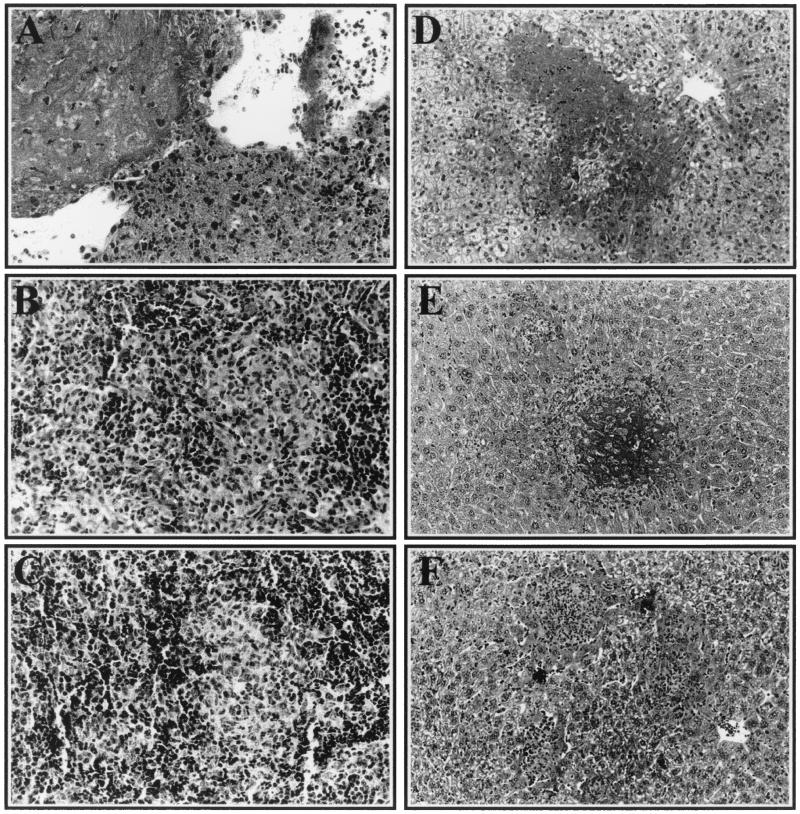
To gain further insight into the progression of S. enterica serovar Typhimurium infection in MOLF/Ei mice and to understand the types of changes that occur in the different hosts during infection, we examined the histology of the lesions in the major sites of replication, as well as in other organ systems. In susceptible C57BL/6J mice, significant inflammatory reactions, marked by infiltration of polymorphonuclear leukocytes (PMN) and macrophages, were visible in both the liver and the spleen as early as day 3 following infection. By day 4, the infiltration in the spleen was predominantly neutrophilic, and the red pulp expansion was more marked and included accumulation of abundant necrotic cell debris and fibrin deposition (Fig. 2A). In the liver, numerous randomly distributed foci of infiltration by PMN were observed, and they were associated with degeneration and necrosis of hepatocytes throughout the parenchyma (Fig. 2D). C57BL/6J mice also had additional necrotic lesions in the heart, kidney, small intestine, and lung (data not shown). The inability of C57BL/6J mice to destroy this pathogen causes an overwhelming, systemic bacterial infection that leads to deleterious tissue injury culminating in the death of the host.
FIG. 2.
Histology of Salmonella-induced lesions in C57BL/6J, MOLF/Ei, and 129/Sv mice. Spleen (A to C) and liver (D to F) tissue samples were isolated from C57BL/6J mice (A and D) on day 4 postinfection and from MOLF/Ei mice (B and E) and 129/Sv mice (C and F) on day 5 postinfection. Following fixation and hematoxylin-phloxin-saffron staining, the spleen and liver sections were photographed at magnifications of ×400 and ×200, respectively.
In contrast, resistant 129/Sv mice showed mild signs of inflammation starting on day 3 in the liver (Fig. 2F) and on day 4 in the spleen (Fig. 2C). The lesions consisted of randomly distributed foci with mild infiltration of both macrophages and PMN. The hepatic inflammatory foci, most abundant between days 4 and 7, were occasionally associated with coagulation necrosis of individual hepatocytes. By day 8, the cellular infiltration was less extensive and was characterized by the presence of a majority of mononuclear cells. No additional lesions were found in the other tissues of 129/Sv mice examined.
In MOLF/Ei mice, the severity of the inflammatory response was intermediate between that seen in resistant 129/Sv and that seen in susceptible C57BL/6J mice. Mild histological changes occurred in the liver by day 3 and in the spleen by day 4. In the liver, the lesions consisted of randomly distributed foci of hepatocyte degeneration and necrosis, infiltrated mostly by PMN and some macrophages at the periphery (Fig. 2E). In the spleen, the red pulp showed progressively increasing expansion, first by macrophages and then on day 5 by a mixture of macrophages and PMN (Fig. 2B). Interestingly, although the C57BL/6J and MOLF/Ei mice were examined at similar stages of the disease (within 24 h of death), the MOLF/Ei mice did not exhibit the same severity of necrosis or degeneration of hepatosplenic tissue structure as the C57BL/6J mice. Additional lesions were found only in the hearts of MOLF/Ei mice, and these lesions consisted of peracute, necrotic changes in the cardiomyocytes. There were no visible lesions in the kidney, small intestine, lung, brain, or colon. The relatively limited tissue lesions that occurred throughout infection with S. enterica serovar Typhimurium in MOLF/Ei mice correlated well with the reduced bacterial loads in these organs.
Cytokine profiles in spleens and livers of MOLF/Ei mice infected with S. enterica serovar Typhimurium.
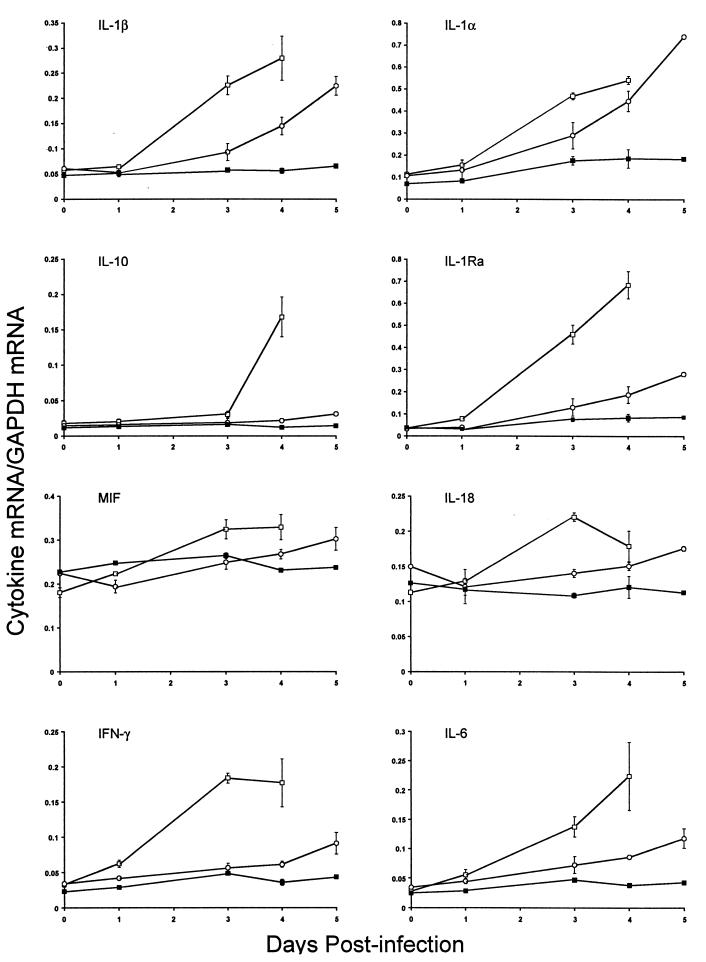
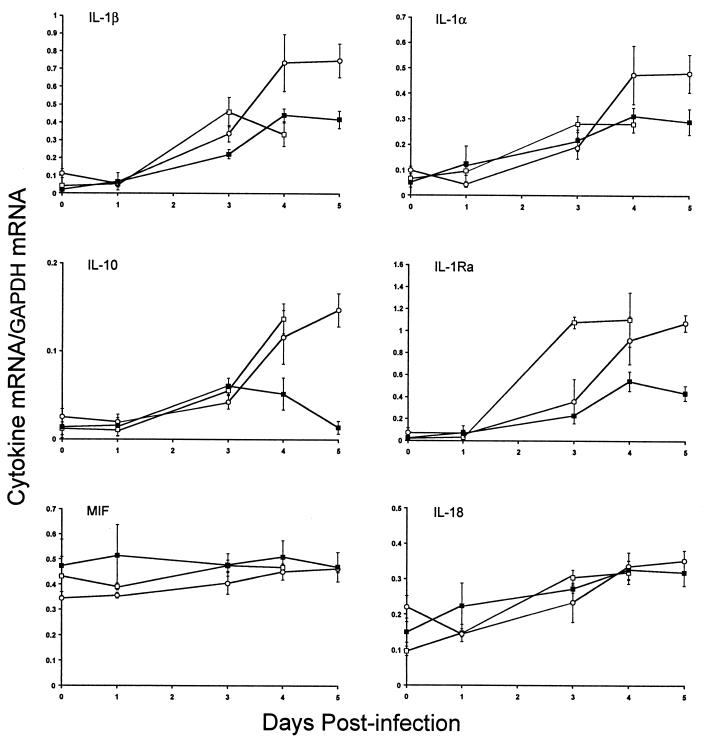
We investigated cytokine gene expression in the RES after S. enterica serovar Typhimurium infection of Salmonella-susceptible MOLF/Ei mice since the production of several key cytokines, such as TNF-α, IL-1, and IL-12, by macrophages has been shown to be crucial in the regulation of inflammation and the severity of the outcome of Salmonella infection (reviewed in reference 15). The cytokine gene expression repertoire in MOLF/Ei mice was determined by an RNase protection assay. Antisense RNA probes that hybridize to the genes encoding IL-12p35, IL-12p40, IL-10, Il-1α, IL-1β, IL-1Ra, IL-18, IL-6, IFN-γ, and MIF, as well as two housekeeping genes, the genes encoding L32 and GAPDH, were analyzed. The changes in cytokine gene expression in the spleens and livers of MOLF/Ei mice were compared to the changes in the spleens and livers of Salmonella-susceptible and -resistant controls (C57BL/6J and 129/Sv mice, respectively). Analysis of cytokine gene expression demonstrated that the levels of mRNA for most cytokines studied were similar in the spleens of MOLF/Ei and 129/Sv mice during the early phase of infection (Fig. 3). In contrast, most cytokines were expressed at higher levels in MOLF/Ei mice than in 129/Sv mice during the late stage of infection. There were substantial increases in expression of mRNA for both Il1α and Il1β as the infection progressed in the spleens of MOLF/Ei mice, with four- to sevenfold increases by 5 days after infection. In Salmonella-susceptible C57BL/6J mice, expression of most spleen cytokines was upregulated during the course of infection. The levels of mRNA for Mif and Il18 did not vary significantly during infection in the three mouse strains examined, and expression of Il12p35 and Il12p40 mRNA was very low in our experimental system (data not shown). In the liver, the situation appeared to be more complex. In MOLF/Ei mice, there was upregulation of Il10, Il18, Il1Ra, Il1b, and Il1a (Fig. 4). Increases in Il18 liver mRNA levels were similar in all three mouse strains during infection, whereas Il10 and Il1Ra mRNA levels increased only in C57BL/6J and MOLF/Ei mice.
FIG. 3.
Cytokine mRNA levels in mouse spleens after infection with S. enterica serovar Typhimurium. Total RNA isolated from spleens of mice was used to determine the cytokine mRNA levels at different times following infection with S. enterica serovar Typhimurium. The amount of each cytokine mRNA is expressed as the ratio of the signal intensity of the cytokine mRNA to the signal intensity of GAPDH mRNA. Symbols: □, C57BL/6J mice; ○, MOLF/Ei mice; ▪, 129/Sv mice. The values are means ± standard errors of the means based on the data for three mice.
FIG. 4.
Cytokine mRNA levels in mouse livers after infection with S. enterica serovar Typhimurium. Total RNA isolated from livers of mice was used to determine the cytokine mRNA levels at different times following infection with S. enterica serovar Typhimurium. The amount of each cytokine mRNA is expressed as the ratio of the signal intensity of the cytokine mRNA to the signal intensity of GAPDH mRNA. Symbols: □, C57BL/6J mice; ○, MOLF/Ei mice; ▪, 129/Sv mice. The values are means ± standard errors of the means based on the data for three mice.
In vitro and in vivo responses of MOLF/Ei mice to LPS.
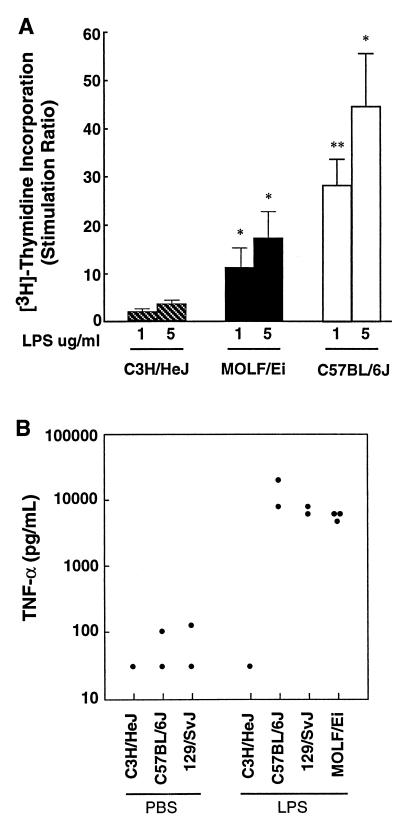
LPS is the major constituent of the outer membranes of gram-negative bacteria, including S. enterica serovar Typhimurium, and is known to have the capacity to initiate a protective systemic inflammatory response in the host that is aimed at elimination of gram-negative bacteria. Paradoxically, an overactive response to LPS may be injurious to the host, as observed in endotoxic shock. Initially, we examined the ability of spleen cells from MOLF/Ei mice to proliferate in response to stimulation with LPS. Spleen cells from these mice showed an intermediate but dose-dependent response to LPS. The stimulation ratio for MOLF/Ei mice was 11 with 1 μg of LPS per ml; in comparison, the stimulation ratios for C3H/HeJ (nonresponder) and C57BL/6J (responder) mice were 1.8 and 28.3, respectively (Fig. 5A). All three mouse strains showed a strong response to the T-lymphocyte mitogen ConA (data not shown). The lower response of splenocytes from MOLF/Ei mice to LPS was not due to differences in the numbers and phenotypes of leukocytes in the spleens, as fluorescence-activated cell sorter analysis revealed that the relative proportions of different populations of macrophages (MAC-1, F4/80), B lymphocytes (B220), T lymphocytes (CD3, CD4, CD8, αβ T-cell receptor, γδ T-cell receptor), natural killer cells (PanNK, NK1.1), and granulocytes (Gr-1) were comparable in C57BL/6J and MOLF/Ei mice (data not shown).
FIG. 5.
In vitro and in vivo responses to LPS. (A) Spleen cells isolated from individual C3H/HeJ, MOLF/Ei, and C57BL/6J mice were stimulated with various doses of LPS (1 to 5 μg/ml). The spleen cell proliferation in response to LPS stimulation was measured by determining the incorporation of tritiated thymidine over a 16-h period. The data are expressed as the ratio of spleen cell proliferation with LPS to spleen cell proliferation without LPS. The values are means ± standard errors of the means based on three or four mice for each group. Triplicate wells were prepared for each mouse. The levels of statistical significance based on comparisons with the data for the C3H/HeJ negative control are indicated (∗, P < 0.05; ∗∗, P < 0.01). (B) Production of TNF-α as measured by an enzyme-linked immunosorbent assay in the sera of mice 90 min after intraperitoneal stimulation with 25 μg of protein-free E. coli K235 LPS or with PBS as a basal level control. Each point represents the data for one mouse.
To determine the in vivo responsiveness of MOLF/Ei mice to LPS, the serum concentrations of TNF-α were measured. Circulating levels of this inflammatory cytokine normally increase substantially in LPS-responsive mice, such as C57BL/6J and 129/Sv mice, treated with LPS (43). The LPS-hyporesponsive strain C3H/HeJ was used as a negative control. Figure 5B shows that as expected, C3H/HeJ mice failed to produce TNF-α (<31 pg/ml), while both LPS-responsive strains responded to LPS by producing TNF-α. In MOLF/Ei mice, the levels of TNF-α in the sera (6,179 pg/ml) reached the levels observed in C57BL/6J and 129/Sv mice (14,964 and 7,090 pg/ml, respectively). These results clearly indicate that the LPS signaling pathway is functional in MOLF/Ei mice and may suggest that TNF-α secretion plays a small role in the susceptibility of MOLF/Ei mice to infection with S. enterica serovar Typhimurium.
Nitric oxide determination in MOLF/Ei mice.
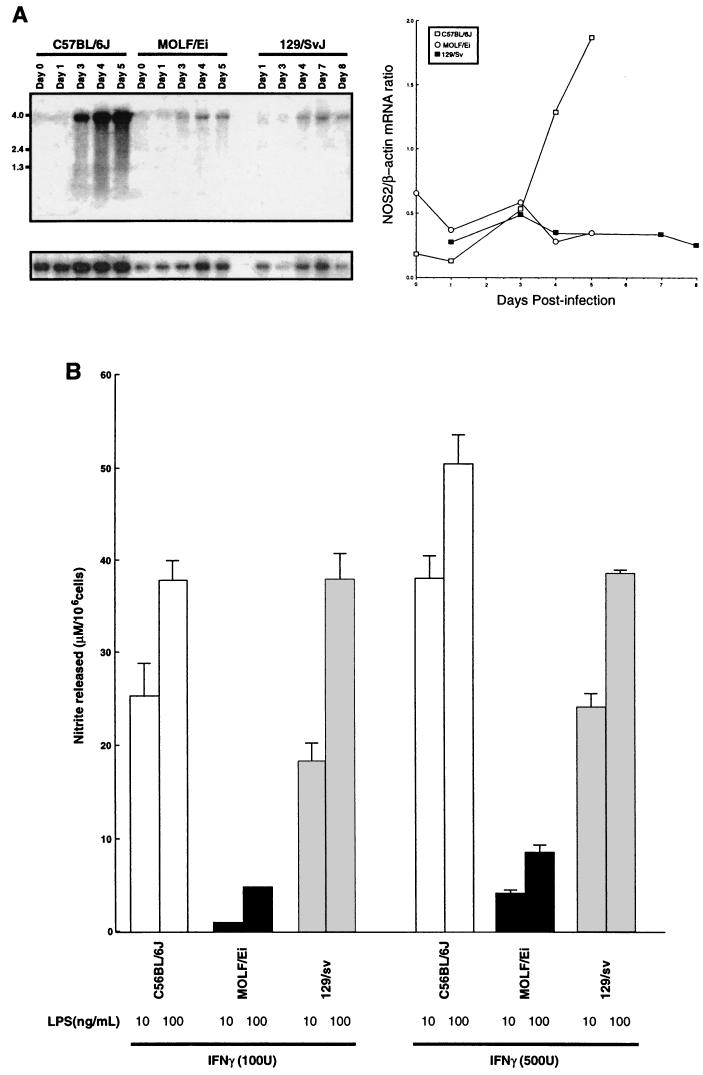
To evaluate NOS2 activity in MOLF/Ei mice, we measured Nos2 mRNA expression during infection and NO production by estimating the levels of the stable NO metabolite nitrite in cell-free supernatants prepared with peritoneal macrophages by using a spectrophotometric method based on the Greiss reaction. As shown in Fig. 6, Northern blot analysis revealed that during infection with S. enterica serovar Typhimurium the Nos2 mRNA levels were not induced to the same extent in MOLF/Ei mice as in C57BL/6J mice. Furthermore, peritoneal macrophages from MOLF/Ei mice were deficient in nitrite production in response to a combination of IFN-γ and LPS. Macrophages from both the Salmonella-susceptible C57BL/6J mice and the Salmonella-resistant 129/Sv mice released nitrite in a dose-dependent fashion (ranges, 25 to 48 μM/106 cells in C57BL/6J and 18 to 39 μM/106 cells in 129/Sv mice). Although the levels of nitrite produced in vitro by 129/Sv and C57BL/6J macrophages did not correlate with the ability to restrict bacterial growth in RES tissues, the decreased capacities of MOLF/Ei mice to induce Nos2 mRNA and to produce NO may play a role in preventing the deleterious inflammatory response seen in C57BL/6J mice.
FIG. 6.
Inducible NOS2 activity in MOLF/Ei mice. (A) Northern blot showing Nos2 expression in splenic mRNA isolated from C57BL/6J, MOLF/Ei, and 129/Sv mice following intravenous infection with 0.5 × 104 to 1.0 × 104 CFU of S. enterica serovar Typhimurium. Each lane shows the data for one mouse. The lower panel shows the results obtained with the β-actin control. The panel on the right shows the ratio of the signal intensity for Nos2 mRNA to the signal intensity for β-actin mRNA in the Northern blot. (B) Thioglycolate-elicited peritoneal macrophages were stimulated with various concentrations of IFN-γ and/or LPS for 48 h. The amounts of NO2− in cell-free supernatants were determined by using the Greiss reagent. The data are expressed as means ± standard errors of the means based on duplicate samples for each combination.
Central nervous system.
Figure 7 shows representative examples of IκBα and Tlr2 gene expression in response to S. enterica serovar Typhimurium infection in the brains of 129/Sv, MOLF/Ei, and C57BL/6J mice. Weak expression of IκBα and Tlr2 mRNA was found in the brains of control mice, which contrasted with the strong induction of both transcripts in response to infection. Infection with S. enterica serovar Typhimurium stimulated transcription of IκBα in the blood vessels and the circumventricular organ, organum vasculusum of the lamina terminalis, subfornical organ, median eminence, and area postrema. The leptomeninges and the choroid plexus also exhibited increased mRNA levels, although the intensities of the signals were significantly different in the different mouse strains. Indeed, the hybridization signal was less intense in the brains of the resistant 129/Sv mice than in the brains of the MOLF/Ei and C57BL/6J mice. Robust hybridization signals were found in several vascular system-associated structures, as well as along penetrating arterioles and capillaries, in both MOLF/Ei and C57BL/6J mice (Fig. 7A).
Strong induction of the innate receptor TLR2 was detected in the brains of infected mice, although the intensity and spreading of the signal varied considerably in the different mouse strains. S. enterica serovar Typhimurium increased TLR2 mRNA levels in the circumventricular organs and other vascular system-associated regions of the central nervous systems of all strains, but C57BL/6J mice had numerous clusters of positive cells within the brain parenchyma (Fig. 7). These clusters of Tlr2-expressing cells that were found only in these animals were probably mixed populations of cells, including parenchymal microglia and infiltrating blood-derived myeloid cells. These data provide clear evidence that there is a differential innate immune response in the brains of S. enterica serovar Typhimurium-infected mice that may play a key role in the survival of susceptible mice.
DISCUSSION
Because of their genetic diversity, wild-derived inbred mice provide undescribed genetic variations in traits that usually exhibit Mendelian inheritance in common mouse strains. This was clearly shown in our previous study of susceptibility to infection with Salmonella in wild-derived MOLF/Ei mice, which is inherited as a complex trait and does not involve the innate immune genes Nramp1 and Tlr4 (39). Our genetic analysis revealed two major chromosomal regions that affect the outcome of Salmonella infection in MOLF/Ei mice: a protective locus on chromosome 11 (Ity2) and a susceptibility locus on distal chromosome 1 (Ity3). These two QTL are located in regions of the genome rich in attractive candidate genes for which a role in host resistance to Salmonella infection is either known or could be envisioned. The candidate genes include those encoding NOS2, GM-CSF (granulocyte-macrophage colony-stimulating factor), and IL-12p40 on chromosome 11 and IL-10, PTGS2 (prostaglandin endoperoxide synthase, a limiting enzyme for the formation of prostaglandins), and TLR5 on chromosome 1 (38, 39). Several of these genes were assessed by expression and sequencing analysis (38; this study). Sequence analysis of C57BL/6J and MOLF/Ei alleles for a given relevant gene revealed a great number of sequence variants which could be easily explained by the genetic divergence of classical and wild-derived mice. However, this genetic divergence complicated the discrimination between functionally important amino acid changes and simple sequence polymorphisms in our analysis. The goal of the present study was to characterize the immunological responses of MOLF/Ei mice during the course of infection in an effort to provide a systematic approach to candidate gene analysis.
Despite the fact that the survival times for MOLF/Ei and C57BL6/J mice are similar, our study clearly showed that the kinetics of the infection and the severity of the histopathological lesions were intermediate in MOLF/Ei mice compared to Salmonella-resistant 129/Sv mice and highly Salmonella-susceptible C57BL/6J mice. MOLF/Ei mice could partially control the replication of Salmonella within RES organs and succumbed to infection with a lower bacterial load than other Salmonella-susceptible mice. Histopathological examination showed that Salmonella-infected MOLF/Ei mice had inflammatory lesions mainly in the spleen and liver and that the size and number of the lesions were significantly reduced compared to the size and number of lesions observed in Salmonella-susceptible C57BL/6J mice. The inflammatory lesions in MOLF/Ei mice were typical of those observed during the transition phase between the rapid exponential growth of the bacteria and the establishment of the plateau phase during sublethal infection in resistant mice (25), and they correlated well with a lower bacterial load and diminished cellular infiltration in response to infection. These intermediate phenotypes are in accordance with the fact that we detected both protective and susceptibility loci in our genetic analysis of susceptibility to Salmonella infection in wild-derived mice.
During infection with S. enterica serovar Typhimurium, cells of the immune system are exposed to LPS, which induces a rapid cytokine response essential for activation of the host defense mechanism. LPS-induced host cell activation is known to be mediated to a large extent by CD14, a differentiation antigen expressed as a glycosylphosphatidylinositol-anchored membrane glycoprotein in myeloid lineage cells or as a soluble form in the serum. The LPS-CD14 complex triggers signaling pathways through Tlr4, culminating in nuclear translocation of NF-κB and activation of activator protein-1, c-Jun, and c-Fos. Activated NF-κB and activator protein-1 stimulate transcriptional activation of several inflammatory cytokines, including IL-1 and TNF-α (reviewed in references 32 and 44). Our work shows that MOLF/Ei mice respond to LPS to the same extent as 129/Sv and C57BL/6J mice in vivo , as measured by LPS-induced systemic TNF-α production, although they have reduced in vitro LPS-induced spleen cell mitogenesis indices.
Overall, we observed attenuated or intermediate cytokine responses in MOLF/Ei mice during the course of Salmonella infection, except for IL-1α and IL-1β mRNA expression. IL-1 is a highly potent proinflammatory cytokine that is produced by macrophages in response to Salmonella infection and LPS. The IL-1 family of genes comprises three members: the proinflammatory IL-1α and IL-1β genes (collectively referred to as IL-1 genes) and the IL-1Ra gene (IL-1 receptor antagonist gene). IL-1Ra is usually released into the circulation and has a negative influence on the production and activity of IL-1. IL-1α and IL-1β are known to signal through the IL-1RI (type I IL-1 receptor). The extracellular domain of IL-1RI is characterized by a modular structure consisting of immunoglobulin domains connected to an intracellular Toll/IL-1R signaling domain (27). Each individual IL-1 molecule forms a trimer with IL-1RI and IL-1RAcP (accessory protein), which allows recruitment of the downstream signaling proteins necessary for activation of NF-κB. IL-1 initiates the transcription of various genes, including those encoding IL-1Ra, PTGS2, and NOS2 in vivo and in vitro (27). Recent in vitro studies performed with gene arrays showed that the S. enterica serovar Typhimurium-induced gene expression profile in RAW 264.7 macrophages derived from Salmonella-susceptible BALB/cJ mice is characterized by strong upregulation of Il1b and Nos2 (36), which is consistent with our in vivo observations for susceptible C57BL/6J mice. Surprisingly, spleen Nos2 and Il1Ra mRNA levels were not upregulated in MOLF/Ei mice. Although increased mRNA expression does not mean that there is increased expression of functional cytokines, the observation that Nos2 and Il1Ra mRNA are not upregulated in the spleens of MOLF/Ei mice despite significant increases in the levels of Il1a and Il1b mRNA may indicate that there is a defective IL-1 response.
We investigated the possible involvement of NO in antimicrobial activity and its contribution to the inflammatory processes in our model because of the obvious possibility that Nos2 is a Salmonella resistance gene based on our QTL analysis and its prior involvement in host resistance against bacterial infection. Studies with Nos2-deficient mice support the hypothesis that NO plays a dual role during virulent Salmonella infection in vivo. Enhanced production of NO provides increased host defense against pathogens but also contributes to inflammation, tissue damage, and even endotoxic shock (18, 20, 40). Although Nos2 knockout mice are able to control the early replication of Salmonella in the RES organs, they are not able to suppress bacterial growth and they eventually die from the infection (20). The kinetics of bacterial replication in MOLF/Ei mice are similar to those observed in Nos2-deficient mice. In our model, the levels of Nos2 mRNA induced during infection did not correlate with the ability to restrict bacterial growth. In contrast, low Nos2 mRNA levels correlate with reduced NO production and a decreased inflammatory response; this finding is consistent with the histological analysis results that showed that there was reduced inflammatory focus formation in spleens and livers, and it may explain the protective effect of the QTL on chromosome 11.
Finally, we studied the impact of systemic Salmonella infection on the brain immune response. Inflammatory mediators, such as LPS and IL-1 released by leukocytes during Salmonella infection, are responsible for eliciting systemic responses mediated by the central nervous system, such as fever, and have a marked effect on the immune response of the brain (12-14). LPS and IL-1β are known to induce robust and transient expression of PTGS2 and IκBα in endothelial cells of the brain (12, 13), which in turn are responsible for the production of a second wave of mediators, such as prostaglandins and NO, that diffuse rapidly from the vascular side of the blood-brain barrier to the brain (8, 13). As observed in the periphery, the cerebral innate immune response to LPS is mediated through TLR4 (14). In MOLF/Ei mice, we observed strong induction of IκBα during the course of infection, which was associated with an increase in Tlr2 expression in the blood-brain barrier. TLR2 participates in the innate recognition of several classes of pathogens (reviewed in reference 1) and appears to be a critical molecule in the activation of cell-mediated immunity by producing IL-12 upon exposure of cells to bacterial lipoproteins (35). TLR2 is found in most tissues, and its expression is regulated by LPS (16, 17, 21, 22, 35). TLR2 has been shown to be involved in the generation and resolution of inflammation in response to bacterial pathogens by inducing apoptosis (3). The impact of these findings on the response of MOLF/Ei mice to Salmonella infection remains to be elucidated; however, it is possible that the high levels of circulating IL-1 are responsible for the elevated expression of both Iκβα and Tlr2 in the brain.
Of interest is the strong NF-κB activity in the choroid plexus of MOLF/Ei mice in response to Salmonella infection. Such activity is generally associated with profound transcription activation of proinflammatory cytokines and different members of the complement system (23, 24). These data, together with the fact that the choroid plexus is the organ responsible for synthesizing the cerebrospinal fluid, indicate that cytokines may be able to diffuse freely and at high concentrations across the central nervous system in MOLF/Ei-infected mice. These molecules may have potent effects on different neurophysiological functions, including those that control the autonomic system. An exaggerated inflammatory response in the cerebral tissue, especially at the level of the choroid plexus, may therefore be associated with profound alteration of the neuronal circuits that control the autonomic outputs. Whether this mechanism contributes to the susceptibility of MOLF/Ei mice to Salmonella infection remains to be firmly established, but high concentrations of proinflammatory molecules in the brain can have profound detrimental effects on neuronal integrity (reviewed in reference 37).
The data presented here provide an overview of the immune response of wild-derived MOLF/Ei mice to infection with S. enterica serovar Typhimurium. These data also demonstrate that examination of the immune response as a whole is an essential step for evaluating the candidacy of genes in QTL analysis, although it is difficult because of the complex inflammatory cascade initiated after challenge with S. enterica serovar Typhimurium. Immunological and genetic approaches are in fact complementary for understanding the host response phenotype. Identification of the molecular basis of the MOLF/Ei mouse host defense systems will be facilitated by the production of congenic mouse lines carrying individual Salmonella susceptibility loci.
Acknowledgments
We thank L. Laroche, L. Larivière, and M. Tam for technical assistance and S. Vidal for critical reading of the manuscript.
This work was supported by grants to D.M. and M.M.S. from the Canadian Institutes of Health Research (CIHR). P.G. is an International Research Scholar of the Howard Hughes Medical Institute (HHMI) and a Senior Scientist of the CIHR. S.R. and D.M. are CIHR Investigators. D.M. is an International Research Scholar of the HHMI (Infectious Diseases and Parasitology).
Editor: R. N. Moore
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 4.Collins, F. M. 1970. Immunity to enteric fever. Infect. Immun. 1:243-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, F. M. 1972. Salmonellosis in orally infected specific pathogen-free C57B1 mice. Infect. Immun. 5:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlan, J. W. 1997. Critical roles of neutrophils in host defense against experimental systemic infections of mice by Listeria monocytogenes, Salmonella typhimurium, and Yersinia enterocolitica. Infect. Immun. 65:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conlan, J. W. 1996. Neutrophils prevent extracellular colonization of the liver microvasculature by Salmonella typhimurium. Infect. Immun. 64:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ek, M., D. Engblom, S. Saha, A. Blomqvist, P. J. Jakobsson, and A. Ericsson-Dahlstrand. 2001. Inflammatory response: pathway across the blood-brain barrier. Nature 410:430-431. [DOI] [PubMed] [Google Scholar]
- 9.Hormaeche, C. E. 1979. Natural resistance to Salmonella typhimurium in different inbred mouse strains. Immunology 37:311-318. [PMC free article] [PubMed] [Google Scholar]
- 10.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 11.Jin, F. Y., C. Nathan, D. Radzioch, and A. Ding. 1997. Secretory leukocyte protease inhibitor: a macrophage product induced by and antagonistic to bacterial lipopolysaccharide. Cell 88:417-426. [DOI] [PubMed] [Google Scholar]
- 12.Lacroix, S., D. Feinstein, and S. Rivest. 1998. The bacterial endotoxin lipopolysaccharide has the ability to target the brain in upregulating its membrane CD14 receptor within specific cellular populations. Brain Pathol. 8:625-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laflamme, N., S. Lacroix, and S. Rivest. 1999. An essential role of interleukin-1beta in mediating NF-kappaB activity and COX-2 transcription in cells of the blood-brain barrier in response to a systemic and localized inflammation but not during endotoxemia. J. Neurosci. 19:10923-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laflamme, N., and S. Rivest. 2001. Toll-like receptor 4: the missing link of the cerebral innate immune response triggered by circulating gram-negative bacterial cell wall components. FASEB J. 15:155-163. [DOI] [PubMed] [Google Scholar]
- 15.Lalmanach, A. C., and F. Lantier. 1999. Host cytokine response and resistance to Salmonella infection. Microbes Infect. 1:719-726. [DOI] [PubMed] [Google Scholar]
- 16.Liu, S., A. N. Salyapongse, D. A. Geller, Y. Vodovotz, and T. R. Billiar. 2000. Hepatocyte toll-like receptor 2 expression in vivo and in vitro: role of cytokines in induction of rat TLR2 gene expression by lipopolysaccharide. Shock 14:361-365. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Y., Y. Wang, M. Yamakuchi, S. Isowaki, E. Nagata, Y. Kanmura, I. Kitajima, and I. Maruyama. 2001. Upregulation of Toll-like receptor 2 gene expression in macrophage response to peptidoglycan and high concentration of lipopolysaccharide is involved in NF-κB activation. Infect. Immun. 69:2788-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacMicking, J. D., C. Nathan, G. Hom, N. Chartrain, D. S. Fletcher, M. Trumbauer, K. Stevens, Q. W. Xie, K. Sokol, N. Hutchinson, et al. 1995. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81:641-650. [DOI] [PubMed] [Google Scholar]
- 19.Mastroeni, P., J. A. Harrison, and C. E. Hormaeche. 1994. Natural resistance and acquired immunity to Salmonella. Fund. Clin. Immunol. 2:83-88. [Google Scholar]
- 20.Mastroeni, P., A. Vazquez-Torres, F. C. Fang, Y. Xu, S. Khan, C. E. Hormaeche, and G. Dougan. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura, T., A. Ito, T. Takii, H. Hayashi, and K. Onozaki. 2000. Endotoxin and cytokine regulation of toll-like receptor (TLR) 2 and TLR4 gene expression in murine liver and hepatocytes. J. Interferon Cytokine Res. 20:915-921. [DOI] [PubMed] [Google Scholar]
- 22.Medvedev, A. E., K. M. Kopydlowski, and S. N. Vogel. 2000. Inhibition of lipopolysaccharide-induced signal transduction in endotoxin-tolerized mouse macrophages: dysregulation of cytokine, chemokine, and toll-like receptor 2 and 4 gene expression. J. Immunol. 164:5564-5574. [DOI] [PubMed] [Google Scholar]
- 23.Nadeau, S., and S. Rivest. 2001. The complement system is an integrated part of the natural innate immune response in the brain. FASEB J. 15:1410-1412. [DOI] [PubMed] [Google Scholar]
- 24.Nadeau, S., and S. Rivest. 2000. Role of microglial-derived tumor necrosis factor in mediating CD14 transcription and nuclear factor kappa B activity in the brain during endotoxemia. J. Neurosci. 20:3456-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakoneczna, I., and H. S. Hsu. 1980. The comparative histopathology of primary and secondary lesions in murine salmonellosis. Br. J. Exp. Pathol. 61:76-84. [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien, A. D., D. L. Rosenstreich, I. Scher, G. H. Campbell, R. P. MacDermott, and S. B. Formal. 1980. Genetic control of susceptibility to Salmonella typhimurium in mice: role of the LPS gene. J. Immunol. 124:20-24. [PubMed] [Google Scholar]
- 27.O'Neill, L. A., and C. A. Dinarello. 2000. The IL-1 receptor/toll-like receptor superfamily: crucial receptors for inflammation and host defense. Immunol. Today 21:206-209. [DOI] [PubMed] [Google Scholar]
- 28.Pang, T., M. M. Levine, B. Ivanoff, J. Wain, and B. B. Finlay. 1998. Typhoid fever—important issues still remain. Trends Microbiol. 6:131-133. [DOI] [PubMed] [Google Scholar]
- 29.Plant, J., and A. A. Glynn. 1976. Genetics of resistance to infection with Salmonella typhimurium in mice. J. Infect. Dis. 133:72-78. [DOI] [PubMed] [Google Scholar]
- 30.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 31.Qureshi, S. T., O. Bronchain, M. Nemer, and D. Malo. 1996. Mapping of the Gata6 gene to mouse chromosome 18. Mamm. Genome 7:705-706. [DOI] [PubMed] [Google Scholar]
- 32.Qureshi, S. T., P. Gros, and D. Malo. 1999. Host resistance to infection: genetic control of lipopolysaccharide responsiveness by TOLL-like receptor genes. Trends Genet. 15:291-294. [DOI] [PubMed] [Google Scholar]
- 33.Qureshi, S. T., L. Lariviere, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson, H. G., and S. I. Vas. 1972. Resistance of inbred mice to Salmonella typhimurium. J. Infect. Dis. 126:378-386. [DOI] [PubMed] [Google Scholar]
- 35.Rock, F. L., G. Hardiman, J. C. Timans, R. A. Kastelein, and J. F. Bazan. 1998. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 37.Rothwell, N. J., and G. N. Luheshi. 2000. Interleukin 1 in the brain: biology, pathology and therapeutic target. Trends Neurosci. 23:618-625. [DOI] [PubMed] [Google Scholar]
- 38.Sebastiani, G., G. Leveque, L. Lariviere, L. Laroche, E. Skamene, P. Gros, and D. Malo. 2000. Cloning and characterization of the murine toll-like receptor 5 (Tlr5) gene: sequence and mRNA expression studies in Salmonella-susceptible MOLF/Ei mice. Genomics 64:230-240. [DOI] [PubMed] [Google Scholar]
- 39.Sebastiani, G., L. Olien, S. Gauthier, E. Skamene, K. Morgan, P. Gros, and D. Malo. 1998. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics 47:180-186. [DOI] [PubMed] [Google Scholar]
- 40.Umezawa, K., T. Akaike, S. Fujii, M. Suga, K. Setoguchi, A. Ozawa, and H. Maeda. 1997. Induction of nitric oxide synthesis and xanthine oxidase and their roles in the antimicrobial mechanism against Salmonella typhimurium infection in mice. Infect. Immun. 65:2932-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal, S., M. L. Tremblay, G. Govoni, S. Gauthier, G. Sebastiani, D. Malo, E. Skamene, M. Olivier, S. Jothy, and P. Gros. 1995. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J. Exp. Med. 182:655-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal, S. M., D. Malo, K. Vogan, E. Skamene, and P. Gros. 1993. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell 73:469-485. [DOI] [PubMed] [Google Scholar]
- 43.Vogel, S. N., D. Johnson, P. Y. Perera, A. Medvedev, L. Lariviere, S. T. Qureshi, and D. Malo. 1999. Cutting edge: functional characterization of the effect of the C3H/HeJ defect in mice that lack an Lpsn gene: in vivo evidence for a dominant negative mutation. J. Immunol. 162:5666-5670. [PubMed] [Google Scholar]
- 44.Zhang, G., and S. Ghosh. 2001. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Investig. 107:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]