Abstract
Individuals with cystic fibrosis (CF) are commonly colonized with Pseudomonas aeruginosa. The chronic infections caused by P. aeruginosa are punctuated by acute exacerbations of the lung disease, which lead to significant morbidity and mortality. As regulators of virulence determinants, P. aeruginosa quorum-sensing systems may be active in the chronic lung infections associated with CF. We have examined the levels of autoinducer molecules and transcript accumulation from the bacterial populations found in the lungs of patients with CF. We detected biologically active levels of N-(3-oxododecanoyl)-l-homoserine (3-oxo-C12-HSL) and N-butyryl-l-homoserine lactone (C4-HSL) in sputum from CF patients. Interestingly, it appears that C4-HSL is less frequently detected than 3-oxo-C12-HSL in the lungs of patients with CF. We also examined the transcription of the autoinducer synthase gene lasI and showed that it is frequently expressed in the lungs of patients with CF. We observed a significant correlation between the expression of lasI and four target genes of the Las quorum-sensing system. Taken together, our results indicate that quorum-sensing systems are active and may control virulence factor expression in the lungs of patients with CF.
Quorum-sensing signaling systems allow bacteria to regulate gene expression in a population-dependent manner. Quorum-sensing regulatory mechanisms are widespread; they have been described in numerous gram-positive (18) as well as gram-negative (9, 42) bacteria. In acyl-homoserine lactone-based systems, growing bacteria produce small signaling molecules (autoinducers) that accumulate in the surrounding environment. At a specific cell density, the concentration of autoinducer becomes sufficient to interact with the autoinducer-dependent transcriptional activator protein and alter gene expression.
In Pseudomonas aeruginosa, quorum-sensing systems have been extensively studied. Two acyl-homoserine lactone-based systems, the las (10) and rhl (25) systems, have been described. These operate in a hierarchical fashion along with a recently described quinolone signaling system (33) to regulate as much as 4% of the genome (45). LasI and RhlI synthesize the autoinducers N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL) (29) and N-butyryl-l-homoserine lactone (C4-HSL) (30), respectively. LasR and RhlR bind DNA to modify transcription of target genes only after a threshold level of their respective autoinducers is reached.
Quorum-sensing systems are believed to be central to the pathogenesis of P. aeruginosa (reviewed by Rumbaugh et al. [34]). One such infection in which quorum sensing may play an important role is the P. aeruginosa lung infections associated with cystic fibrosis (CF). These infections may be the perfect environment for the expression of the quorum-sensing systems, as the lungs are a spatially limited environment and P. aeruginosa can grow to high densities (107 to 108/ml) (40) in sputum. These conditions should be sufficient to induce expression of P. aeruginosa quorum-sensing-regulated genes within the CF lung (41).
Several lines of experimental evidence implicate quorum-sensing systems as important in the pathogenesis of this infection. First, the lasRI 3-oxo-C12-HSL-regulated genes lasB and lasA are transcribed (39, 40, 41) and translated (13, 15, 16, 20) in the lungs of CF patients. Additionally, lasR transcripts were detected in sputum samples, and their accumulation correlated to those of lasA, lasB, and toxA transcripts (41). This suggests a functional link between these genes. Geisenberger et al. (11) also demonstrated that N-acyl-homoserine lactones are produced by CF isolates. It was subsequently demonstrated, in a chronic mouse infection model, that N-acyl-homoserine lactones could be produced in the mouse lung (47) and that mutations in the lasI and rhlI quorum-sensing systems resulted in milder lung infections (46).
Recently, Singh et al. (37) took a novel approach to address the question of quorum-sensing signals being produced by P. aeruginosa in CF. They developed an in situ assay in which a radioactive precursor is added to sputum and incubated. This allows the bacteria to synthesize newly labeled acyl-homoserine lactones, demonstrating that bacteria in sputum can produce autoinducers. They went on to show that bacteria in sputum could produce quorum-sensing signals in ratios indicative of the biofilm mode of growth (37).
A critical question that remains is whether or not autoinducers are produced in the lungs of patients with CF. In this study, our objective was to determine if autoinducers, produced in vivo, could be directly detected in sputum from patients with CF. We also wanted to determine if lasI transcripts are produced in the lungs of patients with CF and to see if their expression correlates with quorum-sensing target genes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All Escherichia coli and Pseudomonas strains were routinely grown at 37°C in PTSB (26) or Luria-Bertani (LB) broth supplemented with tetracycline (100 μg/ml) and carbenicillin (400 μg/ml) when appropriate. All liquid cultures were shaken at 270 rpm. P. aeruginosa CF isolates were identified to species level by the Microbiology Laboratory at the Foothills Hospital in Calgary. Of the 36 isolates used in the autoinducer screens, 6 were stable mucoid strains and the remainder were either spontaneous nonmucoid revertants or nonmucoid isolates.
Agrobacterium tumefaciens A136(pCF218)(pCF372) was grown in AT medium (44) with tetracycline (4.5 μg/ml) and spectinomycin (50 μg/ml) at 30°C. A. tumefaciens strain A136 has no Ti plasmid (no autoinducer synthase) and contains pCF218 (traR expression) and pCF372 (traI-lacZ), allowing the sensitive detection of all acyl-homoserine lactone autoinducers except those with C4-substituted acyl side chains (48).
Plasmids were transformed into P. aeruginosa by electroporation (38). The reporter strains used in this study have been described elsewhere (8, 29, 31). P. aeruginosa strain PAO-JP2 is a lasI rhlI double mutant, which produces no acyl-homoserine lactone autoinducers. Plasmid pKDT17 contains a copy of lasR under the tac promoter, as well as the lasB promoter fused to lacZ, thereby enabling the detection of 3-oxo-C12-HSL. Plasmid pECP61.5 contains rhlR under the tac promoter, as well as an rhlA′-lacZ fusion, enabling the detection of C4-HSL.
Patient population.
Initially the same group of 23 patients as previously described (39, 40, 41) was used in this study. This group was composed of 13 females and 10 males whose median age at the start of the study was 15.5 years. Of these 23 patients a subset of 16 patients were selected for additional study. This subset consisted of seven females and nine males whose age varied from 10 to 19 years. The median age at the beginning of the study for this group was 13 years. For both groups, patients were selected so that there would be an equal representation of mild, moderate, and severe pulmonary disease.
Patients defined as suffering from mild pulmonary disease had a forced expiratory volume in 1 s (FEV1) of >70% of expected values. Those patients classified as having moderate pulmonary disease had an FEV1 which fell in the range between >40% and <70% of predicted values. Severe cases of pulmonary disease consisted of patients whose FEV1 was <40% of expected values. All of the patients selected were receiving treatment at the Alberta Children's Hospital Cystic Fibrosis Clinic. CF isolates were obtained from patients attending either the University of Calgary Medical Clinic or the Adult Cystic Fibrosis Clinic at the Foothills Hospital. Voluntary consent was given by all patients and/or their guardians for participation in these studies. Approval of the design and purpose of this study was granted by the Conjoint Research Ethics Board of the University of Calgary.
Sputum collection and storage.
Sputum samples were obtained during each clinic visit of patients involved in the study. These samples were treated with an equal volume of Sputalysin (Calbiochem-Behring, Horescht) and divided into two portions. The first of the two portions was used for extraction of total RNA and autoinducers. The other portion was used to identify and quantify the bacteria present in the samples.
Extraction of autoinducers from sputum samples and bacterial culture supernatants.
Autoinducers were extracted from sputum samples with 1:2 (vol/vol) dichloromethane/volume of sputum sample. Samples were extracted three times for 1 h at 4°C. The organic phase was collected via centrifugation (500 × g, 1 min, 4°C), and the three extracts were combined. Aqueous residue was removed after freezing the samples for 1 h at −20°C. The solvent was removed via rotary evaporation, and the residue was resuspended in 200 μl of ethyl acetate. The extracts were stored at −20°C. To judge the efficiency of our extractions, we added synthetic C4- or 3-oxo-C12-HSL to separate samples that contained no detectable autoinducers and extracted them as above. For thin-layer chromatographic analysis, sputum samples from a single patient were combined so that the volume (after Sputalysin treatment) was 30 ml, and this volume was extracted with dichloromethane and resuspended in 20 μl of ethyl acetate. Cell-free bacterial supernatants (20 ml) from early-stationary-phase cultures (10 h) were extracted in the same manner and resuspended in 200 μl of ethyl acetate.
Autoinducer bioassays.
E. coli strain MG4(pKDT17) or P. aeruginosa strain PAO-JP2(pECP61.5) was used in liquid assays to measure the autoinducer levels in sputum extracts and culture supernatants as described previously (29, 31). The pKDT17 reporter plasmid in E. coli is reasonably specific for 3-oxo-C12-HSL. It contains a copy of the lasR gene as well as a lasB-lacZ fusion, so that exogenous autoinducer results in induction of β-galactosidase. pECP61.5 was used in P. aeruginosa PAO-JP2 because it resulted in greater sensitivity while remaining relatively specific for C4-HSL. pECP61.5 contains a copy of rhlR and an rhlA-lacZ fusion. β-Galactosidase activity was calculated as described by Miller (23). Individual sputum extracts were divided into two 100-μl aliquots, dried under cold air, and assayed for 3-oxo-C12-HSL or C4-HSL. A P. aeruginosa strain PAO1 culture supernatant (50 μl of a consistent sample) was included in each separate assay in order to standardize the results. A subset of samples were analyzed quantitatively by including synthetic standard curves for 3-oxo-C12-HSL or C4-HSL (Aurora Biosciences) and calculating the concentrations in sputum using the nanomoles of acyl-homoserine lactones detected and correcting for the original volume of the sputum sample.
To rapidly screen P. aeruginosa CF isolates for autoinducer production, a plate assay was used in which CF isolates and E. coli strain MG4 containing either pKDT17 or pECP61.5 were streaked 1.0 cm apart on MacConkey agar plates, and induced β-galactosidase activity in the reporter strain was scored visually (purple color on MacConkey agar).
Thin-layer chromatography.
Reverse-phase thin-layer chromatography as described by Shaw et al. (36) was used to examine the individual acyl-homoserine lactones present in our samples. Sputum sample extracts (20 μl) along with a series of culture supernatants (10 μl) and synthetic standards (Fluka; 3 nmol each) were spotted onto a C18 reverse-phase thin-layer chromatography plate (20 cm by 20 cm; Whatman) and developed with 60:40 (vol/vol) methanol-water. Then 30 ml of an A. tumefaciens strain A136(pCF218)(pCF372) late-exponential-phase culture was added to 150 ml of AT agar (0.7%) with 5-chloro-4-bromo-3-indolyl-β-d-galactopyranoside (X-Gal) at 60 μg/ml and poured over the plate, which was then incubated at 30°C for 24 h.
Isolation and quantification of total RNA from sputum samples.
Extraction, slot blotting of total RNA, hybridization of [32P]dCTP-labeled DNA probes, autoradiography, and subsequent quantitation of signal intensity were performed as previously described (40). The probes used to detect transcript accumulation of algD, lasA, lasB, lasR, and toxA have been described previously (39). For lasI, a 700-bp internal SalI fragment from plasmid pJMC31 (28) was used. This fragment does not contain any of the rsaL gene (2). Probes were labeled with [32P]dCTP using an oligolabeling kit (Pharmacia) in accordance with the manufacturer's specifications.
Positive control RNA was incorporated into each blot of CF sputum samples. For positive control for the lasI transcript, P. aeruginosa strain PAO1 was grown in PTSB and samples were extracted at an optical density at 540 nm of 1.0, 2.5, and 3.0. Negative control mRNA was extracted from P. aeruginosa strain PAO-JP2 at the same optical density. In each case RNA was extracted from 2 × 108 cells as previously described (6, 7).
The half-life of the lasI transcript was determined by growing P. aeruginosa strain PAO1 in PTSB to an optical density (540 nm) of 2.5. Rifampin was then added to the culture to a final concentration of 200 μg/ml. An aliquot of cells was removed for subsequent RNA extraction (time zero). Additional samples of the culture were removed at 10-min intervals. The final sample was removed 40 min after the addition of rifampin. RNA from each sample was immediately extracted upon its removal from the parent culture, as outlined previously (6). The RNA collected from these extractions was then electrophoresed on a glyoxal gel, blotted onto a Nytran membrane (Schleicher and Schuell), probed with the [32P]dCTP-labeled lasI probe, autoradiographed, and scanned for signal intensity using a scanning laser densitometer (Biomed Instruments) (39, 40, 41). The relationships between expression of lasI and the other transcripts in the study were analyzed using Spearman rank correlations as described previously (41).
RESULTS
P. aeruginosa lasI transcript can be detected in sputum from CF patients and its accumulation correlates with toxA, lasB, lasA, and lasR transcript accumulation
In order to assess lasI transcription in relation to transcription of other genes, differences in the half-lives of the mRNAs must be taken into account. Using RNA extracted from P. aeruginosa strain PAO1 at 10-min intervals after the addition of rifampin, the half-life of lasI was determined to be 18 min (data not shown). Half-lives for toxA (10 min), algD (19 min), lasB (11 min), lasA (8.5 min), and lasR (2 min) have been determined previously (39, 40, 41). A relatively long half-life of 18 min for lasI suggested that obtaining valid relationships for these genes would be possible. Thus, we wanted to determine whether the lasI gene is coordinately expressed with the other quorum-sensing genes in CF lung infections.
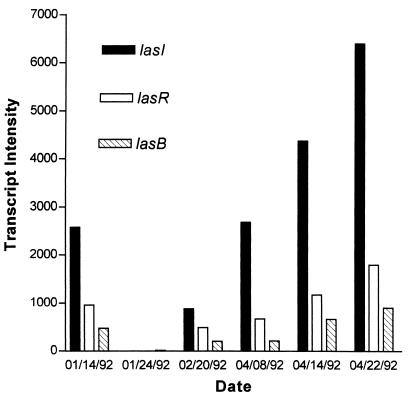
We examined transcript accumulation in samples that contained ≈108 bacteria/ml, taken from a single patient (patient 15) over a 4-month period (Fig. 1). Although we saw wide differences in the levels of transcript accumulation, it appeared that lasI transcript accumulation correlated with the transcript accumulation of lasR and lasB. Thus, further analysis on samples from additional patients would prove useful.
FIG. 1.
Transcript accumulation of lasB, lasR, and lasI in a male pediatric CF patient with severe lung disease (patient 15). The figure displays the relationships between transcript accumulation of lasB, lasR, and lasI. Total RNA was obtained from equivalent numbers of bacteria in the sputum samples, and 10 μg was hybridized with the lasB probe. The lasB probe was then stripped from the membrane, and the procedure was repeated for the lasR and lasI probes. Intensities of the hybridized probes on the autoradiographs were measured using a soft laser scanning densitometer.
We monitored transcript accumulation of lasR, lasI, toxA, lasB, lasA, and algD in a number of patients over a 3- to 5-year period. We detected lasI transcripts in 101 of 135 samples. The levels of lasI transcript accumulation varied from background to levels above the maximal level of expression of lasI in strain PAO1 grown under optimal conditions for expression of lasI. Spearman rank correlations were performed between accumulations of lasI and the aforementioned genes using a number of RNA samples extracted from the sputum of 16 different patients (Table 1). Table 1 shows a subset of our data where P. aeruginosa is the predominant pathogen or where P. aeruginosa is the sole pathogen in the sample. We found statistically significant correlations between lasI and lasR, lasA, lasB, and toxA transcript accumulation. A weak relationship occurred between lasI and algD. However, the level of significance was below our cutoff for these studies.
TABLE 1.
Statistical analysis of associations between the population transcript accumulations for lasA, lasB, lasR, toxA, algD, and lasI during the chronic lung infections associated with CFa
| Comparison |
P. aeruginosa and other species (n = 46)
|
P. aeruginosa onlya (n = 18)
|
||
|---|---|---|---|---|
| rsb | P | rs | P | |
| lasR-lasI | 0.763 | <0.001 | 0.739 | <0.001 |
| lasB-lasI | 0.696 | <0.001 | 0.893 | <0.001 |
| lasA-lasI | 0.661 | <0.001 | 0.774 | <0.001 |
| toxA-lasI | 0.674 | <0.001 | 0.795 | <0.001 |
| algD-lasI | 0.368 | <0.02 | 0.313 | <0.25 |
Samples contained equivalent numbers (108 bacteria/ml) of P. aeruginosa and no other pathogenic strains.
rs, Spearman rank correlation coefficient.
The strong correlations that exist between transcript accumulation of lasI and toxA, lasB, lasA, and lasR suggest that lasI may be involved in the regulation of or be coordinately regulated with the expression of the aforementioned genes in the CF lung. Detection of lasI transcripts and correlation of those transcripts with target genes regulated by the LasI-LasR quorum-sensing system suggested the possibility that autoinducers were being produced during the lung infections associated with CF.
Autoinducers are present in CF patient sputum.
In order to assess the quorum-sensing activity of P. aeruginosa in CF lung infections, we extracted autoinducers from patient sputum samples and assayed these extracts for the ability to induce β-galactosidase activity in the reporter plasmids pKDT17 (detects 3-oxo-C12-HSL) and pECP61.5 (detects C4-HSL). The volume of sputum in our samples was variable. Therefore, we were careful to first calculate our results on the basis of 1 ml of the original sputum sample. pKDT17 induction (3-oxo-C12-HSL) occurred more often (78%) than did pECP61.5 induction (C4-HSL) (26%) in assays of these patient samples. Table 2 displays the results from a large number of samples that contained mixed bacterial populations. Clearly, biologically active autoinducers can be found in the majority of these samples. Curiously, in some samples we did not detect C4-HSL and found only background levels of 3-oxo-C12-HSL.
TABLE 2.
3-Oxo-C12 and C4-HSL detectable in CF sputum samplesa
| Sample no. | β-Galactosidase (Miller units)
|
|
|---|---|---|
| 3-Oxo-C12-HSL | C4-HSL | |
| 8745 | 2,208 | 744 |
| 12622 | 3,645 | ND |
| 38052 | 5,253 | ND |
| 15727 | 1,761 | ND |
| 15085 | 5,793 | 462 |
| 46356 | 1,626 | 196 |
| 44520 | 237 | ND |
| 7135 | 43 | ND |
| 11732 | 40 | 92 |
| 8524 | 32 | 2 |
| 8712 | 30 | 152 |
| 41860 | 31 | 86 |
| 41511 | 36 | ND |
| 38028 | 638 | 0 |
| 42872 | 763 | 0 |
| 49819 | 2,033 | 0 |
| 53656 | 230 | 1,500 |
| 59934 | 951 | 0 |
| 41845 | 328 | 0 |
| 44293 | 635 | 0 |
| 49818 | 381 | 86 |
| 40584 | 171 | ND |
| 5392b | 3,361 | 618 |
| 8762b | 4,493 | 379 |
| 11947b | 1,596 | 0 |
| 8523b | 555 | 0 |
| 5195b | 315 | 45 |
| 15765c | 2,401 | 0 |
| PAO1d | 1,076 | 411 |
β-Galactosidase values were obtained by using the reporter plasmid pKDT17 (lasB-lacZ) to detect 3-oxo-C12-HSL and pECP61.5 (rhlA-lacZ) to detect C4-HSL. ND, value not determined due to insufficient sample volume.
Samples contained P. aeruginosa (108 CFU/ml) and no other pathogenic species.
Sample contained P. aeruginosa (106 CFU/ml) and no other pathogenic species.
Activity in 50 μl of a spent culture supernatant grown to an OD540 of 2.0.
A subset of our sputum samples contained only P. aeruginosa and no other pathogenic species. Of these samples, 5392, 8762, 11947, 8523, and 5195 all contained 108 cells/ml. Sample 15765 contained 106 cells/ml. The samples with 108 bacteria/ml displayed highly variable levels of autoinducer, and the sample with 106/ml contained higher levels than some of the samples with 108/ml. This shows that autoinducer levels in our samples are not merely a function of the number of bacteria. These results also demonstrate that P. aeruginosa must be responsible for the biologically active autoinducers detected in these samples.
We measured the levels of 3-oxo-C12-HSL and C4-HSL in a subset of samples using a comparison to synthetic standards and correcting for the original sputum sample volume (Table 3). These patient samples also showed variability in the observed autoinducer concentrations. Generally, 3-oxo-C12-HSL levels were higher (less than 1 to 22 nM) than C4-HSL (less than 1 to approximately 5 nM) (Table 3). These results also support the hypothesis that biologically active autoinducers can be produced in the lungs of patients with P. aeruginosa infections but that the overall concentrations in these sputum samples were low.
TABLE 3.
C4-HSL and 3-oxo-C12-HSL in sputum samplesa
| Sample no. | Concn (nM)
|
|
|---|---|---|
| C4-HSL | 3-Oxo-C12-HSL | |
| 330 | 0.00 | 1.86 |
| 342 | 0.00 | 1.39 |
| 351 | 5.00 | 14.40 |
| 360 | 1.13 | 1.64 |
| 361 | 2.61 | 2.15 |
| 362 | 0.84 | 21.20 |
| 363 | 0.00 | 1.06 |
| 364 | 1.84 | 0.00 |
| 365 | 1.00 | 0.00 |
| 366 | 0.00 | 14.70 |
| 11938 | 0.00 | 1.13 |
| 13165 | 0.00 | 0.92 |
Concentrations were determined by comparing β-galactosidase levels obtained from known amounts of synthetic 3-oxo-C12-HSL or C4-HSL using reporter plasmids pKDT17 and pECP61.5.
The low levels of autoinducers detected in the sputum samples from CF patients were surprising, given the correlation between lasI and lasR transcript accumulation and that of the genes, such as lasB, that are regulated by the quorum-sensing system. To determine if we were losing autoinducers during our extraction process, control experiments were performed in which we added synthetic autoinducer to sputum samples that contained no autoinducer. After extraction, we recovered 80% of the 3-oxo-C12-HSL and 60 to 65% of the C4-HSL added to these samples (data not shown). This experiment suggested that we were losing some autoinducers in the extraction process but that we should still be able to detect the majority of the autoinducer in the sample.
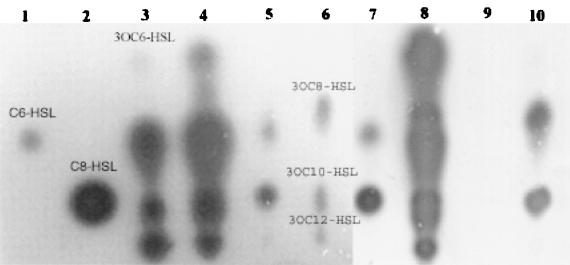
It is possible that the measured concentrations were skewed by the presence of other acyl-homoserine lactones. The reporter plasmids pKDT17 and pECP61.5 are relatively specific for 3-oxo-C12-HSL and C4-HSL, respectively, but will also respond to high levels of some noncognate autoinducers (31). We used thin-layer chromatography and an Agrobacterium reporter strain to address this question. If large amounts of acyl-homoserine lactones other than 3-oxo-C12-HSL or C4-HSL were present in these samples, then we could detect them via this method. We extracted larger volumes (15 ml) of sputum pooled from individual patients. The result from one such sample is shown in Fig. 2, along with culture supernatant extracts from P. aeruginosa strain PAO1 and P. aeruginosa CF isolates 4384, 6106, and 14655.
FIG. 2.
Thin-layer chromatography of spent culture supernatant and patient sputum extracts. Samples were applied to a reverse-phase C18 plate and developed as outlined, and autoinducers were detected by overlaying with a suspension of A. tumefaciens A136(pCF218)(pCF372). Samples in lanes 1 to 10 are C6-HSL (3 nmol), C8-HSL (3 nmol), P. aeruginosa PAO1 supernatant, P. aeruginosa CF isolate 4384 supernatant, P. aeruginosa CF isolate 14655 supernatant, patient 3 sputum extract, B. cepacia K56-2 supernatant, P. aeruginosa CF isolate 4384 supernatant, P. aeruginosa CF isolate 6106 supernatant, and P. aeruginosa CF isolate 14655 supernatant, respectively. Putative identification of spots in samples (from bottom to top): for PAO1 and 4384, 3-oxo-C12-HSL, 3-oxo-C10-HSL, 3-oxo-C8-HSL, and 3-oxo-C6-HSL; for 14655, C8-HSL and 3-oxo-C8-HSL; for sputum samples, 3-oxo-C12-HSL, 3-oxo-C10-HSL, and 3-oxo-C8-HSL; for B. cepacia, K56-2, C8-HSL and C6-HSL.
Three different structures were visible in the sputum sample. These were tentatively identified as 3-oxo-C12-HSL, 3-oxo-C10-HSL, and 3-oxo-C8-HSL based on their Rf values and their teardrop shape, characteristic of 3-oxo derivatives of acyl-homoserine lactones. We repeated these experiments on other sputum samples and generated similar patterns using thin-layer chromatography. Since high levels of other acyl-homoserine lactones were not present in any of our extracts, we are confident that the activity we detected from the pKDT17 and pECP61.5 reporters was due to the presence of 3-oxo-C12 and C4-HSL. The possibility still exists that different acyl-homoserine lactone autoinducers are produced by P. aeruginosa in the CF lung environment, and we are currently examining extracts from multiple patient samples via high-pressure liquid chromatography and mass spectroscopy to address this issue.
Production of autoinducers by P. aeruginosa CF isolates.
An explanation for the low levels of 3-oxo-C12-HSL and C4-HSL produced in the lungs could be variation in the levels of autoinducers produced by CF isolates. Therefore, autoinducer production by CF isolates was scored visually by induction of β-galactosidase in E. coli MG4 carrying the reporter plasmid pKDT17 (responds best to 3-oxo-C12-HSL) or pECP61.5 (responds best to C4-HSL) when plated together on MacConkey agar. Of the 36 isolates tested, 27 were positive for the induction of pKDT17, 25 for pECP61.5, and 22 for both. Five of the 36 isolates did not induce either. Autoinducer production by a portion of these isolates was also measured after growth in liquid medium. pKDT17 and pECP61.5 induction varied considerably among the strains tested, ranging from undetectable to nearly twice the activity induced by the P. aeruginosa laboratory strain PAO1. These results suggested that the majority of CF isolates have the capacity to produce autoinducers but that some do not produce any autoinducer.
We wanted to determine if acyl-homoserine lactones other than 3-oxo-C12-HSL and C4-HSL were produced in vitro by P. aeruginosa CF isolates. We assayed our samples via thin-layer chromatography in conjunction with an A. tumefaciens reporter strain. CF isolate 4384 has been shown to produce large amounts of 3-oxo-C12-HSL and C4-HSL, while strain 14655 produced little or none. As shown in Fig. 2, the autoinducers that could be detected from these strains varied in intensity and in pattern. Strain 4384 had an autoinducer profile similar to that of strain PAO1, as did the majority of isolates we tested (data not shown). However, strain 14655 produced predominantly C8-HSL. Interestingly, another CF pathogen, Burkholderia cepacia strain K56-2, also produces C8-HSL (22). Supernatants from strain 14655 were not able to induce β-galactosidase in pKDT17 or pECP61.5. Taken together, these results suggest that P. aeruginosa CF isolates have the ability to produce autoinducers but that differences exist between strains as to the types and amounts of autoinducers produced.
DISCUSSION
Quorum-sensing systems are believed to be involved in the interactions between many pathogenic bacteria and their hosts. Consequently, numerous approaches have been explored aiming to interfere with these signaling processes, including autoinducer-degrading enzymes (4), inhibition of the autoinducer biosynthetic pathway (14), and use of autoinducer structural analogs (19, 48). Few studies have examined directly whether quorum-sensing systems are active in human infections. This prompted us to examine the role of P. aeruginosa quorum-sensing systems in the chronic lung infections associated with CF. Our working hypothesis is that the quorum-sensing systems are active in the lungs of patients with CF and control the expression of important virulence factors. Previously we have shown that lasR is transcribed and coordinately regulated with several virulence genes in the sputum of infected patients (41). In the present study, we focused specifically on the coordinate regulation of the autoinducer synthase gene lasI with lasR, toxA, lasA, and lasB and the production of autoinducers in the lungs of patients with CF.
In this study we have made several important observations. First, lasI (the gene encoding the 3-oxo-C12-HSL synthase) is transcribed by P. aeruginosa in the lungs of patients with CF. Second, transcript accumulation of lasI correlates with transcription of several genes regulated by the quorum-sensing system. This correlation is apparent in the samples from a single patient as well as from a group of patients over a period of years. Table 1 shows that these correlations are statistically significant in samples where P. aeruginosa is either the predominant organism or the sole pathogen. Third, we were able to detect biologically active 3-oxo-C12 HSL and C4-HSL in sputum samples. These autoinducers were produced during the lung infections rather than in situ. We also detected two additional molecules in sputum samples from CF patients, these being 3-oxo-C10 and 3-oxo-C8-HSL. Fourth, we were able to measure the concentrations of 3-oxo-C12-HSL and C4-HSL in sputum samples and found them to be lower than the concentration produced by strain PAO1 growing under conditions optimal for the production of autoinducers. Finally, we found that the majority of CF isolates produced both autoinducers. Interestingly, not all isolates produced both, and some produced neither C4- nor 3-oxo-C12-HSL.
Several considerations complicate studying the role of regulatory mechanisms such as quorum sensing in the virulence of P. aeruginosa in chronic CF lung infections. For instance, CF affects a wide range of individuals who differ in their genetic background and who also differ in the clinical presentation of their disease (49). Numerous strains of P. aeruginosa with potentially different phenotypes can be involved in infection, a situation made even more complex by a high frequency of hypermutable strains (27). Additionally, the bacteria may modify their activity in response to the CF lung environment (5). Traditionally, analysis of regulatory mechanisms in pathogenic bacteria involves deletion of the regulatory gene followed by examination of the mutant bacterium's activity during growth in the laboratory or in infection models. Our approach was to examine autoinducer production and to look for correlations between the accumulations of transcripts of various genes in the bacterial populations from sputum taken directly from infected patients. Thus, we are avoiding some of the disadvantages of studying a single isolate introduced into an animal model by looking at the activity of the organisms that actually cause the infection.
According to the current model of quorum sensing in P. aerguinosa, autoinducers produced by bacteria accumulate as the cell density grows. At a threshold concentration, autoinducer binding to the transcriptional activator triggers transcription of target genes. Therefore, functional quorum sensing requires transcription of the autoinducer synthase (lasI) and response regulator (lasR) and production of autoinducer. Previous research has suggested that CF isolates have the ability to produce autoinducers both in the laboratory and in situ in sputum (11, 37). Furthermore, the correlation of lasR transcript accumulation to two target genes (lasA and lasB) of the quorum-sensing system suggests that quorum sensing may be functional in the lungs of patients with CF (41). The detection of lasI transcripts in the bacterial populations taken directly from sputum and the correlation of lasI transcript accumulation to those of lasR, lasA, and lasB (Table 1) add further support to the concept that the P. aeruginosa quorum-sensing system is operational and is coordinating gene expression in the lungs of patients with CF.
We needed to determine whether autoinducers were produced in the lungs of patients with CF. We detected functional C4- and C12-HSL directly from patient samples, providing further evidence that the quorum-sensing systems are regulating virulence in these infections (Tables 2 and 3). However, we had some unexpected findings in these studies. Many of the patient samples assessed for autoinducers had previously been or were concurrently analyzed for lasR, lasI, lasA, and lasB transcript accumulation. We expected to observe a correlation of these transcript levels with autoinducer in sputum samples. No such correlation could be observed (data not shown). This may be due to differences in the persistence of autoinducer and bacterial mRNA in patient samples. Variability is likely, given the chronic nature of these infections in a highly selective environment. Changes in the LasR sequence could result in different affinities for 3-oxo-C12-HSL, and consequently the same autoinducer levels may influence transcription in different ways. In addition, the concentrations of autoinducer that we measured in our samples are an average for the entire sample and certainly do not reflect the amounts that individual cells might be exposed to, especially within a microcolony or biofilm environment.
The low concentrations of 3-oxo-C12 and C4-HSL that we observed in patient samples were also surprising. However, a number of factors could account for the low levels of autoinducers. First, we may be underestimating the autoinducer concentration in our samples because our efficiency of extraction is less than 100% (80% for 3-oxo-C12HSL and 60% for C4-HSL) and because the acyl-HSLs may be unstable in this environment. Another factor could be that bacteria in the lung likely exist in microcolonies or biofilms and are not growing homogeneously throughout the lung. Furthermore, the bacteria are most likely growing in stationary phase in a somewhat hostile environment. In contrast, most studies on quorum sensing have been done on rapidly growing cells in nutrient-rich culture media. Bacteria in well-aerated planktonic cultures are believed to be more or less uniform in their gene expression and production of autoinducers, and therefore high levels of autoinducer accumulate at high cell density. However, considerable heterogeneity exists within biofilm communities. Pockets of high autoinducer levels may exist but would not be detected using our methods, as they would be diluted out in sputum samples. This is supported by the findings of Charlton et al. (1), who showed that autoinducers accumulate at very high levels in biofilms but that the effluent has low levels of autoinducers.
Another explanation for the low level of autoinducers in sputum could be differential expression of genes when P. aeruginosa is growing as a biofilm in the lungs of patients with CF. De Kievit et al. (3) recently examined expression of lasI and rhlI in P. aeruginosa biofilms using unstable green fluorescent protein fusions. They showed differential expression of these genes, especially lasI, where cells closest to the substratum exhibit the highest activity. Additionally, expression of lasI is initially high but decreases over 8 days of biofilm development, whereas rhlI expression occurs in a lower percentage of cells and is more stable. Our data indicate higher levels of C12- than C4-HSL in sputum, in agreement with these results. As well, fluctuations in lasI expression during biofilm development match the high variability in lasI and C12-HSL levels from sample to sample that we observed in sputum.
Another aspect of differential expression could occur at the cellular level, since P. aeruginosa CF isolates can vary in their phenotypes, and the CF lung environment can select for traits different from the those of the common laboratory strains. We have shown that, under laboratory conditions, 75% of CF isolates tested produce autoinducers that activate the reporter plasmid pKDT17 (most sensitive to 3-oxo-C12-HSL), 69% produce autoinducers that activate pECP61.5 (C4-HSL), and 61% produced both. Thus, not all strains produce both 3-oxo-C12-HSL and C4-HSL, and some strains produce neither. Geisenberger et al. (11) have also observed that P. aeruginosa isolates from chronic lung infections associated with CF also produce acyl-homoserine lactones. Thus, in the lungs of patients with CF, not all the strains may be producing the autoinducers, but the vast majority may be able to respond to them.
The low levels of C4-HSL detected were also unexpected, especially since Singh et al. (37) had demonstrated higher levels of C4-HSL than 3-oxo-C12-HSL in a biofilm and in an in situ sputum assay. The significance of reduced C4-HSL accumulation in sputum is not clear at this time, but it may suggest that particularly tight regulatory control over genes activated by RhlR is necessary in the CF lung environment. Alternatively, C4-HSL levels may be lower due to differences in how the signals are cleared from the lung. If 3-oxo-C12-HSL but not C4-HSL requires active efflux out of the bacteria (32), then it also may be true that C4-HSL diffuses into lung tissue more readily than 3-oxo-C12-HSL. This might result in higher accumulation of 3-oxo-C12-HSL in sputum.
Finally, antibiotic treatment and expression of efflux pumps may affect the ratio of C12- to C4-HSL in sputum. Kohler et al. recently observed that overexpression of MexEF-OprN results in decreased rhlI and C4-HSL levels but not C12-HSL levels (21). Alterations in two efflux systems, MexCD-OprJ and MexEF-OprN, may be the predominant mechanism of fluoroquinolone resistance in P. aeruginosa strains from the lungs of CF patients. A high percentage of CF strains have mutations that result in high expression of these pumps (17). Thus, in the lung, low levels of C4-HSL might be due to high expression of efflux pumps, especially the MexEF-OprN pump.
Another factor that might explain the low concentrations of autoinducers in sputum is that higher autoinducer levels may result in deleterious consequences to the host. This effect could be due to increased transcription of virulence genes and consequent tissue damage resulting from their action, or could be the result of host response to these products. In addition to their role in activating expression of quorum-sensing-controlled genes, autoinducers have also been shown to have direct effects on human cells (35, 43). Therefore, the autoinducer molecule in and of itself may have direct effects on patient status. It is possible that the severity of a patient's lung disease might correlate with lasR/lasI transcription and autoinducer levels.
The observation that some CF isolates may produce structurally different autoinducers than those produced by strain P. aeruginosa PAO1 (Fig. 2) suggests sequence variation in their luxI homologues. Another CF-associated pathogen, B. cepacia, has recently been shown to exhibit quorum-sensing activity and produces predominantly N-octanoyl-l-homoserine lactone (22). Cocolonization with B. cepacia and P. aeruginosa frequently worsens the prognosis for the patient (12). It is possible that the production and use of autoinducers similar to those in B. cepacia by such strains as P. aeruginosa CF isolate 14655 may result in cross-species communication.
The low level of autoinducers in most samples, the lack of correlation of autoinducers with cell density, and the lack of correlation between the autoinducers and transcript accumulation seem at odds with our significant correlations between lasI, lasR, and the target genes of this quorum-sensing system. Recently, Nilsson et al. (24) developed a model for the kinetics of an acyl-homoserine lactone regulatory system in a biofilm. In this model, they speculate that even at low external concentrations of autoinducers, it may still be possible to get expression of acyl-homoserine lactone-regulated genes (24). Their idea is that in biofilms, slow growth rates coupled with the extracellular matrix that may be slowing or restricting diffusion of the autoinducers results in fast autoinduction. What we may be seeing in the lungs of patients with CF is slow-growing microcolonies or biofilms producing low external concentrations of autoinducers but intracellular concentrations of autoinducers that are high enough to cause induction of the quorum-sensing target genes.
In summary, we have shown that P. aeruginosa isolates from CF patients produce autoinducers when cultured under laboratory conditions and therefore have the potential to regulate gene expression via quorum-sensing mechanisms. As well, we have demonstrated that in human infections, functional autoinducers are produced and therefore could activate the transcription of quorum-sensing-controlled genes. Furthermore, the detection in sputum of biologically active autoinducers coupled with high levels of lasI and lasR transcript accumulation suggest induction of the LasR-LasI quorum-sensing system in the chronic lung infections associated with CF. Finally, the detection of statistically significant relationships between transcript accumulation levels of lasI, lasR, and the target genes in the bacterial population of sputum suggests that this quorum-sensing system may be regulating virulence factor expression.
Acknowledgments
This work was supported by funding from the Canadian Cystic Fibrosis Foundation to D.G.S. and D.L.E.
We thank Barbara Iglewski and Clay Fuqua for providing reporter strains and plasmids containing genes of interest and Pam Sokol and Michael Surette for critical review of the manuscript.
REFERENCES
- 1.Charlton, T. S., R. de Nys, A. Netting, N. Kumar, M. Hentzer, M. Givskov, and S. Kjelleberg. 2000. A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ. Microbiol. 2:530-541. [DOI] [PubMed] [Google Scholar]
- 2.de Kievit, T. R., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 6.Frank, D. W., and B. H. Iglewski. 1988. Kinetics of toxA and regA mRNA accumulation in Pseudomonas aeruginosa. J. Bacteriol. 170:4477-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, D. W., D. G. Storey, M. S. Hindahl, and B. H. Iglewski. 1989. Differential regulation by iron of regA and toxA transcript accumulation in Pseudomonas aeruginosa. J. Bacteriol. 171:5304-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuqua, W. C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 10.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisenberger, O., M. Givskov, K. Riedel, N. Hoiby, B. Tummler, and L. Eberl. 2000. Production of N-acyl-l-homoserine lactones by P. aeruginosa isolates from chronic lung infections associated with cystic fibrosis. FEMS Lett. 184:273-278. [DOI] [PubMed] [Google Scholar]
- 12.Govan, J. R. W. 1995. Burkholderia cepacia in cystic fibrosis. N. Engl. J. Med. 332:819-820. [PubMed] [Google Scholar]
- 13.Granstrom, M., A. Ericsson, B. Strandvik, B. Wretlind, O. R. Pavlovskis, R. Berka, and M. L. Vasil. 1984. Relation between antibody response to Pseudomonas aeruginosa exoproteins and colonization/infection in patients with cystic fibrosis. Acta Paediatr. Scand. 73:772-777. [DOI] [PubMed] [Google Scholar]
- 14.Hoang, T. T., and H. P. Schweizer. 1999. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollsing, A. E., M. Granstrom, M. L. Vasil, B. Wretlind, and B. Strandvik. 1987. Prospective study of serum antibodies to Pseudomonas aeruginosa exoproteins in cystic fibrosis. J. Clin. Microbiol. 25:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagger, K. S., D. L. Robinson, M. N. Franz, and R. L. Warren. 1982. Detection by enzyme-linked immunosorbent assays of antibody specific for Pseudomonas proteases and exotoxin A in sera from cystic fibrosis patients. J. Clin. Microbiol. 15:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jalal, S., O. Ciofu, N. Hoiby, N. Gotoh, and B. Wretlind. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleerebezem, M., L. E. Quadri, O. P. Kuipers, and W. M. deVos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 19.Kline, T., J. Bowman, B. H. Iglewski, T. de Kievit, Y. Kakai, and L. Passador. 1999. Novel synthetic analogs of the Pseudomonas autoinducer. Bioorg. Med. Chem. Lett. 9:3447-3452. [DOI] [PubMed] [Google Scholar]
- 20.Klinger, J. D., D. C. Straus, C. B. Hilton, and J. A. Bass. 1978. Antibodies to proteases and exotoxin A of Pseudomonas aeruginosa in patients with cystic fibrosis: demonstration by radioimmunoassay. J. Infect. Dis. 138:49-58. [DOI] [PubMed] [Google Scholar]
- 21.Kohler, T., D. C. Van, L. K. Curty, M. M. Hamzehpour, and J. C. Pechere. 2001. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183:5213-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1974. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Nilsson, P., A. Olofsson, M. Fagerlind, T. Fagerstrom, S. Rice, S. Kjelleberg, and P. Steinberg. 2001. Kinetics of the acyl-homoserine lactone regulatory system in a model biofilm system: how many bacteria constitute a “quorum”? J. Mol. Biol. 309:631-640. [DOI] [PubMed] [Google Scholar]
- 25.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1253. [DOI] [PubMed] [Google Scholar]
- 28.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbe Infect. 2:1721-1731. [DOI] [PubMed] [Google Scholar]
- 35.Saleh, A., C. Figarella, W. Kammouni, S. Marchand-Pinatel, A. Lazdunski, A. Tubul, P. Brun, and M. D. Merten. 1999. Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone inhibits expression of P2Y receptors in cystic fibrosis tracheal gland cells. Infect. Immun. 67:5076-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. J. Cronan, K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 38.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storey, D. G., E. E. Ujack, I. Mitchell, and H. R. Rabin. 1997. Positive correlation of algD transcription to lasB and lasA transcription by populations of Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Infect. Immun. 65:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey, D. G., E. E. Ujack, and H. R. Rabin. 1992. Population transcript accumulation of Pseudomonas aeruginosa exotoxin A and elastase in sputa from patients with cystic fibrosis. Infect. Immun. 60:4687-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storey, D. G., E. E. Ujack, H. R. Rabin, and I. Mitchell. 1998. Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect. Immun. 66:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Telford, G., D. Wheeler, P. Williams, P. T. Tomkins, P. Appleby, H. Sewell, G. S. Stewart, B. W. Bycroft, and D. I. Pritchard. 1998. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 66:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Témpe, J., A. Petit, M. van Montagu, and J. Schell. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: Possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. USA 74:2848-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, H., Z. Song, M. Givskov, G. Doring, D. Worlitzsch, K. Mathee, J. Rygaard, and N. Hoiby. 2001. Pseudomonas aeruginosa mutations in lasI and rhlI quorum-sensing systems result in milder chronic lung infection. Microbiology 147:1105-1113. [DOI] [PubMed] [Google Scholar]
- 47.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, A. Heydorn, K. Mathee, C. Moser, L. Eberl, S. Molin, N. Hoiby, and M. Givskov. 2000. Detection of N-acylhomoserine lactones in lung tissues of mice infected with Pseudomonas aeruginosa. Microbiology 146:2481-2493. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, J., J. W. Beaber, M. I. More, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zielenski, J., and L. C. Tsui. 1995. Cystic fibrosis: genotypic and phenotypic variations. Annu. Rev. Genet. 29:777-807. [DOI] [PubMed] [Google Scholar]