Abstract
Burkholderia cepacia is an important opportunistic human pathogen that affects immunocompromised individuals, particularly cystic fibrosis (CF) patients. Colonization of the lungs of a CF patient by B. cepacia can lead not only to a decline in respiratory function but also to an acute systemic infection, such as bacteremia. We have previously demonstrated that a CF clinical isolate of B. cepacia, strain J2315, can invade and survive within cultured respiratory epithelial cells. In order to further characterize the mechanisms of invasion of B. cepacia, we screened a transposon-generated mutant library of strain J2315 for mutants defective in invasion of A549 respiratory epithelial cells. Here we describe isolation and characterization of a nonmotile mutant of B. cepacia with reduced invasiveness due to disruption of fliG, which encodes a component of the motor-switch complex of the flagellar basal body. We also found that a defined null mutation in fliI, a gene encoding a highly conserved ATPase required for protein translocation via the flagellar type III secretion system, also resulted in loss of motility and a significant reduction in invasion. Both mutants lacked detectable intracellular flagellin and failed to export detectable amounts of flagellin into culture supernatants, suggesting that disruption of fliG and fliI impaired flagellar biogenesis. The reduction in invasion did not appear to be due to defective adherence of the flagellar mutants to A549 cells, suggesting that functional flagella and motility are required for full invasiveness of B. cepacia. Our findings indicate that flagellum-mediated motility may facilitate penetration of host epithelial barriers by B. cepacia, contributing to establishment of infection and systemic spread of the organism.
Over the last several decades Burkholderia cepacia has emerged as an important opportunistic human pathogen of the lower respiratory tract that affects immunocompromised individuals, particularly cystic fibrosis (CF) patients (18). Chronic colonization of the lungs of a CF patient by B. cepacia can lead not only to a decline in respiratory function, due to the onset of a necrotizing pneumonia, but also to an acute systemic infection, such as bacteremia or septicemia (11, 46). In addition to being an invasive pathogen that is capable of entering deeper tissues and becoming blood borne, B. cepacia can survive in epithelial cells and macrophages (4, 35, 42, 48), which may contribute to the persistence of the organism in the host. The rapid clinical decline due to B. cepacia colonization is known as cepacia syndrome, and this decline leads to mortality in approximately 20 to 35% of chronically colonized individuals (25, 47). Furthermore, colonization by B. cepacia reduces the life expectancy of a CF patient by 50%, from 30 to 15 years (24). The inherent resistance of B. cepacia to multiple antibiotics makes eradication of this pathogen from the lungs of infected individuals especially difficult. Despite the known association of B. cepacia with fatal pulmonary infections in CF patients, the pathogenic mechanisms and virulence determinants responsible for B. cepacia infection are only beginning to be elucidated (38).
B. cepacia currently encompasses at least six genotypically distinguishable genomovars (genomovars I to VI), which are collectively known as the B. cepacia complex (7, 50). Recently, genomovars II, IV, and V have been reclassified into distinct species (Burkholderia multivorans, Burkholderia stabilis, and Burkholderia vietnamiensis, respectively) (50, 51). While strains representing all six genomovars of B. cepacia have been associated with opportunistic infections in humans, strains belonging to genomovar III have been most commonly linked to epidemic outbreaks and fatal infections in CF patients (50, 51).
B. cepacia is a motile organism, and motility is mediated by polar flagella (3, 21). Little is known about the structure and function of B. cepacia flagella, although attempts to type strains based on the elaborated flagellin protein have been made. Two major flagellin types were detected in a survey of B. cepacia strains; these types were distinguished on the basis of molecular mass (45 and 55 kDa) (20). The corresponding flagellar genes, designated fliC, encode proteins that exhibit high levels of homology to the flagellin of Burkholderia pseudomallei, another closely related human pathogen (9).
In Escherichia coli, as well as in other bacteria, expression of functional flagella requires more than 40 structural and regulatory genes, and biogenesis proceeds via a complex, hierarchical pathway (33). The extracellular components of the flagellum (namely, the hook and filament) are extended by addition of new structural subunits at the distal end of the growing organelle. The flagellar structural proteins are translocated from the cytoplasm into the growing structure via a highly conserved secretion system, designated the type III system (32). Components of the flagellar type III system exhibit high levels of homology to the components of type III secretion systems utilized by a number of bacterial pathogens to secrete virulence determinants into host cells (23).
It has been demonstrated previously that flagella and flagellum-mediated motility contribute to the virulence of a number of pathogenic bacterial species. Mutations in flagellar biogenesis genes have been shown to attenuate the virulence of several human pathogens, including Pseudomonas aeruginosa, Proteus mirabilis, Campylobacter jejuni, and Helicobacter pylori (12, 14, 37, 39, 40). In some bacteria, components of the flagellum, including the mucin-binding flagellar cap protein of P. aeruginosa (1) and flagellin proteins of C. jejuni (55), may act as adhesins and implicate flagella in facilitating colonization through direct interactions with host ligands. The adhesive properties of some flagella, however, do not fully account for the observed reduction in virulence of flagellar mutants, and active motility is often required for full pathogenesis. This hypothesis is supported by studies which showed that flagellar motility can enhance invasion of host cells by C. jejuni, P. mirabilis, Vibrio anguillarum, and other pathogenic species (37, 41, 55).
Several laboratories have demonstrated that B. cepacia is capable of invading and surviving within cultured respiratory epithelial cells (4, 28, 35, 48). B. cepacia invasion has also been observed in vivo in a murine model of infection (6). Together, these findings suggest that invasion may be an important virulence factor in the pathogenesis of B. cepacia. Invasion may be a mechanism by which B. cepacia breaches the epithelial barrier to enter deeper tissues and ultimately becomes blood borne, which results in systemic infection. We have previously shown that B. cepacia strain J2315, a clinical isolate associated with epidemic outbreaks and mortality in CF patients, can invade cultured human respiratory epithelial cells (35). In order to further characterize the mechanisms employed by B. cepacia to invade respiratory epithelial cells, we screened a transposon-generated mutant library of B. cepacia strain J2315 for mutants with reduced ability to enter A549 respiratory epithelial cells. Here we describe the isolation and characterization of a nonmotile mutant of B. cepacia defective in invasion due to disruption of fliG, which encodes a component of the motor-switch complex of the flagellar basal body. Additionally, we found that a defined null mutation in fliI, a gene encoding a highly conserved ATPase required for protein translocation via the flagellar type III secretion system, results in a loss of motility, which is coupled with a significant reduction in invasion of A549 respiratory epithelial cells. Our findings indicate that flagellum-mediated motility may play a role in the pathogenesis of B. cepacia by facilitating penetration of the host epithelial cell barriers and contributing to the onset of systemic spread of the organism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. E. coli strains were grown with aeration at 37°C in Luria-Bertani (LB) broth or on LB agar plates supplemented with ampicillin (100 μg/ml), tetracycline (12 μg/ml), or chloramphenicol (30 μg/ml) as necessary. B. cepacia strain J2315 is a CF clinical isolate belonging to B. cepacia genomovar III (17). Strain J2315 was grown with aeration at 37°C in LB broth or peptone-yeast extract (PYE) (13) supplemented with tetracycline (50 μg/ml) or chloramphenicol (30 μg/ml) or on LB agar plates supplemented with tetracycline (500 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (300 μg/ml) as necessary. For quantitative invasion and adherence assays B. cepacia was grown in PYE.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 lacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| S17-1 | Integrated RP4-2, Tc::Mu Km::Tn7 | 44 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lac1qZΔM15 Tn10) | Stratagene |
| B. cepacia strains | ||
| J2315 | CF clinical isolate, genomovar III | 17 |
| D9 | fliG::mini-Tn5Tc derivative of J2315 | This study |
| CM58 | fliI::cat derivative of J2315 | This study |
| CM100 | CM58 with plasmid pJG7 | This study |
| CM340 | D9 with plasmid pMT46 | This study |
| Plasmids | ||
| pBluescript SK(−) | Cloning and single-stranded phagemid | Stratagene |
| pBluescript SK(+) | Cloning and single-stranded phagemid | Stratagene |
| pGEM-T Easy | TA cloning vector | Promega |
| pLAFR-5 | Cosmid vector, Tcr | 27 |
| pCAT1 | Source of cat cassette, Cmr | This study |
| pMR4 | Broad-host-range vector, Tcr | C. Mohr and R. Roberts |
| pUCP18 | Broad-host-range vector, Apr | 52 |
| pCMT1 | Cmr derivative of pUCP18 | This study |
| pCM117 | pLAFR-5 cosmid containing 20-kb B. cepacia genomic DNA insert encoding the fliG locus | This study |
| pAR1 | 3.5-kb XhoI fragment from pCM117 subcloned into pBluescript SK(+) | This study |
| pAR2 | 1.3-kb PstI fragment containing fliI, subcloned from pAR1 into pBluescript SK(+) | This study |
| pAR3 | 1.3-kb XhoI-PstI fragment from pAR2 subcloned into pBluescript SK(+) | This study |
| pAR4 | pAR3 containing fliI gene inactivated with cat cassette, Cmr/Apr | This study |
| pJG7 | 1.6-kb PstI fragment encoding fliI truncated at codon 476, cloned in pMR4 | This study |
| pMT38 | 1.5-kb PCR product encoding fliG cloned into pGEM-T Easy | This study |
| pMT46 | 1.5-kb fliG PCR product cloned into pCMT1 | This study |
Transposon mutagenesis.
The mini-Tn5Tc transposon (8, 22) was introduced into B. cepacia strain J2315 by conjugation using E. coli donor strain S17-1 (44). For mating, 250-μl portions of an E. coli overnight culture and 500-μl portions of a B. cepacia overnight culture were mixed on sterile nitrocellulose filters. Following incubation of the filters on LB agar for 4 to 5 h at 37°C, the bacteria were resuspended in LB broth and plated on LB agar containing 300 μg of tetracycline per ml and 50 μg of kanamycin per ml to select against wild-type B. cepacia and the E. coli donor strain, respectively. Southern blot hybridization analysis of a subset of individual transposon-generated mutants indicated that the mini-Tn5Tc transposon inserted randomly into the B. cepacia chromosome.
Screening for invasion-defective mutants.
Individual transposon-generated mutants of B. cepacia were grown overnight in 200-μl portions of LB broth in 96-well microtiter plates, subcultured into new microtiter plates, and grown to the mid-exponential phase (A630, ∼0.4). Aliquots (20 μl) were used to infect confluent monolayers of A549 respiratory epithelial cells. The A549 cell line (American Type Culture Collection, Manassas, Va.) is a human alveolar epithelial carcinoma cell line. The A549 cells were seeded 24 h prior to infection in 96-well microtiter plates at a level of 8.8 × 104 cells per well. The bacteria were centrifuged onto the monolayers (165 × g for 5 min) and incubated at 37°C in an atmosphere containing 5% CO2 for 30 min to allow bacterial entry. Killing extracellular bacteria and quantification of intracellular bacteria were performed as previously described (35). Approximately 4,000 transposon-generated mutants were screened. The phenotypes of invasion-defective mutants identified in the primary invasion screening analysis were confirmed by quantitative invasion assays.
Quantitative invasion assays.
Invasion assays were performed essentially as described previously (35), with the following modifications. Bacterial strains were grown to the exponential phase in PYE, and where indicated below, the centrifugation step was omitted. The bacterial strains were allowed to invade for 2 h. All quantitative invasion assays were performed in triplicate wells, with two samplings per well.
Cosmid library construction.
Cosmid vector pLAFR-5 (27) was used to construct a cosmid library of B. cepacia strain J2315. Genomic DNA was extracted from strain J2315 with a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.) used according to the manufacturer's instructions. Approximately 3 μg of J2315 DNA was partially digested with Sau3A, and 20- to 40-kb fragments were eluted and combined with 1.5 μg of pLAFR-5 linearized with BamHI and ScaI in a 22-μl ligation mixture. Following overnight incubation at 14°C, 4 μl of the ligation mixture was packaged into bacteriophage lambda by using Gigapack III Plus packaging extract (Stratagene, La Jolla, Calif.). The resulting phage extract was transfected into E. coli XL-1 Blue. Approximately 2,100 cosmids (average insert size, ∼28 kb) were picked into 96-well plates, grown overnight, and replica plated onto LB agar. After overnight growth, the bacterial grids were replica plated onto nylon membranes (Hybond N; Amersham Pharmacia Biotech, Piscataway, N.J.). Dimethyl sulfoxide (10%) was added to each well of the 96-well plates, and the plates were stored at −70°C.
Nucleotide sequencing.
Nucleotide sequencing was performed at the Advanced Genetic Analysis Center at the University of Minnesota, using the dideoxy chain termination method and an ABI 1371A DNA sequencer (Applied Biosystems, Foster City, Calif.). The oligonucleotide primers used for sequencing were standard forward and reverse (T3 and T7) pBluescript primers or custom oligonucleotides synthesized by Integrated DNA Technologies (Coralville, Iowa). The nucleotide sequences of both strands were determined. Double-stranded sequences were aligned and assembled by using the EditSeq and SeqMan components of a demonstration version of the Lasergene sequence analysis software package (DNASTAR, Inc., Madison, Wis.). For nucleotide and amino acid sequence searches and analysis we utilized the BLASTX and BLASTP programs of the National Center for Biotechnology Information.
DNA manipulations.
DNA-modifying enzymes, including restriction endonucleases, T4 DNA ligase, and T4 polynucleotide kinase, were obtained from Roche (Indianapolis, Ind.), New England Biolabs (Beverly, Mass.), and Gibco BRL (Rockville, Md.). Plasmid DNA was isolated by the boiling lysis method (43) or with a QIAprep Spin Miniprep kit (Qiagen Inc., Valencia, Calif.). Recombinant plasmids were introduced into E. coli and B. cepacia strain J2315 by electroporation with a Gene Pulser II (Bio-Rad, Richmond, Calif.), as previously described by Burns and Hedin (5). Southern blot and colony hybridization analyses were generally performed as described by Sambrook et al. (43) using Hybond N nitrocellulose membranes, probes labeled with [α-32P]dCTP (Amersham Pharmacia Biotech), and the random primer method.
To clone the DNA flanking the site of the mini-Tn5Tc insertion in mutant D9, genomic DNA was digested with a series of restriction enzymes and examined by Southern blot hybridization analysis in which probes internal to the mini-Tn5Tc transposon were used. A 4-kb PstI cross-hybridizing fragment encoding mini-Tn5Tc and flanking DNA was identified and cloned by generating a subgenomic library of PstI fragments ranging from 3 to 6 kb long in pBluescript SK(+) and plating the library on LB agar containing 12 μg of tetracycline per ml to select for mini-Tn5Tc-encoded tetracycline resistance.
For complementation of B. cepacia mutant D9, the fliG gene was amplified by performing PCR with oligonucleotide primers fli18 (5′-CAAGGCGGCGGAGGAAC-3′) and fli19 (5′-GATGCGAGATCGTGTTCG-3′), a PCR Sprint thermocycler (Hybaid, Franklin, Mass.), and Taq DNA polymerase (Promega, Madison, Wis.). The PCR was carried out for 35 cycles consisting of 94°C for 1 min, 50°C for 1 min, and 72°C for 1.5 min, concluding with an additional 5 min of extension at 72°C after the 35 cycles. The 1.5-kb fliG PCR product, containing the 3′ end of fliF, fliG, and the 5′ end of fliH, was cloned into the pGEM-T Easy (Promega) TA cloning vector (pMT38) and subsequently excised as an EcoRI fragment and cloned into the pCMT1 broad-host-range vector to generate pMT46. Plasmid pMT46 was introduced into D9 by electroporation, which resulted in strain CM340.
A B. cepacia J2315 fliI mutant strain (CM58) was generated by allelic exchange mutagenesis. Briefly, the chloramphenicol resistance cassette (cat) encoded in plasmid pCAT1 was excised as an EcoRI fragment and inserted into the unique EcoRI site located in the fliI coding sequence on plasmid pAR3. The resulting construct, designated pAR4, was introduced into strain J2315 by electroporation, and the transformants were plated on LB agar containing 300 μg of chloramphenicol per ml. Chloramphenicol-resistant colonies were stabbed into swarm agar, and mutants whose motility was impaired were examined by Southern blot hybridization analysis to confirm that the cat cassette had integrated into the fliI gene and that a double-crossover event had occurred. For complementation of CM58, a 1.6-kb PstI fragment encoding amino acids 1 to 475 of the fliI gene product was cloned into the pMR4 broad-host-range vector, generating pJG7. The pJG7 plasmid was introduced into CM58 by filter mating, which resulted in strain CM100.
Motility assays.
For motility assays LB agar swarm plates containing 0.25% (wt/vol) agar were stab inoculated with overnight cultures of B. cepacia and incubated for 24 h at 37°C.
Immunoblot analysis.
Cytoplasmic and supernatant protein fractions were prepared from exponential-phase cultures as previously described (45). Briefly, B. cepacia strains were grown to the mid-exponential phase in 50 ml of PYE. For whole-cell protein analysis, 1 ml of each culture was harvested by centrifugation, and the bacterial pellets were resuspended in Laemmli buffer (30). Protein samples were boiled, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by immunoblotting. For supernatant analysis, 25-ml portions of mid-exponential-phase cultures were harvested, and the supernatants were filtered through a 0.45-μm-pore-size filter. Each supernatant protein fraction was precipitated overnight at 4°C by adding ammonium sulfate to a concentration of 50% (wt/vol). The precipitated protein samples were centrifuged at 10,000 rpm in a Sorvall SS-34 rotor for 30 min, and the pellets were washed with 70% ethanol, resuspended in Laemmli buffer, boiled, and analyzed by immunoblotting.
Immunoblotting on SDS-polyacrylamide gels was performed as described by Jenal et al. (26). The flagellin-specific rabbit polyclonal antiserum used was a generous gift from D. Woods and was raised against purified B. pseudomallei flagellin (2). Blots were probed with the primary flagellin-specific antiserum at a dilution of 1:10,000 and then with secondary antibody (goat anti-rabbit immunoglobulin G) at a 1:2,500 dilution. Western blots were developed with a Renaissance chemiluminescence kit (DuPont NEN, Boston, Mass.) used according to the manufacturer's instructions. The protein standards used were prestained SDS-PAGE low-range standards (Bio-Rad).
Adherence assays.
Mid-exponential-phase bacteria were added to confluent A549 monolayers at a multiplicity of infection of 10:1. The bacteria were centrifuged onto the monolayers (165 × g for 5 min) and incubated at 37°C in the presence of 5% CO2 for 1 h to allow bacterial adherence. The monolayers were washed five times with phosphate-buffered saline to remove nonadherent bacteria. The adherent bacteria were enumerated by lysing A549 cells with 0.25% Triton X-100 and determining the viable cell counts on LB agar. While this assay does not allow discrimination between adherent and intracellular bacteria, under these conditions the subpopulation of internalized bacteria accounts for approximately 5% of the total number of bacteria recovered from the assay (data not shown). Therefore, the vast majority of the CFU recovered in this assay represent adherent, noninternalized bacteria. All assays were performed in triplicate wells, with two samplings per well.
Statistical analysis.
The statistical significance of the observed differences in mean invasion frequencies was determined by calculating the P values using the two-tailed Student t test for unpaired data sets.
Nucleotide sequence accession number.
The nucleotide sequence of the fliG locus has been deposited in the GenBank database under accession no. AF453480.
RESULTS
Identification of a B. cepacia mutant defective in invasion.
We have previously shown that a CF clinical isolate of B. cepacia, strain J2315, is able to invade cultured A549 respiratory epithelial cells (35). In order to identify the genetic elements required for invasion, we developed a method to screen large numbers of transposon-generated mutants of B. cepacia strain J2315 for defects in the ability to invade A549 cell monolayers. Initially, we screened approximately 4,000 mutants, and the ability of one of these mutants, designated mutant D9, to enter A549 cells appeared to be reduced. In order to confirm the invasion defect, mutant D9 and wild-type B. cepacia strain J2315 were compared using quantitative invasion assays. The mutant D9 was consistently reduced in ability to invade A549 respiratory epithelial cells, with approximately 45% reduced cell invasion compared to the wild-type strain J2315 (Fig. 1A). These findings suggested that the transposon insertion in mutant D9 disrupted functions required for optimal invasion by B. cepacia.
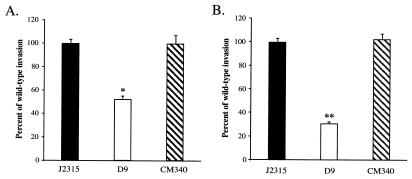
FIG. 1.
Invasion of A549 cell monolayers by B. cepacia strain J2315 (wild type), mutant D9, and D9 complemented with fliG (strain CM340). (A) Invasion of A549 cell monolayers with centrifugation. Quantitative invasion assays were performed in triplicate as described in Materials and Methods. The invasion values were calculated by determining the percentages of the bacterial inocula that survived after 2 h of antibiotic treatment. The values were normalized to the value for wild-type strain J2315, which was arbitrarily set at 100%. The actual invasion frequency for strain J2315 was 0.67% ± 0.03%. The asterisk indicates that the level of invasion by mutant D9 was significantly less than the level of invasion by the parent strain (P < 0.000002). (B) Invasion of A549 cell monolayers without centrifugation. The actual invasion frequency for strain J2315 without centrifugation was 0.31% ± 0.01%. The double asterisks indicate that the P value was <0.0000002.
Characterization of invasion-defective mutant D9.
Southern blot hybridization analysis in which mini-Tn5Tc DNA was used as a probe indicated that there was a single transposon insertion in invasion-defective mutant D9 (data not shown). In order to identify the transposon insertion site, DNA flanking the mini-Tn5Tc transposon was cloned and sequenced. Sequence analysis revealed that the site of Tn5 insertion in D9 disrupted an open reading frame homologous to the fliG gene family encoding components of the flagellar basal body (Fig. 2). The transposon insertion site was determined to be between codons 203 and 204 of the B. cepacia fliG gene. The B. cepacia fliG homolog encodes a putative polypeptide consisting of 331 amino acids, which is 57% identical and 76% similar to FliG of Salmonella enterica serovar Typhimurium (Fig. 2).
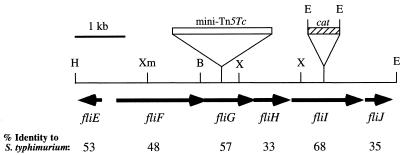
FIG. 2.
Physical map of the B. cepacia fli locus. The open box indicates the site of mini-Tn5Tc insertion in fliG mutant D9, and the cross-hatched box indicates the site of insertion of the cat cassette in fliI null strain CM58. The arrows indicate the directions of transcription. The numbers below the open reading frames indicate the levels of amino acid identity to the corresponding flagellar homologs in S. enterica serovar Typhimurium. Abbreviations: B, BamHI; E, EcoRI; H, HindIII; X, XhoI; Xm, XmaI.
In order to further delimit the extent of the fliG locus, a strain J2315 cosmid library was constructed and screened for clones encoding fliG. Subclones from a fliG cross-hybridizing cosmid were generated and sequenced. Additional genes with high levels of homology to known flagellar genes were identified. The 6.5-kb flagellar locus of B. cepacia, shown in Fig. 2, encodes at least six genes, including fliFGHIJ and fliE in divergent orientations. All of these genes encode homologs of early structural and regulatory components of the flagellar assembly pathway in E. coli and Salmonella spp. (Fig. 2). The FliF, FliG, FliH, and FliI proteins of S. enterica serovar Typhimurium are components of a flagellum-specific type III secretion system that is utilized for export and assembly of flagellar protein substrates (23). The fliE gene encodes a structural component of the flagellar basal body (36), while it has been proposed that the fliJ gene product functions as a molecular chaperone, facilitating flagellar protein export and assembly (45). The genetic organization of the B. cepacia fliE-fliJ locus identified thus far is identical to the gene arrangement in the corresponding loci of E. coli and S. enterica serovar Typhimurium. The B. cepacia fli gene products exhibit high levels of amino acid identity to the S. enterica serovar Typhimurium flagellar proteins (range, 33 to 68%) (Fig. 2), suggesting that the B. cepacia fli locus is also involved in flagellar biogenesis.
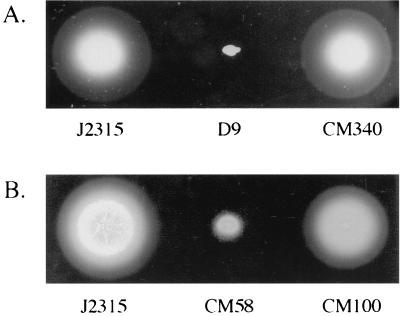
Analysis of the FliG protein in S. enterica serovar Typhimurium has shown that FliG is required for flagellar morphogenesis and plays a central role in mediating interactions within the motor-switch complex of the flagellar basal body (54). Insertional inactivation of fliG in S. enterica serovar Typhimurium renders cells nonmotile and results in a block in flagellar assembly (54). In order to examine the effect of disruption of the B. cepacia fliG gene on motility, we analyzed the D9 mutant by performing swarm agar motility assays. As shown in Fig. 3A, the D9 mutant was nonmotile in swarm agar. Additionally, the nonmotile phenotype of mutant D9 was confirmed by light microscopy. There was no significant difference in growth kinetics between wild-type strain J2315 and the D9 mutant (data not shown), indicating that the observed motility and invasion defects were not due to delayed growth of D9. Motility was restored to mutant D9 by providing a wild-type copy of the B. cepacia fliG gene in trans on plasmid pMT46 (Fig. 3A). This construct also complemented the invasion defect of the D9 mutant (Fig. 1). The plasmid vector alone did not restore motility or wild-type invasion levels to the fliG null strain. Successful complementation of both motility and invasion functions with fliG alone suggests that the observed invasion defect in D9 is due to its nonmotile phenotype and that the B. cepacia fliG gene is essential for flagellar biogenesis.
FIG. 3.
Phenotypes of B. cepacia mutants with impaired motility. Strains were stabbed into semisolid LB medium plates (0.25% agar) and incubated at 37°C for 24 h. (A) Wild-type strain J2315, mutant D9 (fliG::mini-Tn5Tc), and D9 complemented with fliG (strain CM340). (B) Wild-type strain J2315, mutant CM58 (fliI::cat), and CM58 complemented with fliI (strain CM100).
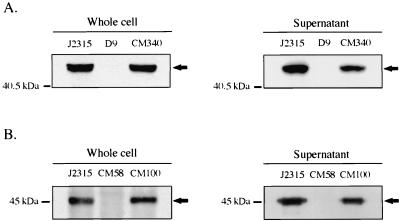
To further define the role of the B. cepacia fliG gene in flagellar biogenesis, whole-cell and supernatant protein fractions of fliG mutant D9 were prepared and examined by immunoblot analysis for the presence of flagellin protein. Since antibodies to the B. cepacia flagellin protein were not available, we utilized a polyclonal antiserum to the flagellin protein of B. pseudomallei, whose amino acid sequence is 77% identical to the amino acid sequence of the 45-kDa flagellin protein of B. cepacia (20). Antibody to B. pseudomallei flagellin recognized a 45-kDa protein in both the whole-cell and supernatant fractions of wild-type B. cepacia strain J2315 (Fig. 4). In contrast, the levels of the 45-kDa protein were dramatically reduced in both the whole-cell and supernatant fractions of fliG mutant D9 (Fig. 4A). The presence of the 45-kDa protein was restored in both the whole-cell and supernatant fractions of the D9 mutant by complementation with the fliG gene on plasmid pMT46 (strain CM340). These data suggest that the nonmotile phenotype of the D9 mutant is probably due to impaired formation of the flagellum structure.
FIG. 4.
Immunoblot analysis of flagellin protein in wild-type and flagellar mutant strains. Equal amounts of protein from whole-cell or supernatant fractions were separated on SDS-12.5% PAGE gels and immunoblotted with anti-flagellin antibody as described in Materials and Methods. (A) Wild-type strain J2315, D9 (fliG::mini-Tn5Tc), and CM340 (D9 complemented with fliG). (B) Wild-type strain J2315, CM58 (fliI::cat), and CM100 (CM58 complemented with fliI). The molecular masses of the protein standards are indicated on the left. The position of the 45-kDa flagellin protein band is indicated by an arrow.
In standard invasion assays, bacteria are typically brought into contact with host cell monolayers by centrifugation. We reasoned that centrifugation may bypass the role of motility in the initial stages of host cell invasion and that in the absence of centrifugation the fliG mutant strain should display a more pronounced invasion defect. When the D9 mutant and the wild-type strain were compared in quantitative invasion assays in the absence of centrifugation, the mutant was approximately 75% less invasive than the wild-type strain (Fig. 1B). Invasion was restored to wild-type levels by complementation of mutant D9 with plasmid pMT46. Therefore, flagellar biogenesis appeared to play an important role in the establishment of contact between B. cepacia and the A549 respiratory epithelial cell monolayers.
Generation and characterization of a B. cepacia fliI null strain.
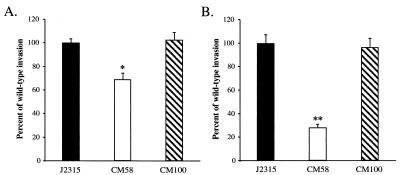
In order to further characterize the role of flagellar biogenesis in B. cepacia host cell invasion, a defined null mutation in the fliI gene was generated (Fig. 2). The fliI gene product is a member of a family of ATP-binding proteins believed to provide energy for the export of flagellar protein substrates, as well as nonflagellar virulence factors, in a wide range of bacterial species. Members of this protein family are central components of bacterial type III secretion systems (23). The B. cepacia FliI amino acid sequence is 68% identical to the FliI amino acid sequence of S. enterica serovar Typhimurium and 43% identical to the amino acid sequence of YscN, the ATPase implicated in virulence protein secretion via the Yersinia spp. type III system (53). Insertional inactivation of the B. cepacia fliI gene impaired the motility of cells (Fig. 3B), and flagellin protein could not be detected in either the whole-cell or supernatant fractions of the fliI null strain, CM58 (Fig. 4B). Compared to the ability of wild-type B. cepacia strain J2315, the ability of CM58 to enter A549 respiratory epithelial cells was significantly reduced; invasion was reduced approximately 30 and 70% when the bacteria were centrifuged and not centrifuged onto A549 epithelial cell monolayers, respectively (Fig. 5). Motility, flagellin protein, and wild-type invasion levels were restored by trans complementation with plasmid pJG7 (strain CM100). None of these phenotypes was restored by the plasmid vector alone. The pJG7 plasmid encodes amino acids 1 to 475 of FliI, and complementation of all three phenotypes by this plasmid suggests that the C-terminal 26 amino acids of FliI are not essential for its function. The successful complementation of mutant CM58 with plasmid pJG7 indicates that impaired motility, loss of flagellin protein, and reduced invasion were due to disruption of fliI and not to polar effects (Fig. 3B, 4B, and 5).
FIG. 5.
Invasion of A549 cell monolayers by B. cepacia strain J2315 (wild type), mutant CM58 (fliI::cat), and CM100 (CM58 complemented with fliI) with (A) or without (B) centrifugation. The invasion values were calculated by determining the percentages of the bacterial inocula that survived after 2 h of antibiotic treatment and were normalized to the value for wild-type strain J2315, which was arbitrarily set at 100%. The actual invasion frequencies for strain J2315 with and without centrifugation were 0.67% ± 0.03% and 0.26% ± 0.02%, respectively. The asterisk indicates that the P value was <0.0007, and the double asterisks indicate that the P value was <0.00004.
Adherence to A549 respiratory epithelial cells.
There is evidence that components of bacterial flagella can act as adhesins and mediate binding to host cells and mucosal surfaces (1, 55). In order to determine if the observed invasion defect of our flagellar mutants was due to reduced binding to A549 respiratory epithelial cells, we compared the adherence of these mutants to that of B. cepacia wild-type strain J2315. Bacteria were centrifuged onto A549 monolayers, and following incubation, the monolayers were washed repeatedly to remove nonadherent bacteria. Enumeration of the remaining adherent bacteria revealed a slight, but not statistically significant, increase in adherence of the flagellar mutants D9 and CM58 compared to the adherence of parent strain J2315 (Table 2). These results suggest that B. cepacia flagella do not function as adhesins in direct binding to A549 cells. Rather, the observed decreases in invasion of the fliG and fliI mutants are probably due to their impaired motility.
TABLE 2.
Adherence of B. cepacia strain J2315 and isogenic nonmotile mutants to A549 respiratory epithelial cells
| Strain | Genotype | % Adherence (mean ± SE) |
|---|---|---|
| J2315 | Wild type | 3.7 ± 0.4 |
| D9 | fliG | 4.4 ± 0.3 |
| CM58 | fliI | 4.1 ± 0.4 |
DISCUSSION
Motility has been shown to play an important role in the invasiveness of a number of bacterial pathogens. Active motility is required for optimal invasion of epithelial cells and translocation across polarized epithelial monolayers by C. jejuni (19, 55). The ability of a P. mirabilis nonmotile mutant to invade cultured human renal epithelial cells has been shown to be significantly reduced, even when the bacteria were centrifuged onto renal cell monolayers (37). Nonmotile mutants of S. enterica serovar Typhimurium have been shown to be less invasive in HeLa cells (29), while aflagellate mutants of S. enterica serovar Enteritidis are both less invasive in cultured intestinal epithelial cells (10, 49) and less adherent to these cells (10). In S. enterica serovar Typhimurium, mutations in flagellar regulatory components have also been shown to affect the expression of other genes required for invasion (31). P. aeruginosa nonmotile mutants have recently been shown to be defective for invasion of rabbit corneal epithelial cells, and similar to the findings reported here, centrifugation did not restore invasion to wild-type levels (15).
Our data are consistent with the results of previously described studies in which it was found that flagellum-mediated motility contributes to the ability of bacteria to invade host cells. The abilities of both of the B. cepacia flagellar mutants generated in this study to invade were reduced even when the bacteria were brought in close contact with A549 cells by centrifugation, thus bypassing the role that motility plays in the initial establishment of contact between B. cepacia and host cells. The reductions in the ability to invade do not appear to be due to defective adherence of the motility mutants to A549 respiratory epithelial cells, although we cannot exclude the possibility that B. cepacia flagella may interact with other types of host cells and/or acellular ligands. Since more than 40 genes are required for flagellar biogenesis in other bacteria, it was somewhat surprising that additional motility mutants were not isolated from the invasion screening analysis. This may have been due to a lack of complete randomness in the targets for mini-Tn5 transposition within the B. cepacia chromosome. Alternatively, some motility mutants may not have been detected due to limited sensitivity of the invasion screening procedure. While the abilities of both the fliG and fliI mutants to invade were impaired, the invasion defect of the fliI mutant was less pronounced than that of the fliG mutant when the bacteria were centrifuged onto the A549 cell monolayers (Fig. 1A and 5A). After prolonged incubation of swarm agar plates inoculated with the fliI mutant, we observed a small but detectable swarm, which we did not observe on swarm agar plates inoculated with the fliG mutant. Motility in the absence of a functional fliI gene has been reported in other bacteria (16), although the genetic basis is not known. It is possible that the B. cepacia fliI mutant retains a low level of FliI activity. Alternatively, there is a second type III secretion homolog of fliI, which can at least partially compensate for the loss of the fliI gene.
Our results indicate that flagellum-mediated motility may be required for optimal invasion of A549 cells during two distinct phases: establishment of contact with the host cell and bacterial entry once contact has been established. The motility of B. cepacia may be particularly important for initiation of contact between the bacteria and epithelial cells and may promote adherence to the airway epithelia of the lung and subsequently invasion of deeper tissues. Additionally, motility may allow B. cepacia to penetrate the viscous mucus that covers the airway epithelia, which is particularly abundant in the CF lung airways due to inefficient mucociliary clearance. The gastric pathogen H. pylori utilizes motility to breach the gastric mucosal barrier and persist within the mucosal lining (56). Similarly, B. cepacia may utilize motility to enter the viscous mucus layer in the CF lung airways, persist within it, or penetrate it to establish contact with the underlying epithelial tissues. Once the epithelial barrier is breached, motility may facilitate invasion of B. cepacia into underlying tissues, thus promoting entry of the bacteria into the bloodstream and systemic dissemination.
Our findings indicate that defective motility due to disruption of the B. cepacia flagellar export and assembly pathway is responsible for the reduced invasiveness of the fliG and fliI mutants. However, at this time we cannot rule out the possibility that other secretion defects contribute to the reduced invasion of the fliG and fliI mutants. It has recently been proposed that the flagellar type III secretion system has two functions, export of flagellar proteins and export of nonflagellar virulence determinants (57). For example, in Yersinia enterocolitica, components of the flagellar type III apparatus are required for export of a virulence-associated phospholipase, YplA (57). It is possible that nonflagellar virulence proteins exported via the B. cepacia flagellar secretion system also contribute to the ability of the organism to interact with and enter host cells.
We have demonstrated that the B. cepacia mutants with impaired motility lack detectable intracellular flagellin and do not export detectable amounts of flagellin into the supernatant. While it is possible that undetectable amounts of B. cepacia flagellin are expressed, it is clear that the levels of the B. cepacia flagellin in both D9 and CM58 are dramatically reduced compared to the level in parent strain J2315. Flagellar gene expression in a number of bacterial species is known to proceed through a hierarchical pathway. The genes encoding early structural components of the flagellum, as well as components of the flagellar type III secretion system, are transcribed first, while transcription of genes encoding later components, such as flagellin, are activated during the final stages of assembly (34). Mutations in early flagellar genes often block expression of genes encoding late flagellar components. Since the B. cepacia flagellar genes identified and characterized in this study are predicted to encode early structural components, as well as components of the export apparatus, it is likely that disruption of these genes in B. cepacia also leads to a block of flagellin gene expression. We cannot, however, exclude the possibility that the B. cepacia flagellin is expressed but subsequently degraded in the absence of either FliG or FliI.
In E. coli, the fliFGHIJ(K) genes are transcribed as an operon, which belongs to class II of the flagellar hierarchy. Transcription of the fliFGHIJ(K) operon in E. coli is induced by the class I FlhDC transcriptional activator complex (34). It is possible that the B. cepacia fliFGHIJ genes are also cotranscribed as an operon, as there are no identifiable internal promoter elements upstream of fliG, fliH, fliI, or fliJ. Additionally, the intergenic regions in the fliFGHIJ locus are 32 bp long (between fliH and fliI) and 5 bp long (between fliI and fliJ), while fliF and fliG overlap by 11 bp and fliG and fliH overlap by 8 bp. Therefore, if internal promoters do exist, they must overlap with the coding region of the gene immediately upstream. Interestingly, we identified a sequence (5′-CACGATAA-3′) that is 70 bp upstream of the B. cepacia fliF predicted start codon and matches at 6 of 8 bp the consensus sequence of the −10 region of E. coli flagellar class II core promoters (34). This observation suggests that the B. cepacia fli gene cluster may be regulated similarly. Studies aimed at elucidating the transcriptional organization and regulation of the B. cepacia flagellar gene locus are under way.
In conclusion, our data provide strong evidence that flagellum-mediated motility is a contributing factor in the ability of B. cepacia to invade respiratory epithelial cells. Thus, motility may be an important virulence determinant of B. cepacia, enhancing the pathogenicity of the organism in the human host. Future studies will focus on further defining the role of motility and flagellar biogenesis in B. cepacia pathogenesis using both chronic and acute animal models of B. cepacia infection and on further characterizing the mechanisms of invasion employed by B. cepacia to enter host cells.
Acknowledgments
We thank Don Woods for providing polyclonal antiserum to the B. pseudomallei flagellin protein and Andrea Rolnicki for constructing the fliI locus subclones. We also thank Sandra Armstrong for critical reading of the manuscript and Tim Leonard for technical assistance.
This work was supported by Minnesota Medical Foundation grant 13700 and by grant CF0I0 from the Cystic Fibrosis Foundation.
Editor: V. J. DiRita
REFERENCES
- 1.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brett, P. J., D. C. W. Mah, and D. E. Woods. 1994. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect. Immun. 62:1914-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkholder, W. H. 1950. Sour skin: a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 4.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns, J. L., and L. A. Hedin. 1991. Genetic transformation of Pseudomonas cepacia using electroporation. J. Microbiol. Methods 13:215-221. [Google Scholar]
- 6.Chiu, C. H., A. Ostry, and D. P. Speert. 2001. Invasion of murine respiratory epithelial cells in vivo by Burkholderia cepacia. J. Med. Microbiol. 50:594-601. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroucke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 8.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 11.Dobbin, C. J., R. Soni, T. Jelihovsky, and P. T. Bye. 2000. Cepacia syndrome occurring following prolonged colonisation with Burkholderia cepacia. Aust. N. Z. J. Med. 30:288-289. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 13.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 14.Feldman, M., R. Bryan, S. Rajan, L. Scheffler, S. Brunnert, H. Tang, and A. Prince. 1998. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 66:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleiszig, S. M. J., S. K. Arora, R. Van, and R. Ramphal. 2001. FlhA, a component of the flagellum assembly apparatus of Pseudomonas aeruginosa, plays a role in internalization by corneal epithelial cells. Infect. Immun. 69:4931-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodfellow, I. G., C. E. Pollitt, and R. E. Sockett. 1996. Cloning of the fliI gene from Rhodobacter sphaeroides WS8 by analysis of a transposon mutant with impaired motility. FEMS Microbiol. Lett. 142:111-116. [DOI] [PubMed] [Google Scholar]
- 17.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 18.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant, C. C., M. E. Konkel, W. J. Cieplak, and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hales, B. A., J. A. W. Morgan, C. A. Hart, and C. Winstanley. 1998. Variation in flagellin genes and proteins of Burkholderia cepacia. J. Bacteriol. 180:1110-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi, S., M. Abe, M. Kimoto, S. Furukawa, and T. Nakazawa. 2000. The DsbA-DsbB disulfide bond formation system of Burkholderia cepacia is involved in the production of protease and alkaline phosphatase, motility, metal resistance and multi-drug resistance. Microbiol. Immunol. 44:41-50. [DOI] [PubMed] [Google Scholar]
- 22.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchison, M. L., and J. R. W. Govan. 1999. Pathogenicity of microbes associated with cystic fibrosis. Microbes Infect. 1:1005-1014. [DOI] [PubMed] [Google Scholar]
- 25.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 26.Jenal, U., J. White, and L. Shapiro. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243:227-244. [DOI] [PubMed] [Google Scholar]
- 27.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 28.Keig, P. M., E. Ingham, and K. G. Kerr. 2001. Invasion of human type II pneumocytes by Burkholderia cepacia. Microb. Pathog. 30:167-170. [DOI] [PubMed] [Google Scholar]
- 29.Khoramian-Falsafi, T., S. Harayama, K. Kutsukake, and J. C. Pechere. 1990. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb. Pathog. 9:47-53. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lucas, R. L., P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 34.Macnab, R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131-158. [DOI] [PubMed] [Google Scholar]
- 35.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minamino, T., S. Yamaguchi, and R. M. Macnab. 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J. Bacteriol. 182:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mobley, H. L., R. Belas, V. Lockatell, G. Chippendale, A. L. Trifillis, D. E. Johnson, and J. W. Warren. 1996. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect. Immun. 64:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohr, C. D., M. Tomich, and C. A. Herfst. 2001. Cellular aspects of Burkholderia cepacia infection. Microbes Infect. 3:1-11. [DOI] [PubMed] [Google Scholar]
- 39.Montie, T. C., D. Doyle-Huntzinger, R. C. Craven, and I. A. Holder. 1982. Loss of virulence associated with absence of flagellum in an isogenic mutant of Pseudomonas aeruginosa in the burned-mouse model. Infect. Immun. 38:1296-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morooka, T., A. Umeda, and K. Amako. 1985. Motility as an intestinal colonization factor for Campylobacter jejuni. J. Gen. Microbiol. 131:1973-1980. [DOI] [PubMed] [Google Scholar]
- 41.Ormonde, P., P. Horstedt, R. O'Toole, and D. L. Milton. 2000. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J. Bacteriol. 182:2326-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 45.Stephens, C., C. Mohr, C. Boyd, J. Maddock, J. Gober, and L. Shapiro. 1997. Identification of the fliI and fliJ components of the Caulobacter flagellar type III protein secretion system. J. Bacteriol. 179:5355-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tablan, O. C., T. L. Chorba, D. V. Schidlow, J. W. White, K. A. Hardy, P. H. Gilligan, W. M. Morgan, L. A. Carson, W. J. Martone, J. M. Jason, and W. R. Jarvis. 1985. Pseudomonas cepacia colonization in patients with cystic fibrosis: risk factors and clinical outcome. J. Pediatr. 107:382-387. [DOI] [PubMed] [Google Scholar]
- 47.Tablan, O. C., W. J. Martone, C. F. Doershuk, R. C. Stern, M. J. Thomassen, J. D. Klinger, J. W. White, L. A. Carson, and W. R. Jarvis. 1987. Colonization of the respiratory tract with Pseudomonas cepacia in cystic fibrosis: risk factors and outcomes. Chest 91:527-532. [DOI] [PubMed] [Google Scholar]
- 48.Tipper, J. L., E. Ingham, J. H. Cove, N. J. Todd, and K. G. Kerr. 1998. Survival and multiplication of Burkholderia cepacia within respiratory epithelial cells. Clin. Microbiol. Infect. 4:450-459. [Google Scholar]
- 49.Van Asten, F. J. A. M., H. G. C. J. M. Hendriks, J. F. J. G. Koninkx, B. A. M. Van der Zeijst, and W. Gaastra. 2000. Inactivation of the flagellin gene of Salmonella enterica serotype Enteritidis strongly reduces invasion into differentiated Caco-2 cells. FEMS Microbiol. Lett. 185:175-179. [DOI] [PubMed] [Google Scholar]
- 50.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 51.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.West, S. E. H., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 128:81-86. [DOI] [PubMed] [Google Scholar]
- 53.Woestyn, S., A. Allaoui, P. Wattiau, and G. R. Cornelis. 1994. YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 176:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi, S., H. Fujita, A. Ishihara, S.-I. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 56.Yoshiyama, H., and T. Nakazawa. 2000. Unique mechanism of Helicobacter pylori for colonizing the gastric mucus. Microbes Infect. 2:55-60. [DOI] [PubMed] [Google Scholar]
- 57.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]