Abstract
Fimbrial adhesins mediate the attachment of pathogenic Escherichia coli to various host tissues leading to the development of disease. The Dr hemagglutinin and F1845 fimbriae belong to the Dr family of adhesins, which is associated with urinary tract infections and diarrheal disease. These adhesins bind to the Dra blood-group antigen present on decay-accelerating factor (DAF). The Dr hemagglutinin is unique in this family since it also binds to type IV collagen and its binding is inhibited by the presence of chloramphenicol. We have purified the major structural subunits of Dr and F1845 fimbriae, DraE and DaaE, as fusions to maltose-binding protein and to oligohistidine tags and examined their binding to erythrocytes, Chinese hamster ovary cell transfectants expressing DAF, and a DAF fusion protein. The DraE and DaaE fusion proteins bind to the DAF receptor in a specific manner resembling the distinct phenotypes of the corresponding Dr and F1845 fimbriae. In contrast to binding studies with the DAF receptor, the DraE fusion proteins did not bind to type IV collagen.
Escherichia coli expresses several proteins on the surface of the bacterial cell that mediate attachment to mammalian receptors. Among these are fimbriae that allow persistence of bacteria in host tissues and play a role in the pathogenesis of various diseases. Several families of fimbriae have been extensively characterized for their expression, composition, and biogenesis. A large group of fimbrial adhesins of E. coli are assembled via the chaperone/usher pathway (12). With respect to their structural composition, fimbriae assembled via the chaperone/usher pathway are divided into two main types: those in which the fimbrial shaft is primarily composed of one major subunit with the adhesive function conferred by a different minor subunit and those in which the major subunit also serves as the adhesive subunit. The first type includes P and type 1 fimbriae in which PapA and FimA are the major structural subunits, but PapG and FimH are the minor adhesive subunits, respectively (14, 16, 17, 20, 23). The second type includes the members of the K88 family of adhesins of porcine enterotoxigenic E. coli, such as CS31A and K88 fimbriae, whose major structural subunits, ClpG and FaeG, also serve as adhesive subunits (3, 7, 39). We examine here the hypothesis that the major structural subunits of two fimbrial members of the Dr family of adhesins, the Dr hemagglutinin and F1845 fimbriae, directly confer adherence.
The Dr family of adhesins of E. coli is associated with urinary tract infections, in particular with cystitis and pregnancy-associated pyelonephritis, as well as with diarrheal disease (1, 9, 13, 19, 28, 30, 35, 41). Members of this family bind to the Dra blood-group antigen present on decay-accelerating factor (DAF; CD55), a complement regulatory and signaling molecule (21, 25, 31). In particular, short consensus repeat 3 (SCR3) of DAF appears to be involved in binding by Dr family adhesins (26). The Dr hemagglutinin is unique among the members of the Dr family that have been examined because it also binds to another receptor, the 7s domain of type IV collagen (40). The binding mediated by the Dr hemagglutinin is also unique among Dr adhesins because it is inhibited by the presence of chloramphenicol (27).
Several genes are involved in the expression and biogenesis of the fimbrial adhesins Dr hemagglutinin and F1845, including draE and daaE, which encode the major structural subunits that compose the respective fimbrial appendages (5, 36). It has been demonstrated that alterations in the sequences that encode the major structural subunits affect receptor-binding phenotypes (36). In particular, inhibition of hemagglutination by chloramphenicol was shown to be associated with the draE sequence. This observation was further analyzed and it was also shown that the product of the draE gene was necessary for binding to type IV collagen (6). Through site-directed mutagenesis of draE, it was shown that various amino acid changes within the mature DraE protein could affect DAF binding, type IV collagen binding, and inhibition by chloramphenicol. However, these studies did not rule out the possibility that other products of these operons are the adhesive subunits. Direct and specific binding by DraE and DaaE to DAF and type IV collagen was not shown. The remaining genes that comprise the operons are highly homologous, and their products could function to directly interact with the receptors. It is possible that alterations in DraE and DaaE affected the conformation of a putative minor adhesive subunit within the fimbrial structure, thus altering the binding indirectly. This phenomenon was observed previously with type 1 fimbriae and the FimH adhesive subunit, where the sequence of the nonadhesive fimbrial subunit can affect the binding properties of the adhesive subunit (24, 38).
Another member of the Dr family of adhesins examined for adhesive properties is the afimbrial adhesin AFAIII and its major structural subunit, AfaE3 (8, 15). This protein is highly homologous to DraE differing by only three residues (18). AfaE3 was expressed and purified as an oligohistidine fusion (8, 10, 15). Although it was shown that this protein could bind to HeLa and Caco-2 cells alone and on the surface of polystyrene beads, specificity of the binding to DAF was not investigated (10, 15). Therefore, it has not been demonstrated that the major structural subunits of the members of this family of adhesins are the adhesive subunits for the DAF receptor.
In this study, we examined directly whether the major structural subunits of the Dr hemagglutinin and F1845 fimbriae, DraE and DaaE, respectively, are also the subunits that provide specific adhesion by these fimbriae.
MATERIALS AND METHODS
Plasmid construction.
The plasmid constructs utilized for this work are listed in Table 1. For construction of maltose-binding protein (MBP) fusion proteins, the draE sequence corresponding to the mature DraE amino acid sequence was PCR amplified by using pCC90 (6) as a template and the DraE5M and DraE3M primers. The amplified gene was inserted into the pMAL-p2 vector (New England Biolabs, Beverly, Mass.) by using the EcoRI and PstI restriction sites in order to generate pMAL-R. The pMAL-p2 vector provides an N-terminal fusion to MBP for targeting to the periplasm and for purification. The daaE sequence corresponding to the mature DaaE amino acid sequence was PCR amplified with pSSS1 (5) as a template and the DaaE5M and DaaE3M primers. The amplified gene was also inserted into the pMAL-p2 vector by using the EcoRI and SalI restriction sites to generate pMAL-A. All oligonucleotide primers were obtained from Life Technologies, Inc. (Rockville, Md.). The finished constructs were transformed into E. coli DH5α for expression (Life Technologies, Rockville, Md.). pMAL-p2 alone was also transformed into E. coli DH5α as a control strain for the purification of MBP.
TABLE 1.
Plasmids, their corresponding protein products, and primers used for their construction
| Plasmid | Expression vector | Fusion protein | Description | Primers (sequences) used in construction | |
|---|---|---|---|---|---|
| pMAL-R | pMAL-p2 | MBP-DraE | DraE fusion to the C terminus of MBP with Factor Xa cleavage site | DraE5M | (5′-CAGAATTCGGGTTCACCCCGAGTGGC-3′), |
| DraE3M | (5′-GCCTGCAGTCATTTTGCCCAGTAACC-3′) | ||||
| pMAL-A | pMAL-p2 | MBP-DaaE | DaaE fusion to the C terminus of MBP with Factor Xa cleavage site | DaaE5M | (5′-CAGAATTCGCCACGTTCCAAGCGAGC-3′), |
| DaaE3M | (5′-GCGTCGACTTAGTTCGTCCAGTAACC-3′) | ||||
| pET-R | pET-27b(+) | DraE-His | DraE fusion to the N terminus of His6 tag and HSV tag | DraE5H | (5′-ACATATGGGGTTCACCCCGAGTGGCACC-3′), |
| DraE3H | (5′-CTCGAGTTTTGCCCAGTAACCCCCGGTCAG-3′) | ||||
| pET-A | pET-27b(+) | DaaE-His | DaaE fusion to the N terminus of His6 tag and HSV tag | DaaE5H | (5′-ACATATGGCCACGTTCCAAGCGAGCGGG-3′), |
| DaaE3H | (5′-CTCGAGGTTCGTCCAGTAACCCCCGGTGAG-3′) | ||||
| pET-Rct | pET-27b(+) | DraECt-His | DraE fusion with a C-terminal 9-amino-acid deletion to the N terminus of His6 tag and HSV tag | DraE5H | (5′-ACATATGGGGTTCACCCCGAGTGGCACC-3′), |
| DraE3HC | (5′-CTCGAGCAGTGTGTAGTTGCCGGGCGG-3′) | ||||
| pET-Act | pET-27b(+) | DaaECt-His | DaaE fusion with a C-terminal 9-amino-acid deletion to the N terminus of His6 tag and HSV tag | DaaE5H | (5′-ACATATGGCCACGTTCCAAGCGAGCGGG-3′), |
| DaaE3HC | (5′-CTCGAGCAGTGTGTATTCACCGGTCGG-3′) | ||||
For construction of oligohistidine fusion proteins, the draE sequence corresponding to the predicted mature DraE amino acid sequence was PCR amplified by using pCC90 as a template and the DraE5H and DraE3H primers. For a nine-amino-acid C-terminal truncation of DraE, pCC90 was used as a template with the DraE5H and DraE3HC primers. The amplified fragments were inserted into the pET-27b(+) vector (Novagen, Inc., Madison, Wis.) by using the NdeI and XhoI restriction sites to generate pET-R and pET-Rct, respectively. The pET-27b(+) vector provides a C-terminal fusion to a six-histidine (His6) tag for purification and a herpes simplex virus (HSV) glycoprotein D epitope tag for detection. The daaE sequence corresponding to the mature DaaE amino acid sequence was PCR amplified by using pSSS1 as a template and the DaaE5H and DaaE3H primers. For a nine-amino-acid C-terminal truncation of DaaE, pSSS1 was also used as a template with the DaaE5H and DaaE3HC primers. The amplified genes were also inserted into the pET-27b(+) vector by using the NdeI and XhoI restriction sites to generate pET-A and pET-Act, respectively. All oligonucleotide primers were obtained from Life Technologies, Inc. The finished constructs were transformed into E. coli BL21(DE3) for expression (Novagen). The final constructs do not include the predicted first two codons encoding the mature protein sequence.
Bacterial strains were grown in Luria-Bertani (LB) broth and Super Broth (SB) at 37°C with 100 μg of ampicillin (for pMAL-p2-derived strains) or 25 μg of kanamycin [for pET-27b(+)-derived strains]/ml. Plasmid DNA was prepared by the Easy Prep method (4). Enzymes were purchased from New England Biolabs and used as recommended by the manufacturer. All constructs were confirmed by sequencing by using Big Dye Terminator method and ABI sequencing (PE Applied Biosystems, Foster City, Calif.).
Protein purification.
The MBP fusions with DraE and DaaE, MBP-DraE and MBP-DaaE, respectively, and MBP alone were purified from the periplasm under nondenaturing conditions as previously described with some modifications (2). Recombinant strains were grown to an optical density at 600 nm (OD600) of 0.6 in SB at 37°C with shaking. The cultures were then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and were incubated for two additional hours at 37°C with shaking. The cells were harvested by centrifugation at 7,000 × g for 10 min and resuspended in 30 mM Tris-HCl-20% sucrose (pH 8.0), and EDTA was added to a final concentration of 1 mM. The suspension was shaken for 10 min at room temperature and then centrifuged at 10,000 × g for 10 min. The pellet was resuspended in ice-cold 5 mM MgSO4 and incubated with shaking on ice for 10 min. The periplasmic fraction was then recovered and adjusted with 1 M Tris-HCl (pH 7.4) to a final concentration of 20 mM. MBP fusions were purified by amylose affinity chromatography (New England Biolabs). Column buffer (20 mM Tris-HCl, pH 7.4; 0.2 M NaCl; 1 mM EDTA) was used to wash the column. Column buffer containing 10 mM maltose was used for elution of the fusion protein. The purified protein eluate was dialyzed against phosphate-buffered saline (PBS), concentrated by microfiltration, and stored at −20°C.
The oligohistidine fusions of DraE and DaaE—i.e., DraE-His, DaaE-His, DraECt-His, and DaaECt-His—were purified under denaturing conditions with 6 M urea as recommended by the vector supplier (Novagen). All of the reagents necessary for purification were purchased from Novagen. Recombinant clones were grown to an OD600 of 0.6 in SB at 37°C with shaking. The cultures were then induced with 1 mM IPTG and incubated for two additional hours at 37°C with shaking. The cells were harvested by centrifugation at 5,000 × g for 5 min, followed by resuspension into 1× binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl; pH 7.9). The suspension was sonicated at medium setting six times for 15 s each time and then centrifuged at 20,000 × g for 15 min. The pellet was then resuspended in 1× binding buffer and centrifuged at 20,000 × g for 15 min. The pellet was resuspended in 1× binding buffer containing 6 M urea and incubated on ice for 1 h. The insoluble material was removed by centrifugation at 39,000 × g for 20 min, and the remaining supernatant was filtered through a 0.45-μm-pore-size filter. This crude extract was further purified by affinity chromatography by using His-Bind resin matrix with immobilized Ni2+ to isolate the oligohistidine fusion proteins, as described by the manufacturer. All chromatography steps were performed under denaturing conditions with 6 M urea. The resulting purified protein preparation was stepwise dialyzed from 6 M urea into PBS at 4°C. The proteins were then concentrated, divided into aliquots, and stored at 4°C.
Purified fusion proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
MBP-DraE, MBP-DaaE, and MBP were cleaved by using Factor Xa (New England Biolabs) as recommended by the manufacturer. Samples were cleaved in cleavage buffer (20 mM Tris-HCl, 2 mM CaCl2, 100 mM NaCl; pH 8.0) at room temperature for a minimum of 3 h and used directly in the binding assays.
Purification of Dr and F1845 fimbriae.
Dr and F1845 fimbriae were purified from strains DH5α(pCC90) and DH5α(pSSS1), respectively (5, 6). Bacteria were grown overnight in LB containing 100 μg of ampicillin/ml. The cultures were centrifuged at 5,000 × g for 15 min, and the pellet was resuspended in 10 mM Tris-HCl (pH 7.0). The bacterial suspension was subjected to mechanical shearing in a blender (Hamilton Beach) to remove the fimbriae, and the cells were pelleted by centrifugation at 12,000 × g for 20 min. The fimbriae were then precipitated from the supernatant with 40% ammonium sulfate and pelleted at 12,000 × g for 20 min. The pellet was resuspended in 50 mM phosphate-50 mM NaCl-2 M urea (pH 7.4) and then dialyzed for 48 h in the same buffer. The fimbriae were then purified by gel filtration chromatography by using a Sepharose CL-4B column (Amersham Pharmacia Biotech, Piscataway, N.J.) in the same buffer. Eluted fractions containing fimbriae were pooled and dialyzed extensively against PBS. Fimbrial samples were stored at 4°C.
Preparation of erythrocyte suspensions.
The human erythrocyte suspension was prepared from blood group O whole human blood donated by a healthy volunteer. The sheep erythrocyte suspension was prepared from whole sheep blood (Hema Resources, Inc., Aurora, Oreg.). The guinea pig erythrocyte suspension was prepared from guinea pig blood (Alsevers anticoagulant; Colorado Serum Company, Denver, Colo.). The whole-blood samples were diluted to 2% erythrocyte suspensions in PBS. Erythrocytes were washed twice in PBS and then resuspended in PBS containing 2% d-mannose. The suspensions were stored at 4°C for up to 2 weeks.
Erythrocyte binding assay.
Purified proteins were diluted in 0.02 M sodium bicarbonate (pH 7.5) to a concentration of 10 μg/ml, and 100 μl was added to each of the wells of a 96-well microtiter plate and incubated for 2 h at 37°C. This concentration of protein was confirmed to provide adequate coating of the plate wells by detection with anti-Dr fimbriae, anti-F1845 fimbriae, anti-MBP, and anti-HSV antibodies. The wells were then washed with PBS and blocked with PBS containing 1% bovine serum albumin (BSA) for 2 h at 37°C. The blocking solution was removed, 100 μl of a 2% human erythrocyte suspension in PBS with 2% d-mannose (Sigma-Aldrich, St. Louis, Mo.) was added to each well, and the plate was incubated with rotation at room temperature for 2 h. The wells were then washed gently once with PBS containing 1% BSA. The remaining bound erythrocytes were lysed by addition of 150 μl of water to each well, and the released hemoglobin was quantified by determination of OD405. To examine the effect of chloramphenicol on binding, chloramphenicol was added to the erythrocyte suspension prior to its addition to the wells at various concentrations. Wells with PBS alone and without protein were used as controls for background binding and were subtracted from the readings obtained for the sample wells.
Maintenance and preparation of CHO cells.
CHO cells transfected with DAF (CHO/DAF/A9) and control Chinese hamster ovary (CHO) cells with vector alone (CHO/SFFV/8G*) were generously provided by Douglas M. Lublin (Washington University, St. Louis, Mo.) (22). Cells were grown in a humidified atmosphere of 5% CO2 at 37°C in Ham's F-12 medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 25 μg of G418 (Geneticin; Life Technologies, Inc., Rockville, Md.)/ml. For preparation of the cells for the CHO cell-binding assay, cells were trypsinized with 0.25% trypsin and 0.02% EDTA. The cells were then centrifuged at 120 × g for 5 min, resuspended in PBS containing 0.2% BSA, and adjusted to a concentration of 3 × 105 cells/ml.
CHO cell-binding assay.
Purified proteins were diluted in PBS to a concentration of 20 μg/ml, and 100 μl of each was added to the wells of a 96-well microtiter plate and incubated for 2 h at room temperature as described above. The wells were then blocked with PBS containing 1% BSA for 2 h at room temperature. The blocking solution was removed, and 100 μl of a suspension of 3 × 105 CHO/DAF/A9 (CHO DAF+) or CHO/SFFV/8G* (CHO DAF−) cells/ml was added to the corresponding wells. The plate was incubated at room temperature for 2 h with occasional rotation to break apart any cell clumps. Unbound cells were then removed, and 100 μl of Ham's F-12 medium containing 0.9% Triton X-100 was added to the remaining cells. The wells were incubated at 37°C for 45 min to allow cell lysis to occur. Samples from each well were then removed, and the levels of lactate dehydrogenase (LDH) released were detected by using Cyto Tox 96 (Promega) as directed by the manufacturer. Wells containing PBS without protein were used to subtract background levels of binding.
DAF234 expression and purification.
Purified DAF containing SCR2, SCR3, and SCR4 and an oligohistidine tag at the C-terminal end (DAF234) was purified from Pichia pastoris expressing DAF234, which was generously provided by Susan Lea (Oxford University, Oxford, England). DAF234 was expressed in P. pastoris and purified from the supernatant of induced cultures by nickel chromatography as described previously (32). Briefly, P. pastoris was grown on yeast extract-peptone-dextrose medium supplemented with 4 mg of G418/ml at 30°C and utilized to inoculate buffered minimal glycerol medium. After overnight culture, the cells were pelleted by centrifugation at 3,000 × g for 20 min. The pellet was then resuspended in buffered minimal methanol medium to an OD600 of 1.0. The cells were incubated with shaking at 30°C for 72 h with the addition of methanol to a final concentration of 0.5% every 24 h for induction. The culture was then adjusted to a final concentration of 20 mM imidazole (pH 8.0) and centrifuged at 3,000 × g for 30 min. The supernatant was collected, and DAF234 was purified by affinity chromatography by using His-Bind resin matrix with immobilized Ni2+ (Novagen). The column was first equilibrated with binding buffer (20 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl; pH 8.0), and then the supernatant was added over the column. The column was washed with binding buffer, and then DAF234 was eluted with 300 mM imidazole-0.5 M NaCl-20 mM Tris-HCl (pH 8.0). The protein was divided into aliquots and stored at −80°C.
DAF and type IV collagen-binding enzyme immunoassays (EIAs).
DAF234 was diluted in PBS to a concentration of 15 or 30 μg/ml, and 100 μl was added to each well of a 96-well microtiter plate and incubated at 4°C overnight. The wells were then washed with PBS and blocked with PBS containing 0.1% Tween 20 for 2 h at 37°C. The blocking solution was removed, and 100-μl portions of the different fusion proteins were added at a concentration of 100 μg/ml in PBS containing 0.05% Tween 20 and incubated for 3 h at 37°C. This concentration had been determined in preliminary experiments to be optimal for maximum binding. After various washes with PBS, the bound fimbrial proteins and the MBP fusion proteins were detected with anti-Dr fimbriae and anti-F1845 fimbriae antibodies (5, 6) at a 1:250 dilution. The MBP control protein was detected with anti-MBP antibodies (New England Biolabs) at a 1:1,000 dilution. The oligohistidine fusion proteins were detected with anti-HSV epitope tag antibodies (Novagen) at a 1:1,000 dilution for 60 min at 37°C. These antibody dilutions were determined to recognize the fimbrial or fusion proteins at the same level. The wells were then washed, and a 1:5,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antibody (Pierce, Rockford, Ill.) was added for 30 min at 37°C. All antibodies were diluted in PBS containing 0.05% Tween 20. The bound antibodies were then detected with Turbo TMB Reagent (Bio-Rad, Hercules, Calif.) as described by the manufacturer. The reaction was stopped with 1 N H2SO4, and the wells were quantified at OD450. To examine the effect of chloramphenicol on binding, chloramphenicol was added to the fusion proteins and incubated for 15 to 30 min at room temperature before addition to the plates. A PBS control was used to subtract background binding values due to the antibodies. The type IV collagen-binding EIA was performed in the same manner except that 20 μg of type IV collagen (from human placenta; Sigma-Aldrich)/ml was used to coat the wells of the microtiter plates. By using anti-DAF (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) and anti-type IV collagen antibodies (Research Diagnostics, Inc., Flanders, N.J.), the concentrations of DAF234 and type IV collagen used were determined to provide maximum and consistent coating of the plate wells.
Rosette agglutination assay.
DH5α(pCC90) and DH5α(pSSS1) were grown on LB agar and resuspended in PBS containing 1% BSA (Sigma-Aldrich) to an OD600 of 0.5. Twofold dilutions of this suspension were made in PBS containing 1% BSA. The different fimbrial and fusion proteins were diluted to 100 μg/ml in PBS containing 1% BSA. Equal volumes of bacterial suspensions or protein dilutions were mixed with a human erythrocyte suspension in 96-well, U-bottom microtiter plates and incubated at room temperature for 30 min. Rosette formation was manifested as settling of the red blood cells as a diffuse carpet in the wells. In the absence of rosette formation, the red blood cells settled in a tight pellet.
RESULTS
Expression and purification of DraE and DaaE.
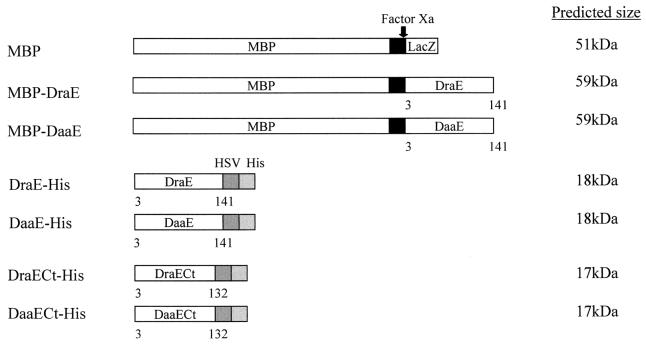
To analyze the binding properties of the major fimbrial subunits of the Dr hemagglutinin and F1845 fimbriae, DraE and DaaE, respectively, two recombinant expression systems were used to create fusions to MBP and an oligohistidine tag. The draE and daaE sequences encoding the mature DraE and DaaE proteins were amplified by PCR, ligated into the expression vectors pMAL-p2 and pET-27b(+), and confirmed by sequencing (Table 1). They were then expressed and purified as fusions to the C-terminal end of MBP (MBP-DraE and MBP-DaaE) and as fusions (DraE-His and DaaE-His) to the N-terminal end of six histidine residues and a peptide tag derived from HSV glycoprotein D (i.e., HSV tag) (Fig. 1).
FIG. 1.
Schematic of expressed fusion proteins and their sizes. Factor Xa, protease Factor Xa; HSV, HSV glycoprotein D epitope tag; His, His6 tag. Numbers below the proteins indicate the amino acids of the mature subunit proteins.
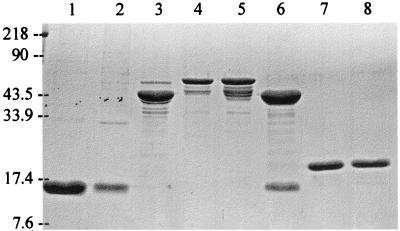
The MBP-DraE and MBP-DaaE fusions were purified in soluble form from the periplasm of the expression host followed by amylose affinity chromatography. MBP-DraE and MBP-DaaE were visualized at their expected sizes of ca. 59 kDa by SDS-PAGE (Fig. 2, lanes 4 and 5). MBP fused to LacZ was also purified from the original expression vector as a control. MBP alone showed an approximate size of 43 kDa, which is smaller than the predicted size because the LacZ domain of the protein is cleaved and degraded (Fig. 2, lane 3) (P. Riggs, New England Biolabs, Inc., unpublished data). Several minor bands of smaller size were also apparent, which are presumed to be breakdown products of MBP and the fusion proteins. Similar degradation has been observed previously with PapG and FimH fusions to MBP (11, 33, 37). The MBP fusion proteins included a site for cleavage by the protease Factor Xa (Fig. 1). Cleavage of the fusion proteins yielded two fragments of the predicted sizes (cleavage of MBP-DraE is shown in Fig. 2, lane 6). Controls of purified Dr fimbriae and F1845 fimbriae showed an expected size of 15 kDa (Fig. 2, lanes 1 and 2).
FIG. 2.
SDS-PAGE of fimbriae and major structural subunit fusion proteins. Purified fimbrial and fusion proteins were prepared as described in the text. After electrophoresis in 15% acrylamide, the proteins were stained with Coomassie blue. Lanes: 1, F1845 fimbriae; 2, Dr fimbriae; 3, MBP; 4, MBP-DaaE; 5, MBP-DraE; 6, MBP-DraE cleaved with Factor Xa; 7, DaaE-His; 8, DraE-His. Size standards are indicated on the left in kilodaltons.
The oligohistidine fusions were purified from inclusion bodies under denaturing conditions with 6 M urea and nickel affinity chromatography. Upon stepwise dialysis for removal of the denaturant, a large fraction of the total protein formed insoluble aggregates. Approximately one-third of the total protein retained solubility, and this was used for further analysis. DraE-His and DaaE-His were visualized at an expected size of ca. 18 kDa by SDS-PAGE (Fig. 2, lanes 7 and 8). Oligohistidine fusions of these proteins were also constructed in which the C-terminal nine amino acids of the fimbrial subunits were removed (DraECt-His and DaaECt-His) (Fig. 1).
The different fimbrial and fusion proteins were also examined by nondenaturing gel electrophoresis. The fimbrial proteins formed high-molecular-weight multimers as expected. The MBP fusions formed multimers similar to the fimbrial proteins, but the oligohistidine fusions only formed low-molecular-weight multimers (data not shown).
Erythrocyte-binding assay for functional analysis of adhesion.
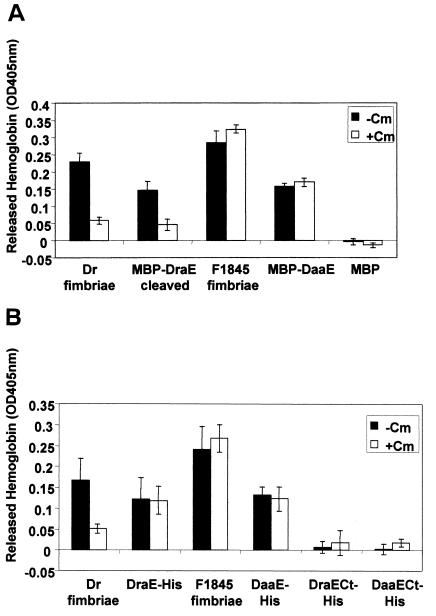
In order to examine the role of the major structural subunits in adhesion, an erythrocyte-binding assay was developed to test whether DraE and DaaE bind to erythrocytes. This assay tested the ability of immobilized fimbrial subunit fusion proteins to bind human erythrocytes. The fusion proteins were bound to microtiter plates and incubated with a suspension of human erythrocytes. Bound erythrocytes were quantified by lysing the cells and measuring released hemoglobin.
In this assay, the fimbriae and all of the full-length fusion proteins bound human erythrocytes, but with distinct properties (Fig. 3). MBP-DraE only bound erythrocytes upon Factor Xa cleavage, whereas MBP-DaaE bound erythrocytes with or without cleavage by Factor Xa (Fig. 3A and data not shown). Erythrocyte binding by Dr fimbriae and MBP-DraE cleaved with Factor Xa was inhibited by chloramphenicol, as has been shown for E. coli expressing the Dr hemagglutinin (6, 29, 36). However, DraE-His erythrocyte binding was not inhibited by chloramphenicol. As expected, erythrocyte binding by F1845 fimbriae and MBP-DaaE was not inhibited by chloramphenicol. Purified MBP and MBP cleaved with Factor Xa did not bind erythrocytes (Fig. 3A and data not shown). The truncated DraECt-His and DaaECt-His proteins were also examined in this assay but, unlike the full-length oligohistidine fusions, these did not bind erythrocytes, demonstrating that binding is not mediated by the oligohistidine tag (Fig. 3B). The binding observed by the fusion proteins was specific for human erythrocytes since none of the proteins tested bound to sheep or guinea pig erythrocytes (data not shown). Additionally, binding of erythrocytes to fimbriae and fusion proteins was inhibited by preincubating human erythrocytes with anti-DAF antibodies specific for SCR3 (data not shown). Taken together, these results demonstrate that DraE and DaaE can bind to human erythrocytes in a specific manner and that this binding is DAF dependent.
FIG. 3.
Erythrocyte-binding assay. Microtiter wells were coated with 1 μg of each purified protein, and 100 μl of a 2% erythrocyte suspension was added to each well. Unbound erythrocytes were then washed off, and the remaining erythrocytes were lysed with 100 μl of water. Released hemoglobin was quantified by determination of the OD405. A final concentration of 3.7 mM chloramphenicol was used. (A) Purified fimbriae and MBP fusion proteins. (B) Purified fimbriae and oligohistidine fusion proteins. Cm, chloramphenicol; MBP-DraE cleaved, MBP-DraE cleaved with Factor Xa. Each protein sample was examined in triplicate in at least three independent experiments. Representative averages from triplicate wells with standard deviations are shown.
CHO cell-binding assay for analysis of specificity of DAF binding.
In order to directly examine whether DraE and DaaE can mediate cell binding via the DAF receptor, the fusion proteins were analyzed for binding to CHO cell transfectants expressing DAF (CHO DAF+) or cells transfected with vector alone (CHO DAF−). Fusion proteins were bound to microtiter plates and incubated with CHO DAF+ and CHO DAF− cells. Bound cells were lysed and quantified by measuring released LDH. Fimbriae and the full-length fusion proteins bound to CHO DAF+ cells but not to CHO DAF− cells (Fig. 4). As with erythrocytes, MBP-DraE bound to CHO DAF+ cells only after cleavage with Factor Xa, whereas MBP-DaaE bound these cells with or without cleavage by Factor Xa (Fig. 4 and data not shown). MBP with or without cleavage by Factor Xa did not bind to CHO DAF+ cells (data not shown and Fig. 4). The MBP fusions showed a low level of binding to CHO DAF− cells, but this binding was not considered specific because MBP alone also showed the same low level of binding to these cells. The truncated DraECt-His and DaaECt-His fusions did not bind CHO DAF+ cells (data not shown). Therefore, DraE and DaaE specifically recognize and bind to DAF on the surface of CHO cells.
FIG. 4.
CHO cell-binding assay. Microtiter plates were coated with 2 μg of each purified protein, and 100 μl of a 3 × 105 cells/ml suspension of CHO cells transfected with DAF (CHO DAF+) or control CHO cells without DAF (CHO DAF−) was added to each well. Bound cells were lysed and quantified by detection of released LDH. MBP-DraE cleaved, MBP-DraE cleaved with Factor Xa. Each protein sample was examined in triplicate in at least three independent experiments. Representative averages from triplicate wells with the standard deviations are shown.
DAF-binding EIA for analysis of DAF binding.
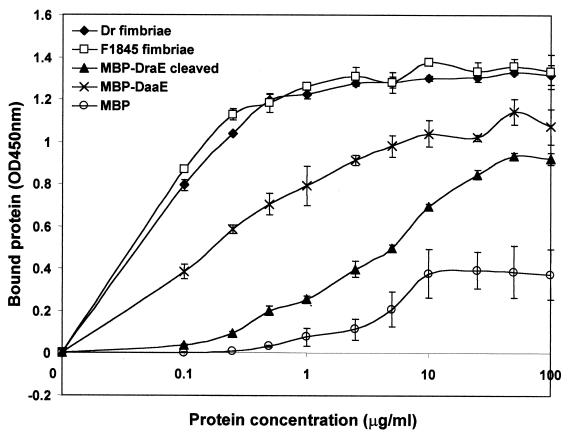
The assays described above demonstrate specific binding of DraE and DaaE to DAF expressed on the surface of cells. To examine binding to DAF in the absence of other cellular components, we employed a purified DAF fusion protein (32). DAF234 contains SCR2, SCR3, and SCR4 of DAF with an oligohistidine tag fused to its C-terminal end. This fusion was bound to the wells of a microtiter plate. Fimbriae or fusion proteins were added, and bound proteins were detected with the corresponding antibodies. As shown in Fig. 5, fimbriae and the full-length fusions bound to DAF234. In contrast to the results of the erythrocyte and CHO cell-binding assays, MBP-DraE bound to DAF234 with or without cleavage by Factor Xa. MBP-DaaE also bound to DAF234 with or without cleavage by Factor Xa (data not shown and Fig. 5). MBP, MBP cleaved with Factor Xa, DraECt-His, and DaaECt-His bound to DAF234 at a very low level. The binding of MBP-DraE cleaved with Factor Xa and Dr fimbriae was partially inhibited by chloramphenicol. F1845 fimbriae, MBP-DaaE, DaaE-His, and DraE-His showed no chloramphenicol inhibition. These results demonstrate that specific binding by DraE and DaaE to DAF does not require additional cellular components. MBP-DaaE, MBP-DraE cleaved with Factor Xa, and the full-length oligohistidine fusions exhibited dose dependent and saturable binding to DAF, although the binding affinity was reduced relative to Dr and F1845 fimbriae (Fig. 6 and data not shown).
FIG. 5.
DAF-binding EIA. Microtiter wells were coated with 3 μg of DAF234, and 10 μg of each of the different purified protein samples was added to each well. The bound proteins were detected with antibodies to the fimbrial adhesins, to MBP, or to the HSV epitope tag. A final concentration of 2.5 mM chloramphenicol was used. Cm, chloramphenicol; MBP-DraE cleaved, MBP-DraE cleaved with Factor Xa; MBP-DaaE cleaved, MBP-DaaE cleaved with Factor Xa; MBP cleaved, MBP cleaved with Factor Xa. The experiment was performed in triplicate. Results shown are the averages of triplicate wells with the standard deviations from a representative experiment.
FIG. 6.
Dose-dependent binding to DAF. Microtiter wells were coated with 1.5 μg of DAF234, and various concentrations of the different protein samples were added to each well. The bound proteins were detected with antibodies to the fimbrial adhesins and to MBP. MBP-DraE cleaved, MBP-DraE cleaved with Factor Xa. The experiment was performed in triplicate. Results shown are the averages of duplicate wells with the standard deviations from a representative experiment.
Rosette agglutination assay for analysis of adherence.
We used the fimbrial subunit fusions to test whether DraE and DaaE can agglutinate human erythrocytes, as has been shown with bacteria expressing Dr and F1845 (5, 6). Bacterial suspensions of DH5α(pCC90) expressing Dr fimbriae and of DH5α(pSSS1) expressing F1845 fimbriae, suspensions of the fimbriae, and fusion protein solutions were incubated with human erythrocytes in U-bottom microtiter plates and examined for rosette formation as an indication of agglutination. The two positive controls, DH5α(pCC90) and DH5α(pSSS1), were positive for rosette formation, but they showed different properties. DH5α(pSSS1) formed rosettes at a 1:8 dilution, whereas DH5α(pCC90) formed rosettes at a 1:2 dilution. These results may be due to differences in the expression levels of the fimbriae on the bacterial cell; therefore, purified fimbriae were examined for rosette formation. Purified F1845 fimbriae formed rosettes; however, purified Dr fimbriae did not under the same conditions. None of the fusion proteins (MBP-DraE, MBP-DaaE, DraE-His, and DaaE-His) was able to form rosettes. The fusion proteins did not inhibit rosette formation by whole bacteria expressing Dr and F1845 fimbriae at concentrations up to 100 μg/ml.
Analysis of type IV collagen binding by the adhesive subunit fusions.
Since the fusion proteins are functional for DAF binding, we investigated whether DraE is able to bind type IV collagen, the second receptor for the Dr fimbriae (6, 40). This is a property unique to this member of the Dr family of adhesins. In order to analyze binding of DraE to type IV collagen, the fusion proteins were examined in a collagen-binding EIA similar to the assay used for analysis of DAF234 binding. With this assay, low levels of binding to type IV collagen were detected by MBP-DraE, MBP-DraE cleaved with Factor Xa, and MBP (Fig. 7). No binding was detected by MBP cleaved with Factor Xa, DraE-His, and DraECt-His. This was in contrast to Dr fimbriae that showed a high level of binding to type IV collagen that was inhibited by the presence of chloramphenicol. The low levels of binding exhibited by the DraE-MBP fusions do not appear to be reflective of the binding properties of the Dr hemagglutinin since F1845 fimbriae and the DaaE fusions also showed a similar, low level of collagen binding (data not shown).
FIG. 7.
Type IV collagen-binding EIA. Microtiter wells were coated with 2 μg of type IV collagen, and 10 μg of each of the different purified protein samples was added to each well. The bound proteins were detected with antibodies to the fimbrial adhesins, to MBP, and to the HSV epitope tag. A final concentration of 2.5 mM chloramphenicol was used. Cm, chloramphenicol; MBP-DraE cleaved, MBP-DraE cleaved with Factor Xa; MBP cleaved, MBP cleaved with Factor Xa. The experiment was performed in triplicate. Results shown are the averages of triplicate wells with the standard deviations from a representative experiment.
An alternate assay in which the fusion proteins were bound to the wells of a microtiter plate and type IV collagen was added and detected also indicated that the DraE fusion proteins did not bind type IV collagen (data not shown). Therefore, although purified DraE subunits are able to bind DAF, they cannot specifically bind type IV collagen.
DISCUSSION
Previous studies with the Dr family of adhesins suggested that the major structural subunits of the different members were also the adhesive subunits, but this has not been directly shown (6, 10, 15, 36). In this study we demonstrated that DraE and DaaE are the adhesive subunits of the Dr and F1845 fimbriae, respectively. We have shown that DraE and DaaE bind to DAF expressed on the surface of cells and to a purified DAF-oligohistidine fusion. This adhesive property is independent of the method of purification, since fusions to both MBP and to oligohistidine tags bound to DAF.
Protein fusions to MBP and to oligohistidine tags have provided valuable tools for the study of fimbrial adhesins. These fusion domains were used successfully with PapG and FimH to examine their adhesive properties (11, 34, 37). In this study, fusions of DraE and DaaE to MBP and an oligohistidine tag also provided soluble proteins that were functional. The MBP fusions were efficiently targeted to the periplasm, which probably allowed proper folding, including the correct formation of the disulfide bond within each protein (6). Although the MBP fusion constructs contained a site for Factor Xa cleavage, we have not been able to purify DraE and DaaE away from MBP after complete cleavage of the fusion proteins. The possible aggregative nature of these fimbrial proteins may trap MBP following Factor Xa cleavage.
The erythrocyte-binding assay that was developed revealed that the fusion proteins bound specifically to human erythrocytes and that binding was inhibited by anti-DAF antibodies. Further evidence that the observed binding specificity is due to binding to DAF is provided by the observation that the fusions bound only to CHO DAF+ cells. The fusions do, however, appear to show different properties. MBP-DraE required cleavage by Factor Xa in order to preserve its cell-binding activity, whereas MBP-DaaE did not. This may correlate with the properties of the native fimbriae in which bacteria expressing F1845 fimbriae are stronger hemagglutinators than are bacteria expressing Dr fimbriae, as demonstrated by the rosette assay. Furthermore, purified F1845 fimbriae formed rosettes, whereas purified Dr fimbriae did not, indicating a different interaction of these fimbriae with erythrocytes. The DraE structure may be weaker and therefore more sensitive to the addition of large protein domains than the DaaE structure. The erythrocyte-binding assay also showed that MBP-DraE binding could be inhibited by chloramphenicol but that MBP-DaaE binding could not, as has been shown for the corresponding Dr and F1845 fimbriae (6, 29, 36). These results further establish that there are functional differences between the Dr and F1845 fimbriae in their binding phenotypes, and these differences are reflected in the binding phenotypes of the individual structural subunits.
The full-length oligohistidine fusions also bound specifically to human erythrocytes. In contrast to Dr fimbriae and the MBP-DraE fusion, DraE-His binding was not inhibited in the presence of chloramphenicol. This result was also observed in the DAF-binding EIA. It is possible that stepwise dialysis may not have yielded correct folding of the adhesin, and this may have affected the interaction with chloramphenicol but not with DAF. This is supported by previous results that have shown that mutations in DraE can affect chloramphenicol inhibition of binding but not the ability to bind DAF, indicating that these are separable phenotypes (6). The mechanism of inhibition by chloramphenicol has not been defined.
Truncated oligohistidine fusions were also generated without the nine C-terminal amino acids of the mature fimbrial subunits. As opposed to the full-length oligohistidine fusions, they did not bind human erythrocytes and they showed low levels of binding to DAF. These results demonstrate that the oligohistidine tag is not responsible for the observed binding. They also suggest that the C-terminal domain of DraE and DaaE may be important for receptor binding. It may contain residues that directly interact with DAF, or it may be important indirectly by providing the proper conformation of the binding site.
The fusion proteins exhibited a lower level of binding than the purified Dr and F1845 fimbriae in both the erythrocyte-binding assay and the CHO cell-binding assay. In particular, this is clearly demonstrated in the DAF-binding EIA, where the fimbriae show saturated binding to DAF at a lower concentration than the fusion proteins. Additionally, the rosette agglutination assays showed that none of the fusion proteins agglutinated human erythrocytes. These assays suggest that the fusion proteins have a lower affinity for DAF. The fusion proteins are most likely monovalent structures that are not capable of hemagglutination. Subunits in fimbriae are in their native conformation and, therefore, may display their maximal binding properties, whereas conformation of the fusion proteins may not be optimal for receptor binding. It is also possible that the presence of additional subunits in the native fimbrial structure may increase the affinity of binding for the DAF receptor.
The DAF-binding EIA shows directly that the DraE and DaaE fusions are able to bind to a DAF fusion protein that contains SCR2, SCR3, and SCR4 and that this binding is dose dependent and saturable. This correlates with previous work showing that SCR2 and SCR3 are important for binding of the Dr and F1845 fimbriae, whereas SCR1 is not necessary (26). This also demonstrates that no additional cellular components are required for binding of the adhesins to DAF. In contrast to the cell-binding assays, MBP-DraE bound to DAF without cleavage by Factor Xa. This may be due to the nature of the assay: the cell-binding assays may require a greater affinity for DAF than the DAF-binding EIA. As in the erythrocyte-binding assay, binding by Dr fimbriae and MBP-DraE was inhibited by chloramphenicol but to a lesser extent. It is possible that chloramphenicol cannot block a higher-affinity interaction of purified DAF and the adhesin in the same manner as with DAF expressed on the surface of cells.
The collagen-binding EIA results showed that, in contrast to the Dr fimbriae, the DraE fusions did not bind at a high level to type IV collagen. Several hypotheses could explain these results. First, binding of the Dr fimbriae to type IV collagen may require a conformational domain comprised of two or more subunits, and this was not obtained through the purification of the DraE fusions. That purified Dr fimbriae were able to bind type IV collagen at a high level in a specific manner in the assays tested supports this possibility. Second, it is possible that an additional protein may be required for type IV collagen binding that is not required for DAF binding. No such candidate protein has been identified thus far. It has been shown that AfaD-III, a homolog of DraD, is expressed on the bacterial surface and functions as an adhesin, but no specific receptor has been assigned for this protein (8, 15). These possibilities remain to be explored.
In summary, the results presented here demonstrate that DraE and DaaE are the adhesive subunits for the DAF receptor of the Dr and F1845 fimbriae, but for type IV collagen a more complex binding structure may be required. Furthermore, DraE and DaaE possess distinct binding properties that are shared by Dr and F1845 fimbriae. The fusion proteins described will provide new tools for structure-function analysis and for further characterization of the binding properties of the Dr family of adhesins.
Acknowledgments
This work was supported by grant DK49862 from the National Institute of Diabetes and Digestive and Kidney Diseases to S.L.M. and grants RO1 AI45820 and RO1 GM60731 from the National Institutes of Health to E.V.S. C.P.V.L.'s financial support was provided by the National Science Foundation Minority Fellowship and the American Society for Microbiology Robert D. Watkins Minority Fellowship.
We thank Douglas Lublin for providing CHO cell transfectants. We thank Susan Lea for providing P. pastoris DAF234. We also thank Steve Lory, Richard Darveau, and Phillip Tarr for critical reading of the manuscript.
REFERENCES
- 1.Arthur, M., C. E. Johnson, R. H. Rubin, R. D. Arbeit, C. Campanelli, C. Kim, S. Steinbach, M. Agarwal, R. Wilkinson, and R. Goldstein. 1989. Molecular epidemiology of adhesin and hemolysin virulence factors among uropathogenic Escherichia coli. Infect. Immun. 57:303-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bakker, D., P. T. Willemsen, L. H. Simons, F. G. van Zijderveld, and F. K. de Graaf. 1992. Characterization of the antigenic and adhesive properties of FaeG, the major subunit of K88 fimbriae. Mol. Microbiol. 6:247-255. [DOI] [PubMed] [Google Scholar]
- 4.Berghammer, H., and B. Auer. 1993. “Easypreps”: fast and easy plasmid minipreparation for analysis of recombinant clones in E. coli. BioTechniques 14:524-528. [PubMed] [Google Scholar]
- 5.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carnoy, C., and S. L. Moseley. 1997. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol. Microbiol. 23:365-379. [DOI] [PubMed] [Google Scholar]
- 7.Di Martino, P., J. P. Girardeau, M. Der Vartanian, B. Joly, and A. Darfeuille-Michaud. 1997. The central variable V2 region of the CS31A major subunit is involved in the receptor-binding domain. Infect. Immun. 65:609-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia, M. I., P. Gounon, P. Courcoux, A. Labigne, and C. Le Bouguenec. 1996. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol. Microbiol. 19:683-693. [DOI] [PubMed] [Google Scholar]
- 9.Giron, J. A., T. Jones, F. Millan-Velasco, E. Castro-Munoz, L. Zarate, J. Fry, G. Frankel, S. L. Moseley, B. Baudry, J. B. Kaper, et al. 1991. Diffuse-adhering Escherichia coli (DAEC) as a putative cause of diarrhea in Mayan children in Mexico. J. Infect. Dis. 163:507-513. [DOI] [PubMed] [Google Scholar]
- 10.Gounon, P., M. Jouve, and C. Le Bouguenec. 2000. Immunocytochemistry of the AfaE adhesin and AfaD invasin produced by pathogenic Escherichia coli strains during interaction of the bacteria with HeLa cells by high-resolution scanning electron microscopy. Microbes Infect. 2:359-365. [DOI] [PubMed] [Google Scholar]
- 11.Haslam, D. B., T. Boren, P. Falk, D. Ilver, A. Chou, Z. Xu, and S. Normark. 1994. The amino-terminal domain of the P-pilus adhesin determines receptor specificity. Mol. Microbiol. 14:399-409. [DOI] [PubMed] [Google Scholar]
- 12.Hung, D. L., and S. J. Hultgren. 1998. Pilus biogenesis via the chaperone/usher pathway: an integration of structure and function. J. Struct. Biol. 124:201-220. [DOI] [PubMed] [Google Scholar]
- 13.Jallat, C., V. Livrelli, A. Darfeuille-Michaud, C. Rich, and B. Joly. 1993. Escherichia coli strains involved in diarrhea in France: high prevalence and heterogeneity of diffusely adhering strains. J. Clin. Microbiol. 31:2031-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, C. H., J. S. Pinkner, R. Roth, J. Heuser, A. V. Nicholes, S. N. Abraham, and S. J. Hultgren. 1995. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 92:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouve, M., M. I. Garcia, P. Courcoux, A. Labigne, P. Gounon, and C. Le Bouguenec. 1997. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect. Immun. 65:4082-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogfelt, K. A., H. Bergmans, and P. Klemm. 1990. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect. Immun. 58:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehn, M. J., J. Heuser, S. Normark, and S. J. Hultgren. 1992. P pili in uropathogenic E. coli are composite fibers with distinct fibrillar adhesive tips. Nature 356:252-255. [DOI] [PubMed] [Google Scholar]
- 18.Le Bouguenec, C., M. I. Garcia, V. Ouin, J. M. Desperrier, P. Gounon, and A. Labigne. 1993. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect. Immun. 61:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine, M. M., C. Ferreccio, V. Prado, M. Cayazzo, P. Abrego, J. Martinez, L. Maggi, M. M. Baldini, W. Martin, D. Maneval, et al. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849-869. [DOI] [PubMed] [Google Scholar]
- 20.Lindberg, F., B. Lund, L. Johansson, and S. Normark. 1987. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature 328:84-87. [DOI] [PubMed] [Google Scholar]
- 21.Lublin, D. M., and J. P. Atkinson. 1989. Decay-accelerating factor: biochemistry, molecular biology, and function. Annu. Rev. Immunol. 7:35-58. [DOI] [PubMed] [Google Scholar]
- 22.Lublin, D. M., and K. E. Coyne. 1991. Phospholipid-anchored and transmembrane versions of either decay-accelerating factor or membrane cofactor protein show equal efficiency in protection from complement-mediated cell damage. J. Exp. Med. 174:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lund, B., F. Lindberg, B. I. Marklund, and S. Normark. 1987. The PapG protein is the α-d-galactopyranosyl-(1-4)-β-d-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 84:5898-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madison, B., I. Ofek, S. Clegg, and S. N. Abraham. 1994. Type 1 fimbrial shafts of Escherichia coli and Klebsiella pneumoniae influence sugar-binding specificities of their FimH adhesins. Infect. Immun. 62:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson-Weller, A., and C. E. Wang. 1994. Structure and function of decay-accelerating factor CD55. J. Lab Clin. Med. 123:485-491. [PubMed] [Google Scholar]
- 26.Nowicki, B., A. Hart, K. E. Coyne, D. M. Lublin, and S. Nowicki. 1993. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J. Exp. Med. 178:2115-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowicki, B., A. Labigne, S. Moseley, R. Hull, S. Hull, and J. Moulds. 1990. The Dr hemagglutinin, afimbrial adhesins AFA-I and AFA-III, and F1845 fimbriae of uropathogenic and diarrhea-associated Escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect. Immun. 58:279-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowicki, B., M. Martens, A. Hart, and S. Nowicki. 1994. Gestational age-dependent distribution of Escherichia coli fimbriae in pregnant patients with pyelonephritis. Ann. N. Y. Acad. Sci. 730:290-291. [DOI] [PubMed] [Google Scholar]
- 29.Nowicki, B., J. Moulds, R. Hull, and S. Hull. 1988. A hemagglutinin of uropathogenic Escherichia coli recognizes the Dr blood group antigen. Infect. Immun. 56:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowicki, B., C. Svanborg-Eden, R. Hull, and S. Hull. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect. Immun. 57:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nowicki, B., L. Truong, J. Moulds, and R. Hull. 1988. Presence of the Dr receptor in normal human tissues and its possible role in the pathogenesis of ascending urinary tract infection. Am. J. Pathol. 133:1-4. [PMC free article] [PubMed] [Google Scholar]
- 32.Powell, R. M., T. Ward, D. J. Evans, and J. W. Almond. 1997. Interaction between echovirus 7 and its receptor, decay-accelerating factor (CD55): evidence for a secondary cellular factor in A-particle formation. J. Virol. 71:9306-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riggs, P. 2000. Expression and purification of recombinant proteins by fusion to maltose-binding protein. Mol. Biotechnol. 15:51-63. [DOI] [PubMed] [Google Scholar]
- 34.Schembri, M. A., H. Hasman, and P. Klemm. 2000. Expression and purification of the mannose recognition domain of the FimH adhesin. FEMS Microbiol. Lett. 188:147-151. [DOI] [PubMed] [Google Scholar]
- 35.Stapleton, A., S. Moseley, and W. E. Stamm. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J. Infect. Dis. 163:773-779. [DOI] [PubMed] [Google Scholar]
- 36.Swanson, T. N., S. S. Bilge, B. Nowicki, and S. L. Moseley. 1991. Molecular structure of the Dr adhesin: nucleotide sequence and mapping of receptor-binding domain by use of fusion constructs. Infect. Immun. 59:261-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thankavel, K., B. Madison, T. Ikeda, R. Malaviya, A. H. Shah, P. M. Arumugam, and S. N. Abraham. 1997. Localization of a domain in the FimH adhesin of Escherichia coli type 1 fimbriae capable of receptor recognition and use of a domain-specific antibody to confer protection against experimental urinary tract infection. J. Clin. Investig. 100:1123-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thankavel, K., A. H. Shah, M. S. Cohen, T. Ikeda, R. G. Lorenz, R. Curtiss III, and S. N. Abraham. 1999. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J. Biol. Chem. 274:5797-5809. [DOI] [PubMed] [Google Scholar]
- 39.Van den Broeck, W., E. Cox, B. Oudega, and B. M. Goddeeris. 2000. The F4 fimbrial antigen of Escherichia coli and its receptors. Vet. Microbiol. 71:223-244. [DOI] [PubMed] [Google Scholar]
- 40.Westerlund, B., P. Kuusela, J. Risteli, L. Risteli, T. Vartio, H. Rauvala, R. Virkola, and T. K. Korhonen. 1989. The O75X adhesin of uropathogenic Escherichia coli is a type IV collagen-binding protein. Mol. Microbiol. 3:329-337. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, L., B. Foxman, P. Tallman, E. Cladera, C. Le Bouguenec, and C. F. Marrs. 1997. Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coli isolates and identification of new subtypes. Infect. Immun. 65:2011-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]