Abstract
Anaplasma (Ehrlichia) phagocytophila and Ehrlichia chaffeensis, the etiologic agents of granulocytic and monocytic ehrlichioses, respectively, are obligatory intracellular bacteria that cause febrile systemic illness in humans. We identified and characterized clusters of genes for a type IV secretion machinery in these two bacteria, and analyzed their gene expression in cell culture and mammalian hosts. Eight virB and virD genes were found in each bacterial genome, and all of the genes were transcribed in cell culture. Although the gene order and orientation were similar to those found in other bacteria, the eight virB and virD genes were clustered at two separate loci in each genome. Five of the genes (virB8, virB9, virB10, virB11, and virD4) were located downstream from a ribA gene. These five genes in both A. phagocytophila and E. chaffeensis were polycistronically transcribed and controlled through at least two tandem promoters located upstream of the virB8 gene in human leukemia cell lines. The virB9 gene of A. phagocytophila was transcriptionally active in peripheral blood leukocytes from human ehrlichiosis patients and experimentally infected animals. Three of the remaining genes (virB3, virB4, and virB6) of both A. phagocytophila and E. chaffeensis were arranged downstream from a sodB gene and cotranscribed with the sodB gene through one or more sodB promoters in human leukocytes. This suggests that transcription of the three virB genes in these two Anaplasma and Ehrlichia spp. is regulated by factors that influence the sodB gene expression. This unique regulation of gene expression for the type IV secretion system may be associated with intracellular survival and replication of Anaplasma and Ehrlichia spp. in granulocytes or monocytes.
Human granulocytic ehrlichiosis (HGE) and human monocytic ehrlichioses (HME) are newly discovered tick-borne febrile illnesses of increasing importance in the United States (1, 7) and have been recognized in several countries in Europe and Africa and in Mexico. The clinical signs of HGE and HME are similar. The patients frequently require hospitalization, and they may die when the treatment is delayed or they are immunocompromised (15, 30). HGE and HME are caused by obligatory intracellular bacteria, Anaplasma (Ehrlichia) phagocytophila (7, 13) and Ehrlichia chaffeensis (1), which have tropism for granulocytes and monocytes, respectively. These agents reside and replicate within membrane-bound inclusions (parasitophorous vacuoles) in the cytoplasm of phagocytes. The 16S rRNA gene sequences of A. phagocytophila and E. chaffeensis are 7.5% divergent (7), and their ultrastructures, antigenic compositions, inclusion compartments, sensitivities to intracellular iron depletion, and properties of the major outer membrane protein gene family are distinct (5, 6, 20, 24, 25, 28, 29, 32, 42-44).
Type IV secretion systems are ancestrally related to the bacterial conjugal system and are thought to function to deliver effector macromolecules produced by parasitic or symbiotic bacteria into eukaryotic target cells. A set of genes orthologous to virB and virD genes for a type IV secretion machinery of Agrobacterium tumefaciens, a phytopathogen, has been found in chromosomes or plasmids of more than a dozen different gram-negative bacterial species to date (8-10, 14). The vir transport system of Agrobacterium tumefaciens mediates the transfer of T-DNA on the Ti plasmid into the nuclei of infected plant cells, and the integration of this DNA into the plant genome leads to a plant tumor (16). The virB operon (containing 11 virB genes) and the virD4 gene encode the components of the type IV secretion machinery, and the assembled gene products serve as a transporter for delivering the substrate complex (T-DNA, VirE2, VirD2, and VirF) from Agrobacterium tumefaciens into recipient plant cells. The T-DNA transport system is related to conjugal systems (Tra, Trw, and Trb) of the pKM101 (IncN), R388 (IncW), RP4 (IncP), and F (IncF) plasmids in Escherichia coli in sequence similarity and structure-function studies (9, 10). For obligate intracellular bacteria, the genes for a type IV secretion machinery have been described only for Rickettsia prowazekii, the agent of epidemic typhus (2); Rickettsia conorii, the agent of Mediterranean spotted fever (23); and Wolbachia spp., symbionts of aphids (19). In R. prowazekii and R. conorii, 16 vir genes (1 virB3, 2 virB4, 5 virB6, 1 virB7, 2 virB8, 2 virB9, 1 virB10, 1 virB11, and 1 virD4) were identified by genome sequencing and further analysis (2, 23, 37), but the transcriptional activities of these genes have not been examined. In Wolbachia spp., five vir genes (virB8, -B9, -B10, -B11, and -D4) were identified, and these genes were polycistronically transcribed (19); however, other vir genes have not been reported, and the promoter region for the wolbachial virB8-D4 transcript has not been determined. In the present study, we identified and characterized (i) eight vir genes (virB3, -B4, -B6, -B8, -B9, -B10, -B11, and -D4) in two distinct and major human ehrlichiosis agents, A. phagocytophila (causing HGE) and E. chaffeensis (causing HME); (ii) the transcriptional mode of the vir genes of these two bacteria in cell culture, human patients, and experimentally infected animals; and (iii) the promoter regions for the transcription of the vir gene clusters. A unique feature found in both A. phagocytophila and E. chaffeensis is the cotranscription of a superoxide dismutase gene (sodB) and three virB genes (virB3, -B4, and -B6) through sodB promoters. This is the first description and expression analysis of the type IV secretion system in the genera Anaplasma and Ehrlichia.
MATERIALS AND METHODS
Organisms and culture.
A. phagocytophila (strains HZ and LL and isolates I-NY31, I-NY36, and I-NY37) was cultivated in HL-60 cells as described elsewhere (32, 44; Q. Lin, N. Zhi, N. Ohashi, H. W. Horowitz, M. E. Auero-Rosenfeld, G. P. Wormser, John Rattalli, and Y. Rikihisa, submitted for publication). Strain HZ was used in all experiments, and four other strains and isolates were used for an experiment for Fig. 3. E. chaffeensis strain Arkansas was grown in DH82 cells (canine monocytic cell line) or THP-1 cells as described previously (6, 31). These organisms were purified from the infected cells by Sephacryl S-1000 chromatography (31) or Percoll density gradient centrifugation (25) for preparation of genomic DNA.
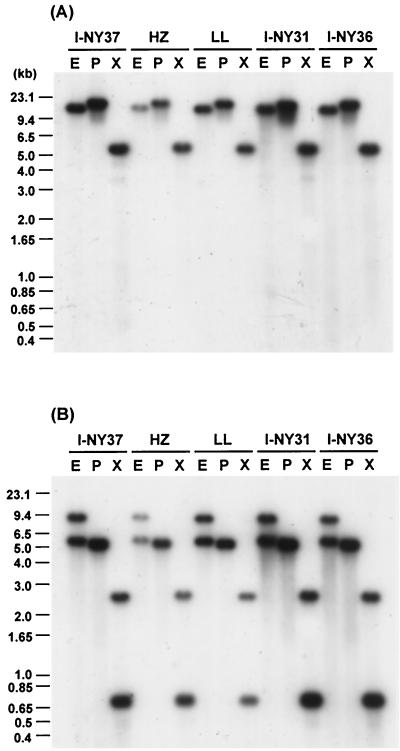
FIG. 3.
Comparative genomic Southern blot analyses of virB and virD loci among strains HZ and LL and three human isolates of A. phagocytophila. (A) virB9 (probe E); (B) virB4 (probe B). The positions of the probes on the maps are shown in Fig. 1. E, EcoRI; P, PstI; X, XbaI. The labels above each panel indicate A. phagocytophila strains (HZ and LL) or isolates (I-NY31, I-NY36, and I-NY37) from which genomic DNAs were prepared.
Cloning of the genes for a type IV secretion machinery in A. phagocytophila and E. chaffeensis.
We designed several degenerate primer pairs based on the conserved sequences of virB4 and virB11 between Agrobacterium tumefaciens (33) and R. prowazekii (2) for PCR. By using these degenerate primers, DNA fragments containing partial sequences of these two genes were amplified from each genomic DNA of A. phagocytophila and E. chaffeensis, cloned into a pCRII vector (Invitrogen-Life Technology, San Diego, Calif.), and sequenced using the dideoxy chain termination method. For assembly of the entire vir region, the overlapping DNA fragments with unknown flanking sequences, which were amplified by the adapter PCR method using the Universal Genome Walker kit (Clontech Laboratories, Palo Alto, Calif.) or by multiplex restriction site PCR (4, 17, 39), were cloned into a pCRII vector and sequenced.
Genomic Southern blot analysis.
PCR products amplified with the primer pairs shown in Table 1 were labeled with [α-32 P]dATP by using the random primer method (Amersham Pharmacia Biotech, Piscataway, N.J.) and used as gene-specific DNA probes. Southern blot analyses with genomic DNAs from purified organisms were performed by a procedure described previously (25). Hybridization was carried out under high-stringency conditions (65°C), and after being washed, the membrane was exposed to a Hyperfilm (Amersham Pharmacia Biotech).
TABLE 1.
Oligonucleotide primers used in RT-PCR, 5′RACE, and Southern blotting
| Target | Amplicon size (bp)a | 5′ primer (5′RACE primer 2) | 3′ primer (5′RACE primer 1) | RT primer | Purpose |
|---|---|---|---|---|---|
| A. phagocytophila | |||||
| virB4 | 268 | 5′-TCTCTTATGTTGTACGTAAAG-3′ | 5′-AACACCCGCGGAGAAAACTT-3′ | Southern blotting (probe B) | |
| virB9 | 358 | 5′-TGGACAACAAGTTATACGTC-3′ | 5′-AGAGAAACGTGAGCACTTC-3′ | Random hexamer | Southern blotting (probe E), RT-PCR |
| virB11 | 190 | 5′-CTCGAGGGAAATTGTCCTAT-3′ | 5′-CCCAGTATTAATAGCCCTCA-3′ | Southern blotting (probe A) | |
| virB8-virD4 | 5,979 | 5′-AAGATCGGAGATTGGTATCATGACAGATAC-3′ | 5′-ACAACCTCATCATCTAAAAGCTCATCCTCA-3′ | 5′-TCTTCTCCAAATTCATCC-3′ | LA-RT-PCR |
| virD4-p44-22 omp | 557 | 5′-GATAGCAGTAACGGAAGAC-3′ | 5′-CCTAGATAACAGGTCTCCA-3′ | Random hexamer | RT-PCR |
| sodB-virB4 | 3,240 | 5′-AGCCGTATATTTCTTCCCATCTCTTAGATC-3′ | 5′-TACCTGACAAAACGTTTATCGTGTCATCGA-3′ | 5′-AACCACACATTTGGATCA-3′ | LA-RT-PCR |
| virB4-virB6 | 1,995 | 5′-GCGAGTCGCATGATGATGTGGCA-3′ | 5′-GCAGCCATCACTGTCTTCTTAAGC-3′ | 5′-TCCCCTGCACTTTGTACC-3′ | LA-RT-PCR |
| Upstream of virB8 | 370 (407), 383 (420), 417 (454) | 5′-TACCTCGTCGGCAGAATACTGT-3′ | ACGAAAGAAGACGTACTCTCGT | 5′-GTTACTAGGACGTATCCA-3′ | 5′RACE |
| Upstream of sodB | 289 (326), 296 (333) | 5′-TCGGCTTCATGGATTGCCAATA-3′ | 5′-TACAAGCCATACCCATCCACTA-3′ | 5′-TTCGCTGTGCTAACAACT-3′ | 5′RACE |
| E. chaffeensis | |||||
| virB4 | 273 | 5′-TTTTGAATATATAGTTCGTAAAG-3′ | 5′-AAGAAAATCCTTGGTGAAAAC-3′ | Southern blotting (probe D) | |
| virB11 | 508 | 5′-TATTTCCTCCAGCCTGTGAA-3′ | 5′-GCCTGTATTAATTGCTCTTAA-3′ | Southern blotting (probe C) | |
| virB8-virD4 | 3,734 | 5′-GTGAGTTCCGTAACTGGATAAGGC-3′ | 5′-GGATTATGCCACCAGTGTTCTACG-3′ | 5′-CTTAGGATACCTATTGGT-3′ | LA-RT-PCR |
| sodB-virB4 | 3,370 | 5′-CATCTGGAGATTTAGGATCTAGAGC-3′ | 5′-TACTGCACCTACTCCCTGTTTCAC-3′ | 5′-ATTTGGATCATCTCCTACTTCT-3′ | LA-RT-PCR |
| virB4-virB6 | 3,258 | 5′-AGAACCAAGACATCCTACAGGATTC-3′ | 5′-GCCTGTTCTCCTACACTACCCTG-3′ | 5′-TGCAGCATCACCAGCTCCT-3′ | LA-RT-PCR |
| virB6-ORFa | 1,939 | 5′-AGGAATTCCTTTGTGGCCTCAAGTT-3′ | 5′-GTCTTCCAGCTAGTACCAAATCCG-3′ | 5′-ACCACATCTTTTGTATTATTAC-3′ | LA-RT-PCR |
| ribA | 294 | 5′-TGGTGACCTACTTAACAGC-3′ | 5′-ATGCCGTGATTCTGAATTTC-3′ | Random hexamer | RT-PCR |
| ribA-virB8 | 354 | 5′-GAAATTCAGAATCACGGCAT-3′ | 5′-ACGACTGACGTGCCAACT-3′ | Random hexamer | RT-PCR |
| pstA | 347 | 5′-ACATGGATCCTATTAGTGAG-3′ | 5′-TTGTTGATATGGAAGTTCCG-3′ | Random hexamer | RT-PCR |
| pstA-sodB | 351 | 5′-TGATAAGATACGATCCTTCC-3′ | 5′-TTGTTGATATGGAAGTTCCG-3′ | Random hexamer | RT-PCR |
| sodB-virB3 | 453 | 5′-TTGGGATTTTGCAGCTCAG-3′ | 5′-GCTTTAACAGTACCTGCCA-3′ | Random hexamer | RT-PCR |
| Upstream of virB8 | 356 (391), 462 (497) | 5′-TGAGTATTGTTTGACTGATACAGG-3′ | 5′-AGGGTCAAAAAGCTCTCTAGATC-3′ | 5′-ACTCACTGTATGTTGTTTG-3′ | 5′RACE |
| Upstream of sodB | 363 (397) | 5′-CCTGTAGGAGATCCTCCACCA-3′ | 5′-CATCCATGCTCCCAAAATCCTC-3′ | 5′-AAACCAACCATACCCATC-3′ | 5′RACE |
Sizes of 5′RACE amplicons are shown as base pairs from the transcriptional initiation site to 5′ end of 5′RACE primer 2. Numbers in parentheses include 35 to 37 bp of a 5′RACE abridged anchor primer.
Specimens from infected animals and humans.
Peripheral blood leukocytes (PBLs) from a mouse and a horse experimentally infected with strain HZ of A. phagocytophila were obtained on days 4 and 8 postinoculation, respectively, as previously described (43). PBLs were prepared from three HGE patients as described elsewhere (Lin et al., submitted).
Transcriptional analysis.
Reverse transcription-PCR (RT-PCR) was performed by a procedure described previously (24, 42). Total RNA was prepared from 5 × 106 A. phagocytophila-infected HL-60 cells (70% infectivity), 5 × 106 E. chaffeensis-infected THP-1 cells (70% infected cells), or the PBL specimens by using the TRIzol reagent (Invitrogen-Life Technologies) or the RNeasy Mini Kit (Qiagen, Valencia, Calif.). After DNase I treatment, the RNA (0.5 to 2.5 μg) was reverse transcribed using Superscript II (Invitrogen-Life Technologies) with random hexamer primers at 42°C for 50 min. The PCR conditions were 30 to 35 cycles consisting of 1 min of denaturation at 94°C, 1 min of annealing at 54°C, and 1 min of extension at 72°C. For cDNA synthesis of long transcripts (approximately 2 to 6 kb), the DNase I-treated total RNA (5 μg) prepared from the tissue culture was reverse transcribed by using a Thermotranscript kit (Invitrogen-Life Technologies) according to the manufacturer's instructions with a gene-specific primer at 54°C for 50 min. The PCR conditions were 35 cycles consisting of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 3 to 7 min of extension at 72°C. The PCR products were electrophoresed and visualized by ethidium bromide staining. The primers used for all RT-PCRs are shown in Table 1.
RACE.
The 5′ rapid amplification of cDNA ends (5′RACE) experiment was performed using the protocol provided by the manufacturer (Invitrogen-Life Technologies). DNase I-treated total RNA (5 μg) was reverse transcribed by using Superscript II with a gene-specific primer. The cDNA was tailed by adding cytosine or adenosine residues at the 3′ end by using terminal transferase and amplified by PCR with a primer set consisting of a second gene-specific primer and an oligo(dG)- or oligo(dT)-linked amplification primer. The PCR conditions were 35 cycles consisting of 1 min of denaturation at 94°C, 1 min of annealing at 54°C, and 1 min of extension at 72°C. The primary PCR products were further amplified by a nested gene-specific primer and the amplification primer without the oligo(dG) or oligo(dT) anchor. The secondary PCR products were purified and cloned. The inserts of 25 to 30 clones in each sample were sequenced. The primers used for all 5′RACE procedures are shown in Table 1.
Sequence analysis.
A database search was carried out with the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Protein localization site and signal sequence were analyzed with the PSORT program (http://psort.nibb.ac.jp/). Multiple alignments were done using the CLUSTAL V method in the DNASTAR program. Phylogenetic analysis was performed with the PHYLIP (Phylogeny Inference Package) version 3.5p. GenBank or EMBL accession numbers of published Vir protein sequences used for the phylogenetic analysis are as follows: Agrobacterium tumefaciens Ti, J03320; Bartonella henselae, U23447 and AF182718; Bordetella pertussis ptl, A47301, B47301, C47301, D47301, E47301, F47301, and G47301; Brucella abortus, AF226278; Brucella suis, AF141604; Legionella pneumophila lvh, Y19029; pKM101, U09868 and AF109305; R. prowazekii, AJ235270, AJ235271, and AJ235273; R. conorii, NC_003103; Wolbachia sp. strain wTai, AB045234; and Wolbachia sp. strain wKueYo, AB045235.
Nucleotide sequence accession numbers.
Sequence data for the virB-D regions of A. phagocytophila and E. chaffeensis determined in the present study have been assigned GenBank accession numbers AF392618 (virB8-p44-22 omp) and AF392616 (pstA-virB6) for A. phagocytophila and AF392617 (ribA-p120) and AF392615 (pstA-ORFa) for E. chaffeensis.
RESULTS
Gene organization and genetic loci of virB and virD regions in genomes of A. phagocytophila (causing HGE) and E. chaffeensis (causing HME).
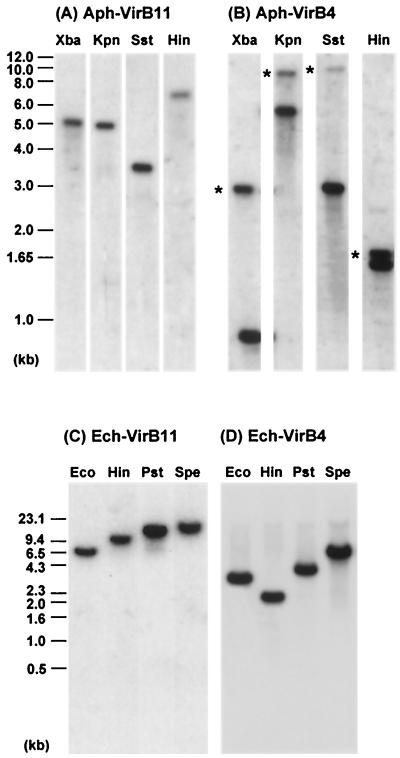
With the degenerate primer pairs, we successfully amplified DNA fragments including partial sequences of virB4 and virB11 genes from each of genomic DNAs of A. phagocytophila and E. chaffeensis. Several other virB and virD genes that were located upstream and downstream from the virB4 and virB11 genes were assembled by cloning and sequencing, using two kinds of genome walking procedures as described in Materials and Methods. By this approach, a total of eight genes orthologous to virB3, virB4, virB6, virB8, virB9, virB10, virB11, and virD4 for a type IV secretion machinery were found in each genome of A. phagocytophila strain HZ and E. chaffeensis strain Arkansas (Fig. 1). The gene organizations of the virB and virD regions in both bacteria were similar to each other, and they were also similar to the corresponding regions of the type IV secretion systems in other gram-negative bacteria (9). However, unlike the single locus of the clustered virB and virD genes in most of other bacteria (9), the vir genes of A. phagocytophila and E. chaffeensis were clustered in two separate loci of each genome, one consisting of five tandem genes (virB8, virB9, virB10, virB11, and virD4) and another consisting of three tandem genes (virB3, virB4, and virB6) (Fig. 1). Four genes orthologous to virB1, virB2, virB5, and virB7 of Agrobacterium tumefaciens were not found in these two loci of either A. phagocytophila or E. chaffeensis. Southern blot analyses supported that these two loci (each) in A. phagocytophila and E. chaffeensis are separated in the respective genomes (Fig. 2). Moreover, the blot result showed that A. phagocytophila strain HZ may have an additional virB4 paralog (asterisks in Fig. 2B) like R. prowazekii (2) and R. conorii (23), because the virB4 probes used did not have XbaI, KpnI, SstI, and HindIII restriction sites, whereas E. chaffeensis strain Arkansas has a single virB4 gene (Fig. 2D). Among strains HZ and LL and three human isolates of A. phagocytophila (Fig. 3), the hybridization patterns with virB9 or virB4 probes were almost identical, suggesting the conservation of genetic loci of the virB and virD genes in strain LL and three isolates of A. phagocytophila. The blot results also revealed the presence of two virB4 paralogs in all of five A. phagocytophila strains and isolates (Fig. 3B).
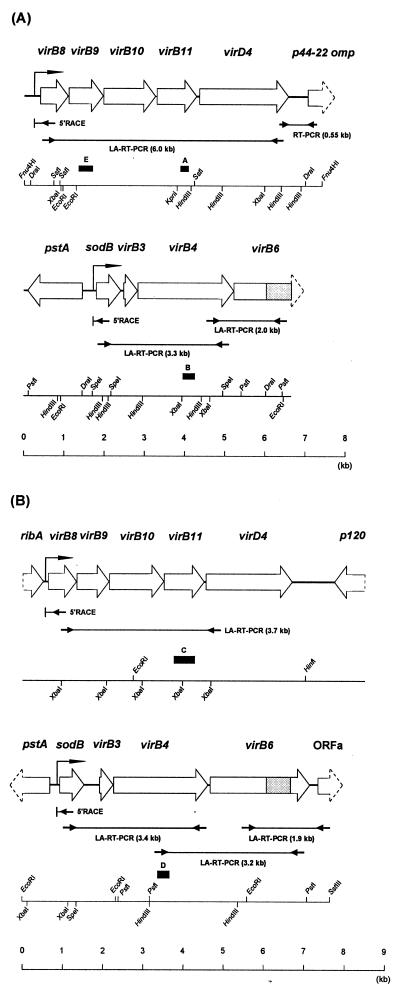
FIG. 1.
Schematic representation of the gene organization of the virB and virD regions in A. phagocytophila HZ (A) and E. chaffeensis Arkansas (B). ORFs are represented as open boxes, with arrowheads indicating their orientation. The region homologous to R. prowazekii RP104 (one of virB6 paralogs) is shaded within the virB6 ortholog from each species. Lines with facing arrows at both ends indicate the LA-RT-PCR or RT-PCR regions in the transcriptional analysis. Bent arrows located upstream of virB8 or sodB represent the transcriptional promoter regions for the respective clustered virB and virD genes which were deduced by 5′RACE analysis (arrows with “5′RACE”). Closed boxes with letters indicate the DNA probes used for genomic Southern blot analysis.
FIG. 2.
Genomic Southern blot analyses of the clustered virB and virD genes in A. phagocytophila HZ (A and B) and E. chaffeensis Arkansas (C and D). Panels A to D show the hybridization patterns with probes A to D, respectively, shown in Fig. 1. Asterisks indicate second, more weakly hybridized fragments. Hin, HindIII; Kpn, KpnI; Pst, PstI; Spe, SpeI; Sst, SstI; Xba, XbaI.
Five genes (virB8 to virD4) within one of the two loci each in A. phagocytophila and E. chaffeensis were clustered upstream from an antigenic protein gene (Fig. 1): in A. phagocytophila a p44 major outer membrane protein gene (designated p44-22 omp) which belongs to a p44 polymorphic multigene family (42, 43) and in E. chaffeensis a 120-kDa antigenic protein gene (p120) in the opposite orientation (41). The intergenic space between virD4 and p44-22 omp (486 bp) in A. phagocytophila was shorter than that between virD4 and p120 (998 bp) in E. chaffeensis (Fig. 1). A ribA gene that encodes a bifunctional enzyme with the activities of both 3,4-dihydroxy-2-butanone 4-phosphate synthase and GTP cyclohydrolase II, which catalyze two essential steps in riboflavin biosynthesis, was located at 134 bp upstream from E. chaffeensis virB8. No open reading frame (ORF) was found within 385 bp upstream from the start codon of the A. phagocytophila virB8 gene. Three genes (virB3, -B4, and -B6) and a sodB gene encoding a superoxide dismutase (SOD) were located downstream from a pstA gene encoding a phosphate ABC transporter permease in the opposite orientation. The sodB gene probably encodes an iron-containing SOD, because the deduced amino acid sequence was significantly more homologous to known iron-containing SODs than to manganese-containing SODs. No gene homologous to a partial ORF located downstream from E. chaffeensis virB6 (designated ORFa) was found in a database search. An additional difference between A. phagocytophila and E. chaffeensis is the length of the intergenic space between the sodB and virB3 genes. The space in A. phagocytophila is shorter (92 bp) than that in E. chaffeensis (396 bp).
Proteins encoded by virB and virD genes of A. phagocytophila and E. chaffeensis.
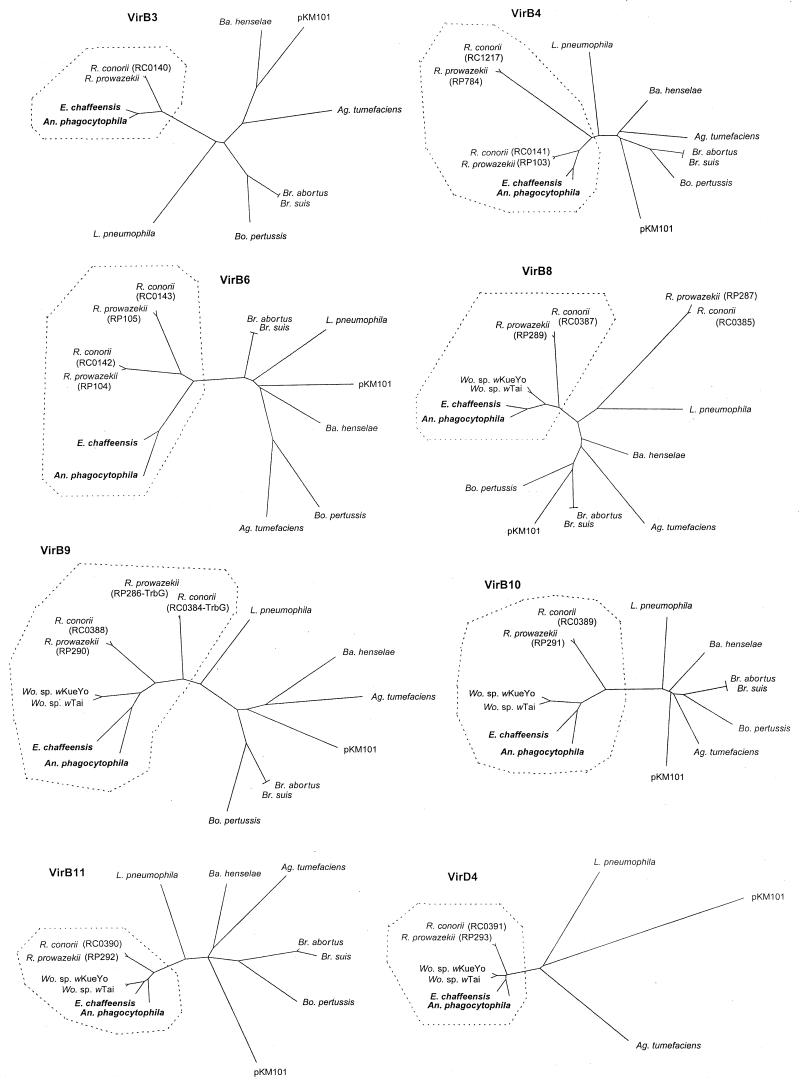
The predicted localization of Vir orthologs in A. phagocytophila and E. chaffeensis deduced by the PSORT program was identical to that of T-DNA transfer system proteins and their orthologs shown by Christie (9), except for that of VirB3 (Table 2). In addition, the PSORT analysis identified putative N-terminal signal sequences of three proteins of eight Vir orthologs in A. phagocytophila and E. chaffeensis (data not shown). The VirB9 proteins of A. phagocytophila and E. chaffeensis as well as those of other bacteria have cleavable signal sequences to translocate into the outer membranes (9). The VirB6 and VirD4 proteins of A. phagocytophila and E. chaffeensis have uncleavable or cleavable signal sequences, probably to anchor and locate in the inner membranes. Comparison with the database available for more than a dozen gram-negative bacteria showed that the VirB and VirD orthologous proteins of these two Anaplasma and Ehrlichia spp. had highest amino acid identities with those of Wolbachia spp. or Rickettsia spp. (Table 2 and Fig. 4). Anaplasma spp., Ehrlichia spp., Wolbachia spp., and Rickettsia spp. are obligatory intracellular bacteria that belong to the α1-proteobacteria. Among the more than one dozen bacteria for which the type IV secretion system has been described, Wolbachia spp., followed by Rickettsia spp., are closest to Anaplasma and Ehrlichia spp. on the basis of 16S rRNA gene sequence comparison (34). Of these obligate intracellular bacterial species, E. chaffeensis had higher identities with A. phagocytophila in three VirB orthologs (VirB8, -B9, and -B10), whereas VirB11 and VirD4 of E. chaffeensis showed the highest identities with those of Wolbachia spp. rather than with those of A. phagocytophila (Table 2). Between A. phagocytophila and E. chaffeensis, the four orthologs of VirB3, -B4, -B11, and -D4 are more conserved (72.6 to 78.1%) than the remaining orthologs of VirB6, -B8, -B9, and -B10 (37.3 to 62.0%). Among the VirB and VirD orthologs, the VirB6 proteins of E. chaffeensis, R. prowazekii, and R. conorii (661 to 1,155 amino acids) were significantly larger than those of Agrobacterium tumefaciens, Bartonella henselae, Bordetella pertussis, L. pneumophila, and Brucella spp. (294 to 436 amino acids) and retained the VirB6-homologous region in a central part of the molecules (Fig. 1) (2, 37). Because the VirB6 protein is involved in assembling the conjugal pore at the inner membrane and may interact with the effector molecules to be delivered (8, 22), the diversity of ehrlichial and rickettsial VirB6 proteins suggests a type of effector molecules different from those of other bacteria.
TABLE 2.
Predicted localization of ehrlichiosis agent Vir proteins, amino acid (aa) identities, and amino acid numbers of Vir orthologs of selected gram-negative bacteria compared with those of E. chaffeensis
| Vir protein | PSORT analysisa | Localizationb | % aa identity
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| E. chaffeensis (aa no.) | A. phagocytophila (aa no.) | R. prowazekii (locus, aa no.) | R. conorii (locus, aa no.) | Wolbachia sp. strain w Tai (aa no.) | Wolbachia sp. strain w TueYo (aa no.) | Agrobacterium tumefaciences (aa no.) | |||
| VirB3 | IM | OM | 100 (97) | 76.3 (97) | 44.2 (NDc, 95) | 42.1 (RC0140, 95) | 16.5 (108) | ||
| VirB4 | IM | Integral IM | 100 (800) | 78.1 (801) | 57.0 (RP103, 805) | 57 (RC00141, 805) | 27.4 (788) | ||
| 21.1 (RP784, 810) | 21.0 (RC1217, 805) | ||||||||
| VirB6 | IM | Polytopic IM | 100 (826) | 37.2 (486, partial) | 12.8 (RP104, 1124) | 11.7 (RC0142, 993) | 15.6 (294) | ||
| 16.8 (RP105, 672) | 17.1 (RC0143, 661) | ||||||||
| 12.5 (RP106, 971) | 12.7 (RC0144, 966) | ||||||||
| 11.3 (RP107, 888) | 12.1 (RC0145, 891) | ||||||||
| 10.5 (RP108, 1155) | 10.4 (RC0146, 1153) | ||||||||
| VirB8 | IM | IM | 100 (237) | 62.0 (234) | 13.9 (RP287, 247) | 13.4 (RC0385, 232) | 50.0 (226) | 45.6 (226) | 16.0 (237) |
| 26.6 (RP289, 202) | 24.9 (RC0387, 243) | ||||||||
| VirB9 | OM | OM | 100 (273) | 46.2 (281) | 31.2 (RP290, 157) | 30.6 (RC0388, 157) | 39.8 (261) | 40.2 (266) | 16.1 (293) |
| 20.8 (R286-TrbG, 250) | 20.4 (RC0384-TrbG, 250) | ||||||||
| VirB10 | IM | Bltopic IM | 100 (447) | 43.1 (434) | 19.2 (RP291, 483) | 20.1 (RC0389, 482) | 33.1 (503) | 35.1 (486) | 16.7 (377) |
| VirB11 | IM | Peripheral IM | 100 (332) | 72.6 (332) | 66.6 (RP292, 334) | 67.5 (RC0390, 334) | 78.6 (330) | 78.5 (330) | 25.6 (344) |
| VirD4 | IM | Integral IM | 100 (714) | 73.9 (740) | 62.9 (RP293, 591) | 63.3 (RC0391, 591) | 74.9 (666) | 74.5 (667) | 22.0 (665) |
Localization of ehrlichiosis agent Vir orthologs predicted by the PSORT program. IM, inner membrane; OM, outer membrane.
Localization of T-DNA transfer system proteins and their orthologs shown by Christie (9).
FIG. 4.
Phylograms of VirB and VirD proteins in A. phagocytophila, E. chaffeensis, and selected gram-negative bacteria. The amino acid sequences were aligned with the CLUSTAL V method, and trees were constructed by using the neighbor-joining method. Two VirB6 proteins of R. prowazekii (RP104 and PR105) and two VirB6 proteins of R. conorii (RC0142 and RC0143) were used as representatives of six paralogs of each rickettsia (Table 2). Vir proteins of the obligate intracellular bacteria are surrounded by dotted lines, except for RP287 of R. prowazekii and RC0385 of R. conorii (virB8 paralogs).
To analyze the relationship among VirB and VirD orthologs from Anaplasma and Ehrlichia spp. and several other bacteria, we constructed phylogenetic trees based on the deduced amino acid sequences (Fig. 4). Sixteen Vir proteins from A. phagocytophila and E. chaffeensis, 10 Vir proteins from Wolbachia spp., and 22 Vir proteins from R. prowazekii and R. conorii made a tight clade segregated from the remaining bacterial species in each of eight phylogenic trees (Fig. 4), except for VirB8 paralogs of R. prowazekii (RP287) and R. conorii (RC0385). However, in the eight trees most of the orthologs from facultative intracellular bacteria and the pKM101 plasmid were promiscuously located. The conservation of Vir proteins among obligate intracellular bacteria and the diversity of Vir proteins among facultative intracellular bacteria and the plasmids suggest that type IV secretion systems in obligate intracellular bacteria play roles different from those in facultative intracellular parasitism.
Transcription of the clustered genes in the virB and virD regions of A. phagocytophila and E. chaffeensis.
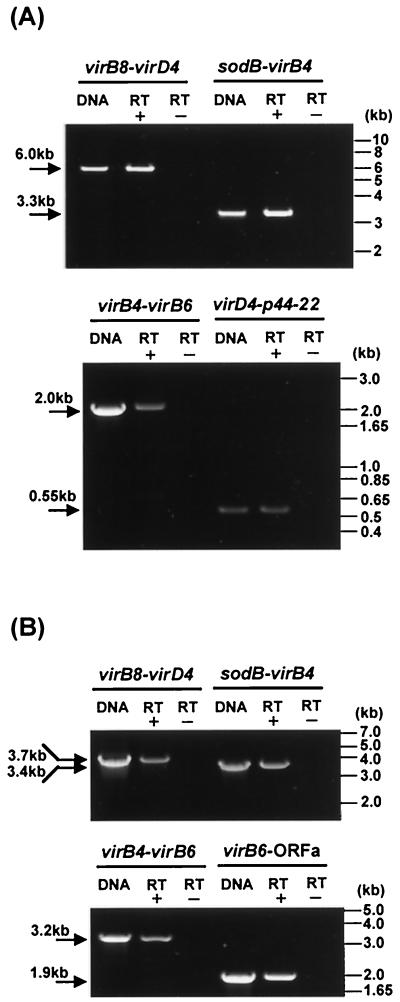
Because of a short intergenic space between the genes within the virB and virD regions of A. phagocytophila and E. chaffeensis, as shown in Fig. 1, it is presumed that these clustered genes are polycistronically transcribed. Therefore, we primarily utilized a long and accurate RT-PCR (LA-RT-PCR) method to analyze the expression of the clustered virB and virD genes by these two bacteria cultivated in a human leukemia cell line. The sequences and positions of the primers and amplicon sizes are shown in Table 1 and Fig. 1. DNA template controls are shown for each primer pair to demonstrate the specificity of the primer to amplify the target sequence (Fig. 5). Without reverse transcriptase, no amplicon was detected in RT-PCR analyses using any of the primer pairs, indicating the absence of contamination of genomic DNA in the RNA preparation. As expected, a 6.0-kb LA-RT-PCR product including virB8, -B9, -B10, -B11, and virD4 within one of the virB-D regions was detected in A. phagocytophila cultured in HL-60 cells, indicating the polycistronic transcription of these genes (Fig. 5A). Furthermore, a 3.3-kb LA-RT-PCR product including sodB, virB3, and virB4 and a 2.0-kb LA-RT-PCR product including virB4 and -B6 within another virB-D region were detected, showing that not only virB orthologs but also the sodB gene were cotranscribed. The transcript of two adjacent genes (virD4 and p44-22 omp) connected by a 487-bp intergenic space at the 3′ end of the virB-D region was detectable, indicating that one of the p44 multigene family members was cotranscribed with the virD genes. Similar results regarding the expression of virB and virD genes were obtained for E. chaffeensis cultivated in THP-1 cells (Fig. 5B). ORFa of E. chaffeensis was cotranscribed with virB6. The results shown in Fig. 5 are diagrammed in Fig. 1. These findings suggest that the transcriptional promoters for the clustered virB and virD genes are present upstream of the virB8 and sodB genes.
FIG. 5.
Transcriptional analysis of the clustered virB and virD genes in A. phagocytophila (A) and E. chaffeensis (B). LA-RT-PCR or RT-PCR was used for transcriptional analysis of the genes in the virB and virD regions. Primers used and their positions are shown in Table 1 and Fig. 1. The DNA template control (0.1 ng of genomic DNA from purified organisms) shows the intensity and specificity of the band detected with each pair of primers. RT+ and RT− indicate the presence and absence of reverse transcriptase in the reaction, respectively. The numbers on the left indicate the respective amplicon sizes.
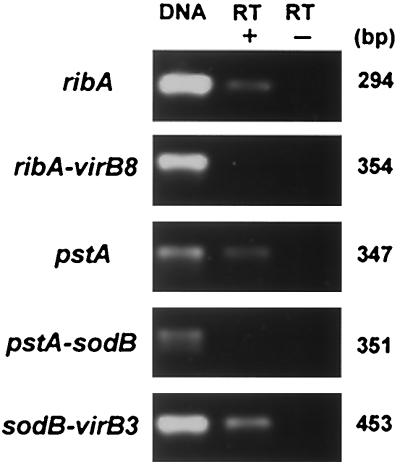
We further examined the transcription of ribA, pstA, and three sets of two adjacent genes (ribA-virB8, pstA-sodB, and sodB-virB3), including their intergenic spaces, in E. chaffeensis cultivated in THP-1 cells by RT-PCR. As shown in Fig. 6, the transcripts of ribA, pstA, and a set of two adjacent genes (sodB-virB3) with a long intergenic space (396 bp) were detected, but two other sets of two adjacent genes (ribA-virB8 and pstA-sodB), including their intergenic spaces, were RT-PCR negative, showing that the sodB and virB3 genes were cotranscribed but that the ribA and virB8 genes or the pstA and sodB genes were not. In addition to the LA-RT-PCR results described above, these results support that the promoters for the clustered virB and virD genes exist within the intergenic spaces between the ribA and virB8 genes and between the pstA and sodB genes but probably do not exist between the sodB and virB3 genes.
FIG. 6.
RT-PCR analysis of ribA, pstA, and sodB and their intergenic spaces in E. chaffeensis. DNA, 0.1 ng of genomic DNA from purified E. chaffeensis; RT+, with reverse transcriptase; RT−, without reverse transcriptase. The primers used are shown in Table 1. The numbers on the right show the respective amplicon sizes.
Characterization of promoter regions for the clustered virB and virD genes in A. phagocytophila and E. chaffeensis.
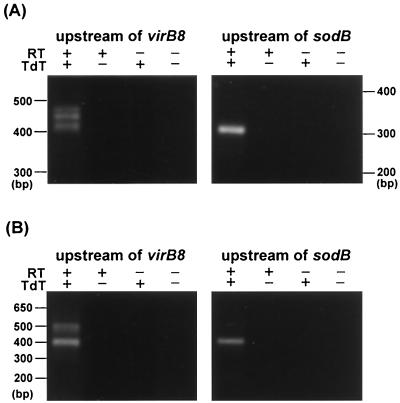
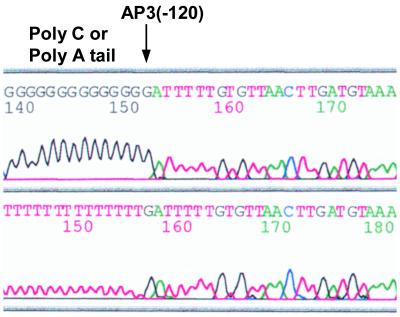
To characterize vir promoter regions, we analyzed a transcriptional initiation site upstream from virB8 and sodB by 5′RACE procedures using the addition of a polymeric dC tail at the 3′ end of the cDNA. Three bands and a single band of the major 5′RACE products for virB8 and sodB, respectively, were detected in A. phagocytophila cultivated in HL-60 cells (Fig. 7A). The same results were obtained when we performed 5′RACE with the addition of a poly(dA) tail at the 3′ end of the cDNA (data not shown). In E. chaffeensis-infected THP-1 cells, two bands and a single band of the major products for virB8 and sodB, respectively, were detected (Fig. 7B). These 5′RACE products were cloned and sequenced to determine the transcriptional initiation site. One of the results is shown in Fig. 8. By sequencing A. phagocytophila 5′RACE clones with both poly(dC) and poly(dA) tails, we determined the initiation site of the AP3 transcript to be a guanine at −120 bp from the start codon of virB8 but probably not to be an adenine at −119 bp (Fig. 8 and 9). In total, we identified three initiation sites at −73 bp, −86 bp, and −120 bp from the virB8 start codon and two initiation sites at −21 bp and −30 bp from the sodB start codon in A. phagocytophila transcripts (Fig. 9). In E. chaffeensis, two initiation sites at −58 or −59 bp and at −165 bp from the virB8 start codon and one initiation site at −69 bp from the sodB start codon were identified by sequencing the 5′RACE clones with poly(dC) tails. Putative sequences of −10 and −35 promoter regions were found upstream of the respective transcriptional initiation sites, which were similar to σ70-type consensus promoter sequences of E. coli (26). One of the promoter regions for virB8 in E. chaffeensis (EP2) is likely to be located within the 3′ coding region of the ribA gene, suggesting that the expression of ribA may influence the transcription of clustered virB8-virD4 genes.
FIG. 7.
5′RACE analysis of virB8 and sodB in A. phagocytophila (A) and E. chaffeensis (B). RT+, with reverse transcriptase; RT−, without reverse transcriptase; TdT+, with terminal deoxynucleotidyl transferase; TdT−, without terminal deoxynucleotidyl transferase.
FIG. 8.
Transcriptional initiation site of a virB8 transcript (AP3) in A. phagocytophila. An arrow indicates the AP3 initiation site that was determined by sequencing the 5′RACE clones with poly(dC) and poly(dA) tails. The location of the AP3 site on the upstream sequence of virB8 is shown in Fig. 9.
FIG. 9.
Alignment of DNA sequences in the promoter regions of virB8 (A) and sodB (B) of A. phagocytophila and E. chaffeensis. The putative −10 and −35 regions of each promoter are underlined. The initiation site (base) of each transcript of A. phagocytophila (AP1 to AP3) and E. chaffeensis (EP1 and EP2) is in boldface and indicated by arrowheads. Sequences with boldface bars in panel B, 5′dATTGTAAT3′ (A. phagocytophila) and 5′dTGTAAT3′ (E. chaffeensis), show putative vir boxes. The numbers in parentheses indicate the position upstream from the start codon of virB8 or sodB (+1). The sequence enclosed by a box shows a 3′-end coding region of ribA. Gaps in the alignment are indicated by dashes.
Transcription of the virB gene in HGE patients and in experimentally infected animals.
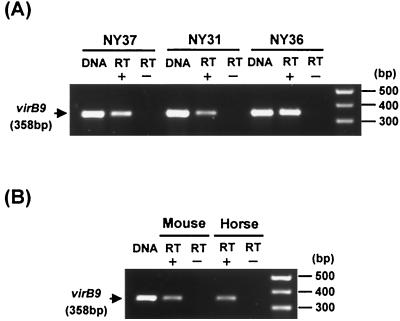
We examined by RT-PCR whether the virB gene is expressed by A. phagocytophila in acute-stage HGE patients or in experimentally infected animals. The clinical signs and laboratory parameters for three patients were described in detail elsewhere (Lin et al., submitted). A transcript of A. phagocytophila virB9 was detected in PBLs from each patient's blood collected at 20 days (patient NY31) and 3 days (patients NY36 and NY37) after recognition of initial clinical signs such as fever (Fig. 10A). The transcript was also detected in PBLs from blood collected on days 4 and 8 after inoculation of A. phagocytophila in an experimentally infected mouse and horse, respectively (Fig. 10B). These findings show that the virB-D region of A. phagocytophila is transcriptionally active in infected humans and animals at the acute stage as well as in cell cultures and suggest that the type IV secretion system is involved in HGE pathogenesis.
FIG. 10.
RT-PCR analysis of a virB9 transcript in PBLs from three human patients (A) and animals (B) infected with A. phagocytophila. The blood samples from three HGE patients are indicated as NY31, NY36, and NY37. DNA, 0.1 ng of genomic DNA from the respective organisms of purified A. phagocytophila isolates (I-NY31, I-NY36, or I-NY37) (A) or strain HZ (B). RT+, with reverse transcriptase; RT−, without reverse transcriptase. Primers are shown in Table 1. Numbers on the left of each panel show the respective amplicon sizes.
DISCUSSION
The present study characterized the genes for the type IV secretion machinery in A. phagocytophila (agent of HGE) and E. chaffeensis (agent of HME) and their transcription. Of primary significance is that these genes were transcribed not only in cell cultures but also in the blood of acute-stage patients and experimentally infected animals. Therefore, the type IV secretion system is expected to have a significant role in both HGE and HME. The type IV systems have been recently classified into two groups (9). One group consists of genes orthologous to the virB and virD genes of Agrobacterium tumefaciens. The type IV systems in most bacteria, including L. pneumophila lvh, belong to this group. Another group contains the dot-icm genes of L. pneumophila (36, 38) and colIb genes (IncI) of Shigella flexneri (9). The Lvh system of L. pneumophila functions as a DNA conjugation system, while the dot-icm system is required for the bacterial virulence (36-38). The type IV secretion systems of A. phagocytophila and E. chaffeensis which were identified in the present study belong to the former group. A number of unique features were found in the type IV secretion systems from these two Anaplasma and Ehrlichia spp. Unlike the single locus of virB and virD genes in extracellular or facultative intracellular bacteria, the virB and virD genes of Anaplasma and Ehrlichia spp. were found in two separate loci like in Rickettsia spp. (three loci). This may allow for an independent transcriptional control of separate loci in Anaplasma and Ehrlichia spp., which ensures that only when two conditions are met is the complete type IV system assembled.
The type IV secretion systems of Anaplasma and Ehrlichia spp. are phylogenetically close to that of Rickettsia spp. but significantly different in virB gene numbers or their genetic loci. Most notably, virB3, -B4, and -B6 in Anaplasma and Ehrlichia spp. were tandemly arranged downstream from the sodB gene and polycistronically transcribed through the sodB promoters. In Rickettsia spp., the virB3 genes are not located downstream from a sodB gene, and between the sodB and virB3 genes in the genome more than 240 other genes are present (2, 23). Therefore, the rickettsial virB3 genes probably could not be cotranscribed with their sodB genes, although transcriptional analysis of these genes has not been done. In Wolbachia spp., sodB, virB3, virB4, and virB6 so far have not been described, but a virB-D gene cluster (ribA, virB8, virB9, virB10, virB11, virD4, and wsp) with a genetic locus and polycistronic transcription similar to those of Anaplasma and Ehrlichia spp. has been identified (19). As far as we know, there has been no report that the sodB and virB genes are polycistronically transcribed in the type IV secretion systems of other bacteria or plasmids so far identified. Therefore, the cotranscription of sodB and virB genes found in the present study is truly unique and conserved for Anaplasma and Ehrlichia spp. This result suggests that the activation of the sodB gene is required for construction of the type IV secretion machinery of Anaplasma and Ehrlichia spp.
The sodB gene of Anaplasma and Ehrlichia spp. probably encodes an iron-containing SOD (FeSOD), based on a database search. The SODs are metalloenzymes that function by catalyzing the dismutation of O2− to H2O2 and O2. There are two additional types of SODs, which can be distinguished by the prosthetic metals present at the active site: manganese (MnSOD [sodA]) or copper or zinc (Cu/ZnSOD [sodC]). Cytoplasmic MnSODs and FeSODs are known to defend bacterial cells against reactive oxygen species generated from bacterial metabolism and protect DNA from oxidative damage, whereas Cu/ZnSOD localized in the periplasm is proposed to play a role in the defense of bacteria against external reactive oxygen. sodB gene expression is known to be controlled by iron uptake systems (12, 18). Therefore, the virB3, -B4, and -B6 genes of Anaplasma and Ehrlichia spp. are expected to be induced under iron-rich conditions such as inside blood-feeding ticks and in eukaryotic cytoplasm.
Analysis of upstream regions of the virB8 and sodB genes of both A. phagocytophila and E. chaffeensis showed that the polycistronic transcription of the vir genes seems to be regulated by multiple σ70-type promoters. However, the transcriptional regulation mechanism of the vir genes still remains to be elucidated. In the Ti plasmids of Agrobacterium tumefaciens, VirA and VirG are members of the family of two-component regulatory systems and are required for induction of all other vir genes. VirA, a signal sensor/histidine kinase transmembrane protein, is autophosphorylated in response to phenolic compounds released by wounded plant cells and then phosphorylates VirG, a transcriptional activator. The phosphorylated VirG activates other vir operons through binding to the consensus sequence (vir box) of the respective promoter regions. The consensus sequence (14 bp) is 5′dPu(T/A)TDCAATTGHAAPy3′ (H = A, C, or T; D = A, G, or T) (11). The vir box of the closely related Ri plasmid vir genes has the 6-bp sequence dTG(A/T)AA(C/T) (3), which is equivalent to the 3′ half of the Ti plasmid vir box. In the present study, the sequence 5′dATTGTAAT3′, equivalent to the 3′ region (8 bp) of the Ti plasmid vir box, was found to overlap with the −35 region of AP1 in the sodB promoter of A. phagocytophila. In addition, the sequence dTGTAAT, equivalent to the Ri plasmid vir box (6 bp), overlapped with the −10 region of EP1 in the sodB promoters of E. chaffeensis. This suggests that a two-component regulatory system similar to that of Agrobacterium tumefaciens Ti plasmids (11) may control the polycistronic transcription of the sodB-virB6 genes in Anaplasma and Ehrlichia spp.
Another unique feature is that antigenic outer membrane protein genes were colocalized with the virB and virD genes in Anaplasma, Ehrlichia, Wolbachia, and Bartonella spp. A p44-22 omp gene of A. phagocytophila, a member of the p44 multigene family (42, 43), was located downstream of virD4 and seems to be cotranscribed with the clustered virB8-virD4 genes. This locus of A. phagocytophila probably is one of the expression sites for the multigene family. A p120 gene of E. chaffeensis was also located downstream of virD4 but with the opposite orientation, suggesting a lack of cotranscription of the p120 and virD4 genes. A wsp gene of Wolbachia spp. was positioned downstream of virD4 like in A. phagocytophila (19). Although transcriptional analysis has not been performed, the wsp and virD4 genes may also be cotranscribed. A 17-kDa antigen gene of B. henselae was located between virB4 and virB6 orthologs, missing virB5, within a single virB locus (27, 35), suggesting the cotranscription of the antigen gene and virB genes. Thus, except in E. chaffeensis, these bacterium-specific outer membrane antigens may be associated with the respective type IV secretion systems.
Eight vir orthologs (virB3, -B4, -B6, -B8, -B9, -B10, -B11, and virD4) present in most bacteria or plasmids that have evolved the type IV secretion systems (9) were found in A. phagocytophila and E. chaffeensis. The virB1, -B2, -B5, and -B7 genes seem to be missing in Anaplasma and Ehrlichia spp.; however, these genes may also be required for the architecture of their type IV secretion machinery (e.g., the VirB2 protein is an F-pilin ortholog). Therefore, the related genes may be present in the genomes of these two bacteria but probably have extremely low homology with virB orthologs of other bacteria.
The etiologic agents of HGE and HME, A. phagocytophila and E. chaffeensis, infect mainly granulocytes and monocytes, respectively, primary host defensive cells with powerful bactericidal activities. Since these agents cannot survive outside host cells, they must enter granuclocytes or monocytes. For intracellular survival, A. phagocytophila and E. chaffeensis form a unique membrane-bound niche that does not fuse with lysosomes (5, 6, 20), and A. phagocytophila (agent of HGE) delays apoptosis of the infected neutrophils (40) and inhibits generation of superoxide anion by neutrophils (21). The type IV secretion system is considered to have evolved in intracellular bacteria to modulate eukaryotic cells for their intracellular survival (9). Further study is required for clarification of the roles of the type IV secretion system for survival of Anaplasma and Ehrlichia spp. in granulocytes or monocytes. The present data on the transcriptionally active type IV secretion systems in two major agents of HGE and HME are expected to facilitate analysis of the type IV secretion systems in the obligate intracellular pathogens.
Acknowledgments
This work was supported by National Institutes of Health grants R01AI30010 and R01AI47407.
Editor: J. T. Barbieri
REFERENCES
- 1.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 3.Aoyama, T., M. Takanami, and A. Oka. 1989. Signal structure for transcriptional activation in the upstream regions of virulence genes on the hairy-root-inducing plasmid A4. Nucleic Acids Res. 17:8711-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect. Immun. 68:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnewall, R. E., N. Ohashi, and Y. Rikihisa. 1999. Ehrlichia chaffeensis and E. sennetsu, but not the human granulocytic ehrlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 67:2258-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnewall, R. E., Y. Rikihisa, and E. H. Lee. 1997. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 65:1455-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, A., and G. J. Pazour. 1989. Delineation of the regulatory region sequences of Agrobacterium tumefaciens virB operon. Nucleic Acids Res. 17:4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumler, J. S., A. F. Barbet, C. P. J. Bekker, G. A. Dasch, et al. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales; unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia, and Ehrlichia with Neorickettsia; description of six new species combinations; and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 14.Gilibert, F., T. M. Finan, S. R. Long, A. Puhler, et al. 2001. The complete genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 15.Hardalo, C. J., V. Quagliarello, and J. S. Dumler. 1995. Human granulocytic ehrlichiosis in Connecticut: report of a fatal case. Clin. Infect Dis. 21:910-914. [DOI] [PubMed] [Google Scholar]
- 16.Hooykaas, P. J. J., and G. M. Beijersbergen. 1994. The virulence system of Agrobacterium tumefaciens. Annu. Rev. Phytopathol. 32:157-179. [Google Scholar]
- 17.Johnston, S. L., M Strausbauch, G. Sarkar, and P. J. Wettstein. 1995. A novel method for sequencing members of multi-gene families. Nucleic Acids Res. 23:3074-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, Y. C., C. D. Miller, and A. J. Anderson. 1999. Transcriptional regulation by iron of genes encoding iron- and manganese-superoxide dismutases from Pseudomonas putida. Gene 239:129-135. [DOI] [PubMed] [Google Scholar]
- 19.Masui, S., T. Sasaki, and H. Ishikawa. 2000. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J. Bacteriol. 182:6529-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mott, J., R. E. Barnewall, and Y. Rikihisa. 1999. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect. Immun. 67:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mott, J., and Y. Rikihisa. 2000. Human granulocytic ehrlichiosis agent inhibits superoxide anion generation by human neutrophils. Infect. Immun. 68:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 23.Ogata, H., S. Audic, P. Renesto-Audiffren, P.-E. Fournier, et al. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 24.Ohashi, N., Y. Rikihisa, and A. Unver. 2001. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi, N., N. Zhi, Y. Zhang, and Y. Rikihisa. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver, D. 1985. Protein secretion in Escherichia coli. Annu. Rev. Microbiol. 39:615-648. [DOI] [PubMed] [Google Scholar]
- 27.Padmalayam, I., K. Karem, B. Baumstark, and R. Massung. 2000. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol. 19:377-382. [DOI] [PubMed] [Google Scholar]
- 28.Popov, V. L., V. C. Han, S.-M. Chen, J. S. Dumler, H.-M. Feng, T. G. Andreadis, R. B. Tesh, and D. H. Walker. 1998. Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J. Med. Microbiol. 47:235-251. [DOI] [PubMed] [Google Scholar]
- 29.Rikihisa, Y. 1996. Ehrlichiae, p. 272-286. In Proceedings of the 5th International Symposium on Rickettsiae and Rickettsial Diseases. International Society for Rickettsiae and Rickettsial Diseases, Slovak Academy of Sciences, Bratislava, Slovak Republic.
- 30.Rikihisa, Y. 1999. Clinical and biological aspects of infection caused by Ehrlichia chaffeensis. Microbes Infect. 1:367-376. [DOI] [PubMed] [Google Scholar]
- 31.Rikihisa, Y., S. A. Ewing, J. C. Fox, A. G. Siregar, F. H. Pasaribu, and M. B. Malole. 1992. Enzyme-linked immunosorbent assay and Western immunoblot analysis of Ehrlichia canis and canine granulocytic Ehrlichia. J. Clin. Microbiol. 30:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rikihisa, Y., N. Zhi, G. P. Wormer, B. Wen, H. W. Horowitz, and K. E. Hechemy. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York State. J. Infect. Dis. 175:210-213. [DOI] [PubMed] [Google Scholar]
- 33.Rogowsky, P. M., B. S. Powell, K. Shirasu, T. S. Lin, P. Morel, E. M. Zyprian, T. R. Steck, and C. I. Kado. 1990. Molecular characterization of the vir regulon of Agrobacterium tumefaciens: complete nucleotide sequence and gene organization of the 28.63-kbp regulon cloned as a single unit. Plasmid 23:85-106. [DOI] [PubMed] [Google Scholar]
- 34.Roux, V., and D. Raoult. 1995. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res. Microbiol. 146:385-396. [DOI] [PubMed] [Google Scholar]
- 35.Schmiederer, M., and B. Anderson. 2000. Cloning, sequencing, and expression of three Bartonella henselae genes homologous to the Agrobacterium tumefaciens VirB region. DNA Cell Biol. 19:141-147. [DOI] [PubMed] [Google Scholar]
- 36.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, J. P., H. L. Andrew, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 39.Weber, K. L., M. E. Bolander, and G. Sarkar. 1998. Rapid acquisition of unknown DNA sequence adjacent to a known segment by multiplex restriction site PCR. BioTechinques 25:415-419. [DOI] [PubMed] [Google Scholar]
- 40.Yoshiie, K., H.-Y. Kim, J. Mott, and Y. Rikihisa. 2000. Intracellular infection by the human granulocytic ehrlichiosis agent inhibits human neutrophil apoptosis. Infect. Immun. 68:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, X.-J., P. Crocquet-Valdes, and D. H. Walker. 1997. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene 184:149-154. [DOI] [PubMed] [Google Scholar]
- 42.Zhi, N., N. Ohashi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828-17836. [DOI] [PubMed] [Google Scholar]
- 43.Zhi, N., N. Ohashi, T. Tajima, J. Mott, R. W. Stich, D. Grover, S. R. Telford III, Q. Lin, and Y. Rikihisa. 2001. Transcript heterogeneity of p44 multigene family in human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 70:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhi, N., Y. Rikihisa, H. Y. Kim, G. P. Wormser, and H. W. Horowitz. 1997. Comparison of major antigenic proteins of six strains of the human granulocytic ehrlichiosis agent by Western immunoblot analysis. J. Clin. Microbiol. 35:2606-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]