Abstract
Glucans synthesized by glucosyltransferase enzymes of oral streptococci facilitate bacterial accumulation on surfaces. The Streptococcus gordonii glucosyltransferase gene, gtfG, is positively regulated by rgg, which encodes a putative cytoplasmic protein. The gtfG promoter and ribosomal binding sequences are located within a DNA inverted repeat immediately downstream of rgg. Polycistronic rgg-gtfG as well as rgg- and gtfG-specific transcripts are associated with this chromosomal region. Previous studies have shown that the rgg product acts in trans near the gtfG promoter to increase the level of gtfG transcript, but it does not affect the level of rgg-gtfG transcript. To further analyze regulation by rgg, a series of strain Challis derivatives was constructed and glucosyltransferase activities were determined. Strains in which rgg was separated from gtfG by integrated vector sequences had decreased levels of glucosyltransferase activity; plasmid-borne rgg could not increase activity to parental levels. As expected, strains with chromosomal deletions involving the rgg structural gene and either the rgg or gtfG promoter also showed decreased glucosyltransferase activity. Plasmid-borne rgg could increase glucosyltransferase activity only in strains which had a 36-bp chromosomal region beginning 72 nucleotides upstream of the gtfG transcriptional start site. Results suggest that these nucleotides, located within the 3′ end of rgg, are necessary, either by direct involvement in binding or by indirectly affecting secondary structure, for Rgg to increase glucosyltransferase activity. Surprisingly, the presence of the rgg promoter upstream of this 36-bp region significantly increased the effects of plasmid-borne rgg. Implications for glucosyltransferase regulation and applicability to other rgg-like determinants are considered.
The glucan polymers synthesized from sucrose by streptococcal glucosyltransferase (GTF) enzymes facilitate the accumulation of a microbial biofilm on the tooth surfaces. Although some oral streptococci have multiple GTF enzymes within one organism, the commensal species Streptococcus gordonii has only one GTF, which can synthesize both α-1,3- and α-1,6-linked glucans (10, 11). The sucrose-promoted colony phenotype (Spp) conferred by these glucans to S. gordonii growing on sucrose agar is related to the level of GTF activity (26). Strains with parental levels of GTF activity are Spp+, whereas strains with up to ca. 40% of the parental level of GTF activity are Spp− (26). Spp+ and Spp− phenotypes have been associated with the ability of growing S. gordonii cells to attach and detach, respectively, from substrata in in vitro dental plaque models (31). Thus, the ability to control expression of GTF protein and subsequent glucan production may provide ecological advantages for colonization of surfaces in vivo.
The expression of the S. gordonii strain Challis GTF structural gene, gtfG (35), is controlled by the positive regulatory determinant rgg, which encodes a predicted 34-kDa cytoplasmic protein (25). Although there were no genes with sequence homologies when rgg was first identified (25), genomic sequencing projects have identified multiple rgg-like genes in Streptococcus pyogenes (7), Streptococcus pneumoniae (27), and Streptococcus mutans (http://www.genome.ou.edu). Indeed, two rgg-like genes in addition to rgg have been identified in S. gordonii (12, 38). Genes with similarity to rgg have also been described in Streptococcus sanguis, Streptococcus oralis, (33, 9) and Lactococcus lactis (23). Clearly, rgg-like determinants appear to make up a fairly widely occurring gene family in pathogenic and commensal bacterial species. Those rgg-like determinants that have been characterized appear to share positive regulatory functions (4, 16, 20, 23, 24).
The putative promoter and ribosomal binding site of gtfG are located within a DNA inverted repeat immediately downstream of rgg. Previous studies have described important aspects of this regulation (24, 25). Northern blotting and primer extension analyses have demonstrated rgg- and gtfG-specific, as well as polycistronic rgg-gtfG mRNA transcripts in the parental strain CH1 (24). Providing rgg in trans on the streptococcal plasmid pAMS57 increased levels of gtfG-specific transcript, resulting in increased levels of GTF activity; pAMS57 did not influence the level of the polycistronic rgg-gtfG transcript, indicating that the rgg product does not act at its own promoter (24).
It has been hypothesized previously that potential nucleic acid secondary structures resulting from the DNA inverted repeat sequence between rgg and gtfG could influence gtfG expression (25). The mRNA in this region may form a stem-loop that inhibits gtfG translation; translation of rgg may be necessary for efficient translation of gtfG, as is seen in translational coupling (19). Previous studies (24) have also shown that a 136-bp EcoRI-NsiI fragment containing the 3-prime end of rgg, the rgg-gtfG intergenic region, and the first 27 nucleotides of gtfG can function as a promoter in Escherichia coli to control expression of a promoterless lacZ. Plasmid-borne rgg increased lacZ activity, indicating that the encoded Rgg increased transcription of the gtfG promoter. However, results obtained using streptococcal promoters cloned in E. coli must be interpreted with caution. E. coli may recognize AT-rich streptococcal DNA sequences as promoters even if these regions do not function as promoters in streptococci (6). Conversely, factors necessary for expression of gram-positive genes, such as sigma factors, polymerases, etc., may not be present in E. coli. To further investigate these possibilities, the studies presented here were undertaken to gain further insights into the relative contributions to GTF activity by the rgg and gtfG promoters in the S. gordonii genetic background. The potential influence of the predicted stem-loop structure between rgg and gtfG was examined, and the chromosomal site of trans-acting rgg product was further defined. Effects on GTF activity and sucrose-promoted colony phenotype resulting from nucleotide changes in these chromosomal regions were examined. The results provide insights into the molecular basis underlying GTF activity in S. gordonii and may be relevant for understanding the general regulatory mechanism(s) shared by the newly recognized family of rgg-like determinants in streptococci and related bacteria.
MATERIALS AND METHODS
Bacteria, medium, and culture conditions.
Bacterial strains and plasmids used in these studies are listed in Table 1. Strains were stored at −70°C in 50% glycerol. S. gordonii strains were grown in Todd-Hewitt medium (TH; Difco) or in defined FMC medium (28) and incubated at 36°C in an anaerobic chamber with a gas mixture of 85% N2, 10% H2, and 5% CO2. The sucrose-promoted colony phenotype (Spp+ for parental strain Challis CH1), a hard colony phenotype which has been associated with glucan synthesis (26), was determined on 3% sucrose TH agar plates incubated for 48 h in 5% CO2. E. coli strain DH5α (Invitrogen) was grown in Luria-Bertani medium at 37°C with aeration. Selection and growth for strains carrying antibiotic resistance determinants were done at levels of 5 μg of erythromycin/ml and/or 250 μg of spectinomycin/ml for S. gordonii strains and with 300 μg of erythromycin/ml, 50 μg of spectinomycin/ml, or 100 μg of ampicillin/ml for the E. coli strains.
TABLE 1.
Bacterial strains and plasmids used in these studies
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. gordonii strains | ||
| CH1 | Parental strain Challis | 26 |
| DS512 | CH1 derivative with chromosomal premature translation stop in rgg | 25 |
| CH8923 | CH1 derivative with chromosomally-integrated pMI8923; Eryr | This study |
| CH8924 | CH1 derivative with chromosomally-integrated pMI8924; Eryr | This study |
| CH8925 | CH1 derivative with chromosomally-integrated pMI8925; Eryr | This study |
| CH8927 | CH1 derivative with chromosomally-integrated pMI8927; Eryr | This study |
| CHΔR1 | CH1 derivative with chromosomal internal deletion of rgg | This study |
| CHΔ35 | CH1 derivative with chromosomal deletion of 3′ end of rgg | This study |
| CHΔ10 | CH1 derivative with chromosomal deletion of gtfG promoter region | This study |
| CHΩN | CH1 derivative with chromosomal deletion of rgg promoter region | This study |
| CHΩR1 | CH1 derivative with chromosomal deletion of rgg promoter region | This study |
| CHR3 | recA-deficient derivative of strain CH1; Spcr | 32 |
| DS512R | recA-deficient derivative of strain DS512; Spcr | This study |
| CHΔR1R | recA-deficient derivative of strain CHΔR1; Spcr | This study |
| CHΔ35R | recA-deficient derivative of strain CHΔ35; Spcr | This study |
| CHΔ10R | recA-deficient derivative of strain CHΔ10; Spcr | This study |
| CHΩNR | recA-deficient derivative of strain CHΩN; Spcr | This study |
| CHΩR1R | recA-deficient derivative of strain CHΩR1; Spcr | This study |
| E. coli strain | ||
| DH5α | F− Φ80d lacZΔM15Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rκ-mκ+) supE44λ1thi-1 gyrA96 relA1 | Invitrogen |
| Plasmids | ||
| pVA891 | E. coli vector, no streptococcal origin of replication; Emr | 18 |
| pMI22Ω | EcoRV- and NcoI/EcoR1-flanked Ω terminator in pBluescript II KS+; Apr | This study |
| pMI891Ω | Derivative of pVA891 with Ω termination region | This study |
| pMI8923 | 3′ end of rgg through 5′ end of gtfG cloned in pMI891Ω | This study |
| pMI8924 | Mid-rgg through 5′ end of gtfG promoter cloned in pMI891Ω | This study |
| pMI8925 | rgg start codon through 5′ end of gtfG cloned in pMI891Ω | This study |
| pMI8927 | rgg promoter region cloned in pMI891Ω | This study |
| pVA749 | Streptococcal cloning vector; Emr | 17 |
| pMISP49 | Derivative of pVA749; Spr, Ems | This study |
| pAMS57 | rgg cloned in pVA749; Emr | This study |
| pMISP57 | rgg cloned in pMISP49; Spr | This study |
| pAM6200 | S. gordonii recA internal fragment in E. coli vector; Spr | 32 |
| pGEM-7Zf | E. coli cloning vector; Apr | Promega |
| pGEM-7:spR | Spectinomycin-resistant determinant in pGEM-7Zf; Apr Spr | 14 |
| pMI2201 | 2.255-kb ApaI-SstI S. gordonii fragment in pGEM-7Zf; Apr | This study |
DNA isolation and manipulations.
DNA preparation and manipulations were done according to standard molecular biology procedures (2). Modifications for S. gordonii strains included growing bacteria in medium supplemented with 0.5% glycine and incubating cell pellets with mutanolysin and lysozyme to facilitate lysis, as previously described (24). Plasmid DNA was prepared using Qiagen purification columns (Valencia, Calif.) according to the manufacturer's directions. Double-stranded PCR products were obtained using Elongase enzyme (Invitrogen Life Technologies). To facilitate cloning, in some cases oligonucleotide primers were designed with engineered restriction sites and random flanking 5′ nucleotides to ensure efficient digestion of products. DNA fragments were isolated by electrophoresis followed by elution from agarose gels with a Qiaex II kit (Qiagen) according to the manufacturer's directions. For cloning, DNA fragments were digested with appropriate restriction enzymes, ligated with T4 DNA ligase into convenient restriction sites of the vector, and transformed into either CaCl2-competent E. coli DH5α or S. gordonii cells made competent (13) with horse serum.
For Southern hybridization analyses, S. gordonii chromosomal DNA was digested with appropriate restriction enzymes, electrophoresed on 0.7% agarose gels, and transferred to Hybond-N membranes (Amersham) by standard procedures (22). Double-stranded DNA probes were labeled with digoxigenin-dUTP, hybridized to the membranes, washed under stringent conditions, and detected by chemiluminescence with the Genius System (Roche Molecular Biochemicals), according to the manufacturer's directions.
Construction of S. gordonii strains with separation of rgg and gtfG by intervening vector DNA.
A schematic representation of the relevant S. gordonii chromosomal regions in these studies is shown in Fig. 1. The method for constructing strains with separation of rgg and gtfG involved a single DNA crossover, resulting in plasmid integration into the strain CH1 chromosome, as is outlined in Fig. 2. To preclude the possibility of readthrough from vector DNA into cloned sequences, a transcriptional and translational terminator omega fragment (Ω) (8) was amplified by PCR with primers 5′-TAGATATCTGATCCGGTGGATGACCTT-3′ and 5′-TAGAATTCCATGGTGATTGATTGAGCAAGCTTTATG-3′ to create compatible cloning sites (underlined). This PCR product also had an engineered NcoI site (italicized), to be used for additional genetic constructs (see construction of strain CHΩN, below). The resulting ca. 150-bp product was digested with EcoRV and EcoRI and directionally cloned into pBluescript II KS(+) (Stratagene) to create pMI22Ω. After the nucleotide sequence analysis, the confirmed product was removed from pMI22Ω by digestion with EcoRV and EcoRI and cloned into the compatible PvuII and EcoRI sites of the E. coli plasmid pVA891 (18), which is unable to replicate in streptococci, to create pMI891Ω.
FIG. 1.
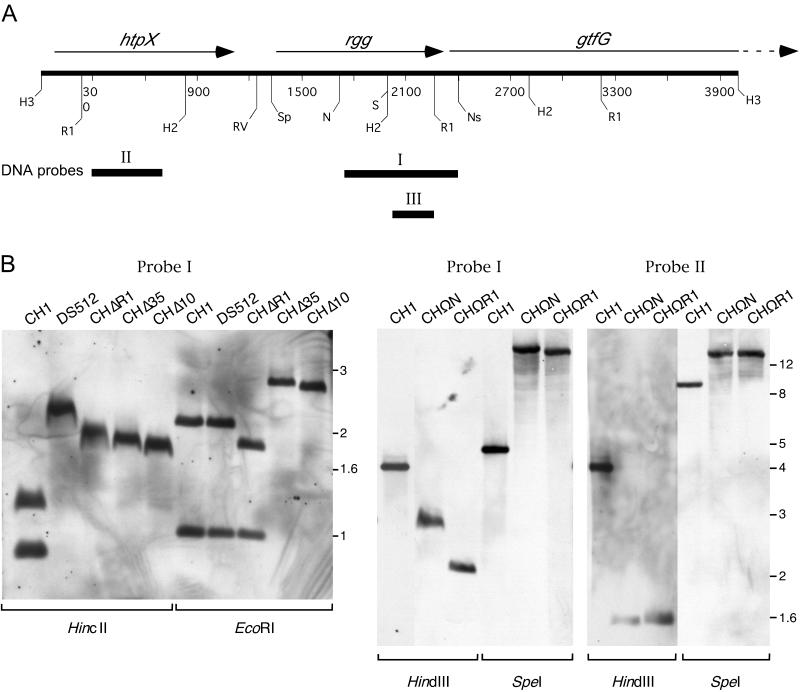
Diagram of features of the S. gordonii strain CH1 chromosomal region containing the 3′ end of htpX (37), rgg, and the 5′ end of gtfG cloned in pMI2201. Relevant nucleotide sequences are detailed and relative positions of restriction sites are shown. Nucleotides are numbered from the ApaI site (designated number 1) within the htpX gene to the SstI site within gtfG. Both rgg and gtfG have putative −35 and −10 promoter regions as underlined. DNA inverted repeats are designated by inverted arrows. Previously identified start sites for rgg and gtfG transcripts are designated by asterisks (24). Putative Shine-Delgarno ribosomal binding sites (SD) and ATG start codons are delineated by complete and dotted boxes, respectively. The 5′ annealing sites for the forward oligonucleotide primers used for constructing strains CHΔR1, CHΔ35, and CHΔ10 are depicted with arrows.
FIG. 2.
Schematic diagram of the construction of S. gordonii strain CH8923. A ca. 150-bp PCR fragment containing translational stops and the T4-transcription termination signal from an omega termination fragment (8) was directionally cloned into PvuII/EcoRI sites of pVA891 to create pMI891Ω. The region surrounding the junction of rgg and gtfG was amplified by PCR using oligonucleotide primers with an engineered translational stop and a BamHI site in the downstream primer to facilitate cloning and prevent potential readthrough from the gtfG ribosomal binding site into vector sequences. The DNA inverted repeat sequestering the gtfG putative promoter (−35 and −10) and ribosomal binding site (SD) is designated by large inverted arrowheads. This PCR fragment was then cloned into the EcoRI and BamHI sites of pMI891Ω. The resulting plasmid was transformed into the parental strain CH1. The erythromycin-resistant transformant strain CH8923 was confirmed by Southern hybridization and nucleotide sequence analysis (data not shown) to have duplicated gtfG promoters with rgg separated from gtfG by the intervening vector. Strains CH8924 and CH8925, which contained larger regions of rgg cloned into pMI891Ω, were similarly constructed.
A series of nested DNA fragments containing the gtfG promoter and increasingly larger regions of rgg were made by PCR using the pAMS57 template. PCR primers were designed with flanking compatible restriction sites to facilitate cloning. The downstream primer for each fragment was the same and was designed to anneal to the NsiI site 27 bp downstream from the start codon of gtfG. This primer, 5′-TAGGATCCTAATGCATTTTATAATGAAC-3′ was also designed to encode an engineered translational stop (boldface) immediately after the NsiI site (italics). The upstream primers were designed to amplify increasingly larger regions of rgg and were either designed to include the EcoRI site near the 3′ end of rgg (Fig. 1, nucleotide 1401) or, for larger fragments, had engineered MunI sites for compatible cloning. To construct pMI8923 (Fig. 2), primer 5′-GAGCGAATTCAACTAACAGTAG-3′, designed to anneal to the rgg region flanking the EcoRI site, was used. To construct pMI8924, primer 5′-ATCAATTGTTAAATACCATTGTGATC-3′, designed to anneal between the NcoI and SalI sites within rgg (Fig. 1), was used. For pMI8925, 5′-ATCAATTGATGCTTATCGTAAAGTCGTC-3′ was designed to anneal at the rgg start codon (boldface). The resulting PCR products were digested with appropriate restriction enzymes and cloned into the EcoRI and BamHI sites of pMI891Ω. As a control for effects of the integrated vector, the DNA region containing the rgg promoter was amplified with primers 5′-CGCAATTGCAATCTCTGAGCGG-3′, designed to anneal 36 bp upstream of the htpX stop codon, and 5′-ATGGATCCACCTCACTTGCATATATG-3′, which annealed 1 bp upstream of the rgg start site. The resulting 422-bp fragment was similarly cloned into pMI891Ω to create pMI8927.
The integrative plasmids were each transformed into the parental strain CH1, and erythromycin-resistant putative transformants were selected. Expected integration of pMI8923, pMI8924, and pMI8925 would result in the chromosomal region encoding rgg and the first 32 bp of the 5′ end of gtfG upstream of the inserted vector in the resulting strains CH8923, CH8924, and CH8925. However, the expected region downstream of the integrated vector differed in each strain, with increasing larger regions of rgg and the gtfG promoter duplicated before the complete gtfG gene. For each strain, the site of chromosomal integration was confirmed by Southern hybridization analyses using both pVA891 and the cloned insert as probes. Constructs with expected patterns on Southern hybridization analysis (data not shown) were confirmed by direct sequencing of PCR products amplified with oligonucleotide primers that flanked the inserted plasmids by using chromosomal template from each strain.
Construction of spectinomycin-resistant streptococcal plasmids.
In order to measure the effect of trans-acting rgg on strains with integrated erythromycin-resistant pMI891Ω, it was necessary to construct a stable streptococcal plasmid with a selectable antibiotic resistance determinant. A spectinomycin-resistant determinant with its own promoter and transcriptional terminator (14) cloned into the SmaI site of pGEM-7Zf− (Promega) was removed from the vector by digestion with EcoRI and BamHI, leaving a HindIII site from the multiple cloning region immediately downstream of the spectinomycin transcriptional terminator. The resulting ca. 1.1-kb fragment was blunted with Klenow fragment and cloned into the SnaBI site of the erythromycin resistance determinant in the streptococcal plasmid pVA749 (17). Digestion with HindIII, to remove the upstream region of the erythromycin resistance determinant, and recircularization resulted in plasmid pMISP49. The same cloning method was used to replace the SnaBI-HindIII fragment of pAMS57 with the spectinomycin resistance determinant, resulting in pMISP57 (Fig. 3). Although 163 bp of the downstream region of the erythromycin resistance structural gene remained, S. gordonii strains carrying pMISP49 and pMISP57 were spectinomycin resistant and erythromycin sensitive. Because these plasmids contained no E. coli origin of replication, all construction was done in S. gordonii strain CH1. The presence of plasmids in all putative transformant strains in these studies was confirmed by plasmid isolation and examination of restriction digest fragment patterns on agarose gels (2).
FIG. 3.
Maps of plasmids derived from the erythromycin-resistant streptococcal plasmid pVA749 (17) used in these studies to determine trans effects of rgg. pAMS57, which has been shown to increase gtfG transcription (24), carries a ca. 1.4-kb rgg fragment. To construct pMISP57, the ca. 1.6-kb SnaBI-HindIII fragment was removed from pAMS57 and replaced with a ca. 1.1-kb spectinomycin resistance determinant (14), as described in Materials and Methods. For control experiments, the spectinomycin-resistant vector pMISP47 was similarly constructed from pVA749. Reading directions of rgg and erythromycin resistance (Emr) and spectinomycin resistance (Spr) determinants are shown by arrows.
Construction of S. gordonii strains with chromosomal deletions of the 3′ end of rgg and various regions of the rgg-gtfG intergenic region.
S. gordonii strain CH1 was transformed with linear DNA that was constructed in E. coli from plasmids which contained the desired nucleotide sequence. Homologous recombination between similar chromosomal and linear DNA on both sides of the deletion site resulted in allelic exchange and integration of the desired mutation into the S. gordonii chromosome. Constructs were made in pMI2201, which carries a 2.255-kbp ApaI-SstI insert fragment with the parental 3′ end of htpX, rgg, and the 5′ end of gtfG (Fig. 1). A series of PCR products containing the desired deletions were designed to be cloned into compatible restriction sites in pMI2201 to facilitate replacement of parental nucleotides with the desired mutations.
To construct S. gordonii strain CHΔR1, which had a deletion between the SalI and EcoRI sites of rgg, the forward primer 5′ TCCTCGAGGGTAGAATTCAACTAACAGTAGTTGC-3′ was designed with a 5′ XhoI site (underlined) compatible with SalI, an in-frame engineered translational stop (boldface) designed to anneal to the EcoRI site (italics) of rgg, and the continuing downstream sequence (Fig. 1). Similarly, to construct strain CHΔ35, which has an additional 36-bp deletion of the 3′ end of rgg (nucleotides shown in italics in Fig. 1), the forward primer 5′-ATCTCGAGAATAGAACCTCCTCTGAAAAAG-3′ was used. This primer was designed to anneal 2 bp upstream of the rgg stop codon (boldface) and the gtfG putative −35 sequence (Fig. 1). To construct strain CHΔ10, which in addition to the deletion of the 3′ end of rgg also has a deletion of the putative gtfG promoter, the forward primer 5′-TACTCGAGGGTAAAACTTTATACTATTTTC-3′, designed to anneal to the sequence designated with the CHΔ10 arrow in Fig. 1, was used. Each forward primer was used in PCR with pMI2201 template and the same reverse primer 5′-TTGAGCTCAACTGCAAAGTT-3′ which contained the gtfG SstI site of pMI2201 (Fig. 1). The resulting PCR products were digested with XhoI and SstI and cloned into SalI-SstI-digested pMI2201, thereby replacing the 1.131-kb parental fragment. The nucleotide sequence of the entire resulting insert in each plasmid was determined to confirm the correct expected sequence.
To prepare DNA for construction of the S. gordonii strains, the pMI2201 derivative plasmids with the three different deletions were each digested with EcoRV and SstI. The resulting ca. 1.5-kb linear EcoRV-SstI fragments had 763 bp upstream of the rgg SalI site and a minimum of 750 bp of gtfG available for recombination with homologous chromosomal DNA on either side of the deletions. These fragments were eluted from agarose gels and transformed into strain CH1. Since the strains were all designed to encode truncated Rgg proteins as in strain DS512 (25), it was hypothesized that they would have similar Spp− colonies. Therefore, transformed cells were plated on TH agar with 3% sucrose. Spp− putative transformants were examined by Southern hybridization analysis, and strains with appropriately sized fragments hybridizing to rgg-gtfG probes (see Results) were chosen for nucleotide sequence analysis. Chromosomal DNA from each selected strain was used as the template in PCR with primers designed to anneal ca. 100 bp upstream of the ApaI site (5′-GGCAGCTACCTTGGTTCAGTTG-3′) and ca. 575 bp downstream of the SstI site (5′-AAGCAGACGATATCTAGAATTGGC-3′). The nucleotide sequences of the resulting products were determined directly to confirm the expected chromosomal sequences for each strain.
Construction of S. gordonii strains with chromosomal replacement of the rgg promoter and the 5′ end of rgg with an Ω fragment.
A similar approach using allelic replacement was used to construct strains CHΩN and CHΩR1. To ensure that transcription of rgg did not occur due to readthrough from the upstream lemA-htpX transcript (37), the Ω transcriptional terminator fragment in pMI22Ω was digested with EcoRV and NcoI and directionally cloned into the EcoRV site immediately downstream of the htpX gene and NcoI site in rgg in pMI2201 (Fig. 1). Similarly, the Ω fragment was removed from pMI22Ω by digestion with EcoRV and EcoRI and cloned into the corresponding sites downstream of htpX and within rgg in pMI22Ω. This resulted in replacement of the rgg promoter and the 5′ end of rgg up to the NcoI site or the EcoRI site with an Ω terminator. After digestion of the resulting plasmids with ApaI and SstI, the ca. 1.762-kb and 1.219-kb linear fragments, corresponding to the Ω replacement of the EcoRV-NcoI and EcoRV-EcoRI parental fragments in pMI2201, respectively, were each transformed into strain CHI. Spp− colonies were selected for further characterization by Southern hybridization and nucleotide sequence analysis to confirm the expected chromosomal sequences.
Construction of recA-deficient strains.
To minimize the potential of recombination between plasmid-borne and chromosomal copies of rgg in complementation studies, recA-deficient strains were constructed. After confirmation of the desired rgg and gtfG chromosomal sequences, strains were transformed with pAM6200, a spectinomycin-resistant streptococcal integrative plasmid carrying a ca. 300-bp internal fragment of the S. gordonii recA gene (32). Expected integration of pAM6200 into the streptococcal chromosome was confirmed by Southern hybridization using the internal 300-bp recA fragment as a probe. Sensitivity to UV light, as an additional indicator of recA deficiency, was detected by inhibition of growth on agar plates, as previously described (32). The recA-deficient strain CHR3 (32), a similarly constructed mutant of the parental strain CH1, was used as the positive control.
Determination of GTF activity.
Relative amounts of GTF protein were determined by measuring the enzyme activity in polyacrylamide gels as previously described (26, 34). Briefly, strains were grown to the same mid- to late-log stage as determined by cell density (optical density at 520 nm [OD520] in FMC medium, or OD600 in TH medium) and equal volumes of cell-free supernatants were run on sodium dodecyl sulfate-8.75% polyacrylamide gel electrophoresis (SDS-PAGE) gels. After electrophoresis, gels were incubated overnight in 3% sucrose, 0.5% Triton X-100 in 10 mM sodium phosphate, pH 6.8, at 37°C, and the resulting glucan bands were treated with periodic acid and pararosanaline. The intensity of the stained bands reflects the relative amounts and activities of the GTF protein (26, 36). Relative amounts of GTF activity were determined via laser densitometric scanning (Ultrascan XL; LKB) of at least four independent gels. Activities are reported as a percentage of the parental activity (set at 100%) on the same gel. For strains in which the recA gene was disrupted by integration of pAM6200, the activity was compared to that of strain CHR3, i.e., the parental strain CH1 with integrated pAM6200 (32). Previous studies (26, 34) have shown that this method of measuring S. gordonii GTF activity is comparable in sensitivity to that of assays using [14C-glucose]sucrose incorporation into glucan polymers.
RESULTS
Separation of rgg and gtfG results in Spp− colonies.
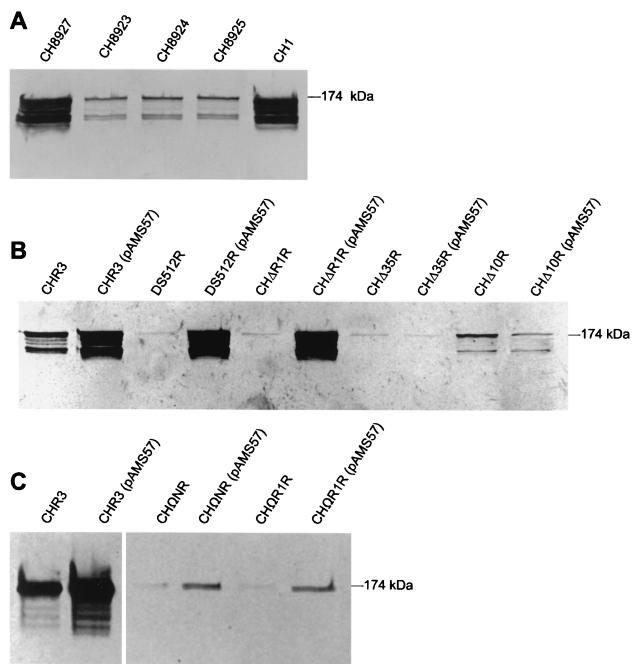
To examine effects of separating rgg from gtfG by intervening vector sequences, strain CH8923 was constructed and its GTF activity was examined. The resulting colonies were Spp− on sucrose agar plates, suggesting a decreased level of GTF activity (Fig. 4). This was confirmed by GTF activity gels which showed that strain CH8923 had only 24.75% ± 8.1% (mean ± standard deviation) of the parental level of GTF activity (Fig. 5A). These results indicated that in S. gordonii gtfG was transcribed from its own functional promoter and did not require the polycistronic rgg-gtfG transcript from the rgg promoter for gtfG expression. Nevertheless, although strain CH8923 had a complete chromosomal copy of rgg upstream of the integrated vector and a complete putative promoter and gtfG downstream of the integrated vector, this strain did not exhibit parental levels of GTF activity. Since the duplicated 136-bp region containing the gtfG promoter could potentially act as a titration site for trans-acting products of rgg, strain CH8923 was transformed with the multicopy plasmid pMISP57, which carried rgg (Fig. 3), in order to overcome any potential titration effects. Unexpectedly, transformants had no change in the Spp− colony phenotype (Fig. 4) or increase in GTF activity (23.2% ± 7.6% of the parental level of GTF activity). Although previous results in E. coli had suggested that the 136-bp EcoRI-NsiI DNA region duplicated in strain CH8923 was sufficient to allow plasmid-borne rgg to increase gtfG transcription (24), it was possible that in S. gordonii additional DNA upstream of the EcoRI site was necessary for the product of rgg to increase gtfG transcription and produce Spp+ colonies. To examine this possibility, strains with increasingly larger regions of DNA upstream of the rgg EcoRI site were similarly constructed.
FIG. 4.
Schematic representation of S. gordonii chromosomal regions downstream of the integrated pMI891Ω and the sucrose-promoted colony phenotypes of each strain. The parental rgg promoter and structural gene and the first 32 bp of gtfG are upstream of the integrated vector in strains CH8923, CH8924, and CH8925. The control strain, CH8927, has only the rgg promoter region directly upstream of the integrated vector. The sizes (base pairs) and relevant regions of the DNA fragments cloned into pMI891Ω and duplicated downstream of the chromosomally integrated vector in each strain are shown with gray shading. The vector with its Ω termination region is indicated in black. Relative positions of engineered MunI sites cloned into the EcoRI vector site (RI/M) and the NcoI (N), SalI (S), and EcoRI (R1) restriction sites within rgg are shown. DNA inverted repeats are designated with large arrowheads. The downstream region of strain CH8925 contains the ATG start codon of rgg. The downstream region of strain CH8927 contains the duplicated rgg promoter (−35 and −10) and ribosomal binding site (SD) as well as the single chromosomal copies of rgg and gtfG. Sucrose-promoted colony phenotype (Spp) as an indicator of GTF activity was determined for each strain when plasmid-free or when transformed with pMISP49 (vector control) or pMISP57 (carrying rgg). Results are expressed relative to the established phenotypes of the Spp+ parental strain CH1, the similar Spp+ colonies of strain CH1(pVA749) and CH1(pMISP49), and the Spp+++ colonies of CH1(pAMS57) (25) and CH1(pMISP57).
FIG. 5.
GTF activity gels. Cells were grown at 36°C to the same mid- to late-log stage as determined by OD520 and OD600 readings in FMC or TH medium, respectively. Equal volumes of cell-free culture supernatants were run on SDS-PAGE gels, incubated in sucrose and Triton X-100, and treated with periodic acid, and the resulting glucan bands were stained with pararosanaline. Band intensity is proportional to GTF activity. The position of the ca. 174-kDa native GTF protein is indicated. Beneath the native protein band are lower-molecular-mass bands with various levels of GTF activity. These lower-molecular-mass forms of GTF are thought to be due to the degradation of the native GTF enzyme by endogenous proteases, and they occur even in the presence of protease inhibitors (10) in both FMC and TH media (36). Gels shown in panels A and B are from strains grown in FMC. The gel in panel C is from strains grown in TH medium. Both media gave similar proportions of GTF activity for all strains (data not shown). Results shown are representative of a minimum of four independent gels. (A) Comparison of the parental strain CH1 to strains with regions of rgg and gtfG separated by the integrated vector pMI891Ω. Each strain has one intact chromosomal copy of the parental rgg promoter and adjacent rgg structural gene. (B) Comparison of the parental recA-disrupted parental strain CHR3 to plasmid-free recA-disrupted strains with deletions in the 3′ end of rgg and the rgg/gtfG intergenic region, and to transformant strains carrying rgg in pAMS57. (C) Effects of pAMS57 on GTF activities of strain CHR3 and recA-disrupted strains with replacement of the rgg promoter and 5′ end of rgg with a transcriptional and translational termination fragment.
Additional DNA immediately upstream of the gtfG promoter is necessary for plasmid-borne rgg to increase transcription.
Although strains CH8924 and CH8925, which contained an additional 506 and 854 nucleotides immediately upstream of the rgg EcoRI site, respectively, had the parental rgg promoter and structural gene upstream of the integrated vector, these strains had Spp− colonies and decreased levels of GTF activity (28.2% ± 9.1% and 26.3% ± 12.8% of the parental level, respectively), similar to that of strain CH8923 (Fig. 5A). Furthermore, transformation of these strains with plasmid-borne rgg was unable to confer the Spp+ phenotype. However, the control strain CH8927, which had a duplication of the rgg promoter and ribosomal binding site upstream of the integrated vector and the rgg promoter, ribosomal binding site, and structural gene adjacent to gtfG downstream of the integrated vector (Fig. 4), had Spp+ colonies and GTF activity similar to the Spp+ parental strain CH1 (Fig. 5A). This confirmed that the integrated vector did not interfere with GTF activity. Additionally, strain CH8927(pMISP57), which carried plasmid-borne rgg, had Spp+++ colonies similar to those of the Spp+++ parental strain CH1(pMISP57) (Fig. 4). Spp+++ colonies are very hard and associated with levels of GTF activity higher than parental levels (25). These findings suggested that transcription and/or translation of chromosomal rgg adjacent to gtfG was necessary for parental levels of GTF activity. Furthermore, this gene arrangement appeared to be necessary for the product of plasmid-borne rgg to increase GTF activity to the level that the synthesized glucans conferred the Spp+++ colony phenotype.
Strains with chromosomal deletion of the 3′ end of rgg and the gtfG promoter region are Spp−.
To overcome the difficulties in interpretation due to potential titration effects and/or recombinations between similar plasmid-borne and chromosomal flanking DNA, a series of strains was constructed via allelic replacement that had chromosomal deletions without integrated vectors. Strains CHΔR1, CHΔ35, and CHΔ10 were designed with nested deletions of the 3′ end of rgg and the rgg-gtfG intergenic region (Fig. 1). These strains had engineered translational stop codons similar to that of strain DS512, which has a premature translational stop site downstream of the SalI site of rgg (25). Thus, the constructed strains would encode truncated Rgg similar to that of strain DS512 and would be expected to exhibit low levels of GTF activity. Accordingly, putative transformants were selected by their Spp− colonies on sucrose agar and analyzed.
Southern analysis of strain DS512 confirmed the previously reported (25) loss of the internal rgg SalI site, which destroys the overlapping HincII site (Fig. 6). Strains CHΔR1, CHΔ35, and CHΔ10 also showed loss of the HincII site, as expected from compatible annealing of the SalI- and XhoI-digested DNA used to construct these strains. Strains CHΔ35 and CHΔ10 showed the loss of the rgg EcoRI site. Chromosomal DNA of these strains was also probed with a DNA fragment containing the common region of desired deletion shared by strains CHΔR1, CHΔ35, and CHΔ10 (Fig. 6, Probe III). Although Probe III hybridized to fragments of expected sizes in strains CH1 and DS512, no hybridization to chromosomal DNA of strains CHΔR1, CHΔ35, and CHΔ10 was detected (data not shown), indicating that the desired deletion had occurred in each of these strains. Nucleotide sequence analysis confirmed these results.
FIG. 6.
(A) Restriction digest map of strain CH1 chromosomal DNA and positions of DNA probes used in Southern hybridization analysis of strains constructed by chromosomal allelic replacement. Arrows above the base-pair markers indicate the positions and reading directions of htpX, rgg, and gtfG. Restriction digest sites are indicated as HindIII (H3), EcoRI (RI), HincII (H2), EcoRV (RV), SpeI (Sp), NcoI (N), SalI (S), and NsiI (Ns). DNA probes made by PCR are designated with roman numerals. The 641-bp Probe I made with primers 5′-TAAATACCATTGTGATCAGG-3′ and 5′-AATGCATTTTATAATGAAC-3′ corresponded to the region 35 bp downstream of the NcoI site within rgg through the NsiI site of gtfG. Probe II contained a 392-bp internal region of htpX (37). Probe III was made with primers 5′-CTACTTAATGTTGTG-3′ and 5′-GCTCAAATTGTTCTTTGAG-3′ to amplify the 253-bp internal rgg region located 14 bp downstream of the SalI site through the EcoRI site. (B) Southern blots of chromosomal DNA from each strain digested with restriction enzymes (designated below each blot), transferred to nylon membranes, and hybridized with Probe I or Probe II to confirm the integration of transformed linear DNA containing replacement and/or deletion of chromosomal sequences.
Opening of the potential stem-loop sequestering the gtfG putative promoter and ribosomal binding site increases GTF activity.
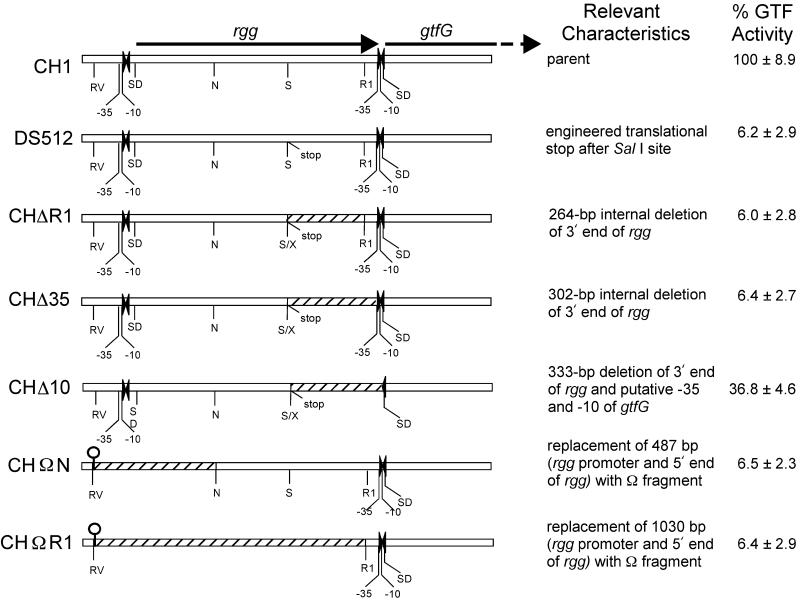
The ca. 6% level of GTF activity compared to that of the parental strain CH1 of strain DS512 (Fig. 7) was consistent with the previously reported 3 to 5% levels reported for this strain (25). As expected, strains CHΔR1 and CHΔ35 had similar low levels of GTF activity (Fig. 7). Thus, the deletion of the DNA downstream of the SalI site in strains CHΔR1 and CHΔ35 did not affect the expression of gtfG compared with the level seen in strain DS512, which encoded a similarly truncated Rgg.
FIG. 7.
Schematic representation of chromosomal regions of S. gordonii strains and relative levels of GTF activity. The white bar indicates existing DNA, whereas the hatched areas indicate deletions. Engineered translational stops, resulting in a similarly encoded truncated Rgg in strains DS512, CHΔR1, CHΔ35, and CHΔ10, are indicated. Putative −35 and −10 promoter regions and Shine-Delgarno ribosomal binding sites (SD) are indicated within regions of dyad symmetry (inverted arrowheads). Strains CHΩN and CHΩR1 have an omega terminator (black loop) replacing the rgg promoter and 5′ end of rgg. Thus, none of these engineered strains can express the product of the parental rgg. Relevant restriction sites are indicated for EcoRV (RV), EcoRI (RI), NcoI (N), SalI (S), and the compatible XhoI (X) site. Relative percent GTF activity (± standard deviation) is shown for each strain as a percentage of strain CH1 activity, determined by densitometric scanning of activity gels (n ≥ 4). Values for the parental strain CH1 were set at 100% for each gel.
The deletion of nucleotides in the inverted repeat in the rgg-gtfG intergenic region in strain CHΔ10 was designed to examine effects of potential nucleic acid secondary structures on gtfG expression. Although strain CHΔ10 had a deletion of the 3′ end of rgg similar to that of strains CHΔR1 and CHΔ35, it showed a significant increase (P ≤ 0.00001; one-tailed Student's t test) in GTF activity compared with these strains (Fig. 7). The strain CHΔ10 nucleotide deletions, which disrupted the region of dyad symmetry, removed the putative gtfG promoter; the putative ribosomal binding site remained. Although it is possible that the DNA deletion in this AT-rich region resulted in the creation of a stronger artificial promoter in strain CHΔ10, it is also possible that because of the nucleotide deletions, the rgg-gtfG polycistronic transcript could not form the potential mRNA stem-loop that sequestered the gtfG ribosomal binding site. Such increased accessibility of the ribosomal binding site could lead to increased translation. Nevertheless, the strain CHΔ10 level of GTF activity was only ca. 36% of the parental strain CH1 level and resulted in Spp− colonies.
Deletion of the rgg promoter and the 5′ end of rgg results in decreased GTF activity.
To examine the role of the rgg promoter in gtfG expression, strains were constructed in which this region was deleted and replaced with an Ω terminator fragment. Additional DNA was deleted in the 5′ end of rgg in order to determine any potential stabilization or secondary structure roles that upstream DNA might play in gtfG expression. Since it was expected that transformants with the desired deletions in the 5′ end of rgg would not synthesize parental levels of glucans, Spp− colonies were selected as putative transformants. Southern hybridization analysis indicated that the desired chromosomal deletions of the rgg promoter up to the NcoI and EcoRI sites within rgg were present in strains CHΩN and CHΩR1, respectively (Fig. 6). Southern blotting demonstrated that these strains had a loss of the SpeI site within the parental rgg putative promoter region, as well as the addition of a HindIII site which is present within the Ω terminator fragment (8), thereby suggesting that the rgg promoter had been replaced by the integrated Ω terminator. Nucleotide sequence analysis confirmed these results. Examination of the GTF activities of strains CHΩN and CHΩR1 indicated that these strains had only ca. 6% of the parental level of GTF activity (Fig. 7). These GTF levels were similar to those of strains DS512, CHΔR1, and CHΔ35, which had both rgg and gtfG putative promoters. The relative strengths of the rgg and gtfG promoters in these engineered strains, which did not express the complete rgg structural gene, were not directly measured in these studies. GTF activities seen in strains CHΩN and CHΩR1 supported results observed with strain CH8923, which had a transcriptional terminator immediately upstream of the putative gtfG promoter. These results strongly suggest that the gtfG promoter alone, without the upstream rgg promoter, is sufficient for a low level of GTF expression in the S. gordonii genetic background.
Determination of the chromosomal region necessary for trans-acting rgg to increase gtfG transcription.
In order to gain insights into the chromosomal region necessary for the rgg product to increase gtfG expression, recombination-deficient strains were constructed. The recA gene was disrupted in strains DS512, CHΔR1, CHΔ35, CHΔ10, CHΩN, and CHΩR1 (data not shown) to minimize the potential for recombination between plasmid-borne and chromosomal copies of rgg. The resulting recombination-deficient strains, designated by the addition of the letter R to the strain name, were transformed with pAMS57, which carries rgg, and the GTF activities were determined. The results from the recA-deficient parental derivative CHR3 and from strain DS512R were consistent with previous findings with the corresponding recA+ strains (25, 32). Transformation with a plasmid carrying rgg resulted in similar GTF levels in strains CHR3(pAMS57) and DS512R(pAMS57) that were ca. threefold higher than that seen in the plasmid-free parental derivative CHR3; this level was a ca. 90-fold increase above the activity level of strain DS512R (Fig. 5B).
Strain CHΔR1R had an increase in GTF activity similar to that of strain DS512 (Fig. 5B). Strains CHR3 and DS512R and CHΔR1R all had the same level of GTF activity (within 10%) when transformed with pAMS57. However, transformation with pAMS57 did not increase the GTF activity of strains CHΔ35R and CHΔ10R. These results indicated that the 36 nucleotides at the 3′ end of the chromosomal rgg in strain CHΔR1R, which were not present in strains CHΔ35R (shown in italics in Fig. 1), were necessary for plasmid-borne rgg to increase GTF activity.
Strains CHΩNR and CHΩR1R also had these 36 chromosomal nucleotides at the 3′ end of rgg. However, transformation of these strains with pAMS57 did not result in the same magnitude of increased GTF activity as seen in strains DS512R(pAMS57) and CHΔR1R(pAMS57). Rather than the ca. 90-fold increase which resulted in a GTF activity level identical to that of CHR3(pAMS57), strains CHΩNR(pAMS57) and CHΩR1R(pAMS57) had only a 2- to 3-fold increase in GTF activity compared to that in the plasmid-free strains (Fig. 5C). These results suggest the possibility that the presence of an upstream promoter to allow transcription of the 36 nucleotides at the 3′ end of rgg was necessary for the trans-acting rgg product to increase GTF activity to the maximum extent seen in strains CHR3, DS512R, and CHΔR1R.
DISCUSSION
The ability of oral streptococci to regulate genes involved in extracellular polysaccharide production can have important implications for the oral microbial ecology. The production of glucans by oral streptococci has been associated with their ability to colonize and accumulate on tooth surfaces (15). Furthermore, the glucans synthesized by the early colonizers of dental plaque can act as substrata for later colonizing bacterial species, such as those with glucan-binding surface proteins. Thus, regulation of GTF enzymes could affect the microbial composition of the biofilm on tooth surfaces and influence the state of health or disease. The results of the present studies confirm that control of GTF expression in the early colonizing commensal species, S. gordonii, is complex. Previous studies indicate that the product of the regulatory determinant, rgg, acts in trans near the gtfG promoter to increase gtfG transcription (24). The present studies support this model, but they also suggest that regulation of gtfG by rgg involves additional molecular mechanisms.
Based on nucleotide sequence analysis, it has been previously hypothesized that translation of rgg is necessary for parental levels of gtfG expression (25). In the parental strain CH1, a stem-loop corresponding to the factor-independent transcription termination site of rgg, but containing the gtfG ribosome binding site, may form in the polycistronic rgg-gtfG mRNA and influence the efficiency of gtfG expression (25). In the parental strain, the gtfG ribosomal binding site may become available when secondary structure is melted by the translation of rgg, as occurs in translational coupling (19). The decreased levels of GTF activity seen in strains CH8923, CH8924, and CH8925, which have intervening vector sequences between rgg and gtfG, suggest that rgg and gtfG expression must be adjacent to each other to provide parental levels of GTF activity. The likelihood of cis effects of rgg on gtfG expression is further supported by the GTF levels seen in strain CHΔ10. The deletion of the putative gtfG promoter in this strain also results in loss of nucleotides involved in the potential mRNA secondary structure of the polycistronic transcript. The disruption of the potential stem-loop between rgg and gtfG presumably would increase the accessibility of the gtfG ribosomal binding site to allow increased translation of gtfG. The increased levels of GTF activity seen in strain CHΔ10, compared with GTF levels seen in the other rgg deletion strains CHΔR1 and CHΔ35, support this possibility. Although such translational coupling, in which efficient translation of a gene is dependent on the translation of the gene immediately preceding it, is fairly well described in E. coli (21), the optimal configuration of the translational stop and start codons is not known in streptococci. Changes in the distance between these sites can increase or decrease the level of gene expression (30). Further studies to examine these parameters in S. gordonii gtfG regulation are in progress.
The present studies have also provided insights into the mechanism by which Rgg increases GTF activity. A 36-bp region within the 3′ end of rgg, extending from the internal EcoRI site to two nucleotides upstream of the translational stop, has been demonstrated to be essential for plasmid-borne rgg to increase GTF activity. This implies that the 3′ end of rgg is involved, either directly as a binding site or indirectly, by playing a role in secondary structure, in the interaction of Rgg with the gtfG promoter. Increased GTF activity in the presence of plasmid-borne rgg was seen in strains with this chromosomal region, whether both rgg and gtfG promoters were present, as in strains DS512 and CHΔR1, or if only the gtfG promoter was present, as in strains CHΩR1 and CHΩN. Thus, Rgg appears to interact with this 36-bp region at the DNA level. Such a role for Rgg is supported by computer structural predictions (3) that amino acids 11 through 64 have a helix-turn-helix structural motif suggestive of a DNA binding function (39).
These genetic studies with S. gordonii have implications for understanding regulatory functions of other rgg-like determinants which have been shown to positively regulate transcription of adjacent genes. In S. pyogenes, an rgg-like gene designated as either ropB (16) or rgg (4) is a positive transcriptional regulator of the speB gene encoding the streptococcal erythrogenic toxin B. In S. mutans, the rgg-like mutR regulates expression of the mutA structural gene in the lantibiotic mutacin II operon (20). The rgg-like gadR of L. lactis encodes a chloride ion-inducible positive regulator of the gadC/B operon involved in pH homeostasis (23). As seen in the S. gordonii rgg-gtfG intergenic region, it has been noted that these other characterized rgg-like genes are separated from the genes they regulate by DNA inverted repeats (16, 20, 23). However, these inverted repeats do not share sequence similarity with each other. Computer analysis (3) of these encoded Rgg-like proteins (38) indicates that despite their varied target genes, they share a similar helix-turn-helix motif with potential DNA binding function (39). Comparison of these characterized rgg-like gene sequences using Clustal analysis (29) indicates that despite overall homology, the 3′ ends of these genes and the carboxyl-terminal regions of their encoded proteins are less conserved (38). This suggests that functional specificity of Rgg-like proteins may involve amino acids in this region. Accordingly, the 36-bp region identified in the present studies as playing an essential role for plasmid-borne rgg to increase gtfG expression is located within the 3′ end of rgg immediately upstream of the DNA inverted repeat sequestering the gtfG putative promoter and ribosomal binding site. Sequences similar to these 36 nucleotides are not found in the 3′ end of S. mutans mutR, L. lactis gadR, or S. pyogenes ropB(rgg), nor is this 36-bp sequence found near the inverted repeats upstream of target genes in these species. Furthermore, nucleic acid database searches (1) did not identify conservation of the S. gordonii 36-bp sequence in other bacteria. This does not preclude the possibility that specific nucleotides within this 36-bp region are conserved among gram-positive species as binding sites for Rgg-like proteins. Nor does it preclude the possibility that additional nucleotides near the gtfG promoter region interact with Rgg. Additional studies will be necessary to identify these specific nucleotides.
A more unexpected result from the present studies was that the magnitude of Rgg interaction with this 36 bp to increase GTF activity was significantly greater if the rgg promoter, ribosomal binding site, and portions of the structural gene were directly upstream of gtfG. This was first evident when comparing GTF levels of strains constructed via plasmid integration. Plasmid-borne rgg could not produce Spp+ colonies in strains with rgg separated from gtfG by integrated vector sequences; intervening nucleotides prevented a polycistronic rgg-gtfG transcript in these strains. This finding was supported in strains DS512 and CHΔR1, constructed by allelic replacement. Due to engineered premature translational stops, the 36-bp region implicated in the Rgg binding interaction could not be translated in either of these two strains. However, both strains had an rgg promoter so that an rgg-gtfG transcript was possible. The level of GTF activity in strains DS512R(pAMS57) and CHΔR1R(pAMS57) was ca. 90-fold higher than that in strains CHΩNR(pAMS57) and CHΩR1R(pAMS57). The chromosomes of the latter two strains each had the 36-bp potential Rgg binding site, but due to the replacement of the rgg promoter with the Ω termination fragment, these strains could not make rgg-specific and rgg-gtfG transcripts. The 36-bp chromosomal region implicated as the Rgg binding site begins 72 bp upstream of the gtfG transcriptional start site previously determined by primer extension analysis (Fig. 1) (24). The basis for the difference in GTF activity levels in these strains is not known. However, taken together, the data raise the intriguing possibility that in addition to its DNA binding function, Rgg may also interact with the polycistronic rgg-gtfG transcript to increase levels of gtfG expression. It can be speculated that this may involve mRNA processing or stabilization. Although computer database searches (1, 3) did not identify recognized RNA binding motifs within rgg or the immediate downstream region, this does not preclude the existence of an as-yet-unrecognized motif. Additional studies will be necessary to investigate this hypothesis.
The potential role the S. gordonii Rgg plays with the rgg-gtfG transcript would be possible for other Rgg-like proteins if the regulatory and target genes were transcribed in the same reading direction; this occurs with S. mutans mutR and L. lactis gadR (20, 23). However, the S. pyogenes ropB (rgg) and its target speB are transcribed in divergent reading directions, precluding the existence of an ropB (rgg)-speB polycistronic transcript (16). Furthermore, recent studies have indicated that S. pyogenes ropB (rgg) may influence a variety of distally located extracellular proteins in addition to SpeB (5). Thus, not all aspects of S. gordonii rgg regulation of gtfG may be generally applicable to all rgg-like determinants. Nevertheless, the present studies have provided important insights into the specificity of gtfG regulation by rgg, and they have provided the bases for additional studies aimed at elucidating the complex molecular mechanisms underlying the control of GTF activity in S. gordonii.
Acknowledgments
This work was supported by U.S. Public Health Service grant DE11090 from the National Institutes of Health.
Editor: V. J. DiRita
REFERENCES
- 1.Altschul, S. G., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., B. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 3.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillard, J. P., and J. Yother. 1991. Analysis of Streptococcus pneumoniae sequences cloned into Escherichia coli: effect of promoter strength and transcription terminators. J. Bacteriol. 173:5105-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey, J., and H. M. Krisch. 1985. Ω mutagenesis in gram-negative bacteria: a selectable interposon which is strongly polar in a wide range of bacterial species. Gene 36:143-150. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara, T., T. Hoshino, T. Ooshima, S. Sobue, and S. Hamada. 2000. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 68:2475-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grahame, D. A., and R. M. Mayer. 1984. The origin and composition of multiple forms of dextransucrase from Streptococcus sanguis. Biochim. Biophys. Acta 786:42-48. [DOI] [PubMed] [Google Scholar]
- 11.Haisman, R. J., and H. F. Jenkinson. 1991. Mutants of Streptococcus gordonii Challis over-producing glucosyltransferase. J. Gen. Microbiol. 137:483-489. [DOI] [PubMed] [Google Scholar]
- 12.Kilic, A. O., M. C. Herzberg, M. W. Meyer, X. Zhao, and L. Tao. 1999. Streptococcal reporter gene-fusion vector for identification of in vivo expressed genes. Plasmid 42:67-72. [DOI] [PubMed] [Google Scholar]
- 13.Lawson, J., and H. Gooder. 1970. Growth and development of competence in the group H streptococci. J. Bacteriol. 102:820-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeBlanc, D. J., L. N. Lee, and J. M. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macrina, F. L., J. A. Tobian, K. R. Jones, and R. P. Evans. 1981. Molecular cloning in the streptococci, p. 195-210. In A. Hollaender, R. DeMoss, S. Kaplan, J. Konisky, D. Savage, and R. Wolfe (ed.), Genetic engineering of microorganisms for chemicals. Plenum Publishing Corp., New York, N.Y.
- 18.Macrina, F. L., R. P. Evans, J. A. Tobian, D. L. Hartley, D. B. Clewell, and K. R. Jones. 1983. Novel shuttle plasmid vehicles for Escherichia-Streptococcus transgeneric cloning. Gene 25:145-150. [DOI] [PubMed] [Google Scholar]
- 19.Oppenheim, D. S., and C. Yanofsky. 1980. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics 95:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi, G., P. Chen, and P. W. Caufield. 1999. Functional analysis of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rex, G., B. Surin, G. Besse, B. Schneppe, and J. E. G. McCarthy. 1994. The mechanism of translational coupling in Escherichia coli. J. Biol. Chem. 269:18118-18127. [PubMed] [Google Scholar]
- 22.Sambrook, J., E. R. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 24.Sulavik, M. C., and D. B. Clewell. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J. Bacteriol. 178:5826-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardif, G., M. Sulavik, G. W. Jones, and D. B. Clewell. 1989. Spontaneous switching of the sucrose-promoted colony phenotype in Streptococcus sanguis. Infect. Immun. 57:3945-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 28.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van de Guchte, M., J. Kok, and G. Venema. 1991. Distance-dependent translational coupling and interference in Lactococcus lactis. Mol. Gen. Genet. 227:65-71. [DOI] [PubMed] [Google Scholar]
- 31.Vickerman, M. M., D. B. Clewell, and G. W. Jones. 1991. Ecological implications of glucosyltransferase phase variation in Streptococcus gordonii. Appl. Environ. Microbiol. 57:3648-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vickerman, M. M., D. G. Heath, and D. B. Clewell. 1993. Construction of recombination-deficient strains of Streptococcus gordonii by disruption of the recA gene. J. Bacteriol. 175:6354-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vickerman, M. M., M. C. Sulavik, and D. B. Clewell. 1995. Oral streptococci with genetic determinants similar to the glucosyltransferase regulatory gene rgg. Infect. Immun. 63:4524-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickerman, M. M., M. C. Sulavik, P. E. Minick, and D. B. Clewell. 1996. Changes in the carboxyl terminal repeat region affect extracellular activity and glucan products of Streptococcus gordonii glucosyltransferase. Infect. Immun. 64:5117-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vickerman, M. M., M. C. Sulavik, J. D. Nowak, N. M. Gardner, G. W. Jones, and D. B. Clewell. 1997. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene gtfG. DNA Sequence 7:83-95. [DOI] [PubMed] [Google Scholar]
- 36.Vickerman, M. M., and D. B. Clewell. 1997. Deletions in the carboxyl terminal region of Streptococcus gordonii glucosyltransferase affect cell-associated enzyme activity and sucrose-associated accumulation of growing cells. Appl. Environ. Microbiol. 63:1667-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vickerman, M. M., N. M. Mather, P. E. Minick, and C. A. Edwards. 2002. Initial characterization of the Streptococcus gordonii htpX gene. Oral Microbiol. Immunol. 17:22-31. [DOI] [PubMed] [Google Scholar]
- 38.Vickerman, M. M., P. E. Minick, and N. M. Mather. 2001. Characterization of the Streptococcus gordonii chromosomal region immediately downstream of the glucosyltransferase gene. Microbiology 147:3061-3070. [DOI] [PubMed] [Google Scholar]
- 39.Wintjens, R., and M. Rooman. 1996. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262:294-313. [DOI] [PubMed] [Google Scholar]