Abstract
Arginine carboxypeptidase was isolated from the cytoplasm of Porphyromonas gingivalis 381 and purified by DEAE-Sephacel column chromatography, followed by high-performance liquid chromatography on DEAE-5PW and TSK G2000SWXL. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the purified enzyme revealed the presence of three major bands at 42, 33, and 32 kDa with identical N-terminal sequences. By Western blotting analysis and immunoelectron microscopy, the arginine carboxypeptidase was found to be widely distributed in the cytoplasm and on the surface of the outer membrane. The open reading frame corresponding to the N-terminal amino acids of the arginine carboxypeptidase was detected by a search of the sequence of the P. gingivalis W83 genome. This sequence showed homology with mammalian carboxypeptidases (M, N, and E/H) and included a zinc-binding region signature, suggesting that the enzyme is a member of the zinc carboxypeptidase family. The purified enzyme was inhibited by EGTA, o-phenanthroline, dl-2-mercaptomethyl-3-guanidinoethylthiopropanoic acid, and some metal ions, such as Cu2+, Zn2+, and Cd2+. On the other hand, Co2+ activated the enzyme. The enzyme released arginine and/or lysine from biologically active peptides containing these amino acids at the C terminus but did not cleave substrates when proline was present at the penultimate position. These results indicate that the arginine carboxypeptidase produced by P. gingivalis is an exo type of metallocarboxypeptidase. This enzyme may function to release arginine in collaboration with an arginine aminopeptidase, e.g., Arg-gingipain, to obtain specific amino acids from host tissues during the growth of P. gingivalis.
Porphyromonas gingivalis is an anaerobic, gram-negative rod that is frequently isolated from periodontal pockets of patients with adult periodontitis. The pathogenic properties of this bacterium have been extensively studied, especially the proteolytic enzymes, such as collagenase, trypsin-like enzymes (Arg-gingipain and/or Lys-gingipain), elastase, and other peptidases (1, 3, 13, 21, 25, 35). These enzymes exhibit their nutritional roles in deep periodontal pockets, where proteinaceous material is abundant. P. gingivalis obtains energy for growth by metabolizing amino acids that are released by these enzymes. Previously, we have reported arginine consumption in culture medium by P. gingivalis (19). In contrast to the arginine concentration, citrulline and ornithine concentrations increased up to late log phase. We also found that P. gingivalis cell extracts clearly demonstrated enzyme activities for the arginine deiminase pathway and ATP production (19). A similar arginine deiminase pathway for arginine catabolism has also been reported in other bacteria (6). This pathway is directly related to energy production, and citrulline and ornithine are intermediate products. Since some of the proteolytic enzymes described above have strong activity for arginine-containing peptides, it is hypothesized that arginine plays an important role in obtaining energy for the growth of P. gingivalis. The release of arginine from peptides is suggested to result from a two-step mechanism: first, an aminopeptidase like Arg-gingipain is needed to split the bond on the carboxyl side of arginine, and second, a carboxypeptidase is needed to cleave the bond on the amino side of arginine. However, little is known of the carboxypeptidase produced by putative periodontopathogenic bacteria. We have previously reported the production of this enzyme by P. gingivalis (17, 18). This report describes the purification and characterization of the arginine carboxypeptidase (RCP) produced by P. gingivalis.
MATERIALS AND METHODS
Chemicals.
Hippuryl-histidyl-leucine, angiotensin I (Asp-Arg-Val-Tyr-His-Pro-Pro), and fibronectin fragment (Gly-Arg-Asp-Ser) were purchased from the Peptide Institute Inc. (Osaka, Japan). Hippuryl-arginine, hippuryl-lysine, hippuryl-phenylalanine, Gly-Phe-Tyr-Arg, Gly-Gly-Tyr-Arg, tufsin (Thr-Lys-Pro-Arg), laminin fragment (Tyr-Ile-Gly-Ser-Arg), anaphylatoxin C3a fragment 72-77 (His-Leu-Gly-Leu-Ala-Arg), anaphylatoxin C3a fragment 70-77 (Ala-Ser-His-Leu-Gly-Leu-Ala-Arg), bradykinin (Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg), Kentsin (Thr-Pro-Arg-Lys), Val-His-Leu-Thr-Pro-Val-Glu-Lys, Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK), p-bromophenacryl bromide, and gel filtration molecular size markers were obtained from Sigma Chemical Co. (St. Louis, Mo.). Phenylmethylsulfonyl fluoride, n-ethylmaleimide, dithiothreitol, bovine DNase I, bovine pancreas RNase A, and EDTA were purchased from Nakarai Tesque Inc. (Kyoto, Japan). EGTA and dl-2-mercaptomethyl-3-guanidinoethylthiopropanoic acid (MGTA; Plummer's inhibitor) were obtained from Dozin Laboratory (Kumamoto, Japan) and Calbiochem-Novabiochem Co. (San Diego, Calif.), respectively.
Bacterial strains and culture conditions.
Strains of P. gingivalis 381 and ATCC 33277, Fusobacterium nucleatum ATCC 23726, Actinobacillus actinomycetemcomitans ATCC 29522, Bacteroides forsythus ATCC 43037, and Treponema denticola ATCC 35405 were obtained from Sunstar Inc. (Osaka, Japan) through the courtesy of Y. Yamamoto. P. gingivalis W50, W83, and hara-1 and Prevotella intermedia 163 were generously provided by K. Okuda, Department of Microbiology, Tokyo Dental College, Chiba, Japan. The other strains of P. gingivalis used were our own isolates from subgingival plaque of patients with adult periodontitis. These bacteria were maintained in our laboratory by weekly transfer on Trypticase soy agar supplemented with 0.1% yeast extract, 0.001% vitamin K1, 0.0005% hemin, and 5% sheep blood in an anaerobic chamber under an atmosphere of 85% N2, 10% H2, and 5% CO2. The bacteria were cultured to early stationary phase in brain heart infusion broth supplemented with 0.5% yeast extract, 0.001% vitamin K1, and 0.0005% hemin under the anaerobic conditions described above, except for B. forsythus and T. denticola. B. forsythus and T. denticola were grown in Basal Medium (4) supplemented with 5% inactivated calf serum and 0.0015% n-acetylmuramic acid and on TYGVS (Tryptone-yeast extract-gelatin-veal heart infusion broth) medium (23), respectively.
Assay of RCP activity.
The assay of RCP activity was performed in accordance with the procedure of Shibata et al. (30, 31), with slight modifications. The reaction mixture (0.5 ml) consisted of an aliquot of the enzyme sample, 2 mM hippuryl-arginine, and 0.04 M sodium acetate buffer (pH 6.0). After 1 h at 37°C, the reaction was stopped by the addition of 0.5 ml of 20% trichloroacetic acid and then the assay mixture was centrifuged at 3,000 × g. The supernatant was used for determination of the released arginine by the ninhydrin colorimetric method (37). The released arginine was alternatively determined with an amino acid analyzer (L-8500; Hitachi, Tokyo, Japan) after dilution of the sample with 3 volumes of 0.1 M sodium acetate buffer (pH 6.0). One unit of enzyme activity was defined as the amount of enzyme that released 1 μmol of arginine from the substrate under the conditions specified. The protein concentration of the sample was determined by measuring the A280 or by the Hartree method (12) with bovine serum albumin (BSA) as the standard.
Cell fractionation.
P. gingivalis cell fractionation was performed in accordance with the method of Grenier (11) by using mid-log-grown cells from a 400-ml culture. The bacterial cells were collected by centrifugation at 10,000 × g for 30 min, washed three times in 50 mM Tris-HCl buffer (pH 7.8) containing 0.03 M NaCl, and then suspended in 50 mM Tris-HCl buffer (pH 7.8) containing 30% sucrose and 1 mM EDTA. Cells were allowed to plasmolyze at room temperature for 15 min. After centrifugation at 13,000 × g for 10 min, the cells were treated for osmotic shock by dispersion of the cell pellet with gentle shaking in ice-cold distilled water for 10 min. The supernatant obtained by centrifugation at 13,000 × g for 10 min was kept as the periplasmic fraction. The pellet was resuspended in 50 mM Tris-HCl buffer (pH 7.8) containing 10% glycerol, 0.002 M MgCl2, DNase at 0.2 mg/ml, and RNase at 0.2 mg/ml. This suspension was subjected to ultrasonic treatment in ice with an Astrason W-385 ultrasonic processor (Heat-Systems Ultrasonics) with the variable pulse rate adjusted to 45% and the duty cycle timer adjusted to 2 s at 120 W. The unbroken cells were removed by centrifugation at 6,000 × g for 15 min, and the supernatant was centrifuged at 200,000 × g for 2 h. The resulting supernatant was kept as the cytoplasmic fraction. The pellet was again suspended in 50 mM Tris-HCl buffer (pH 7.8) containing 2% Triton X-100 and 1 mM MgCl2 and then centrifuged at 200,000 × g for 2 h. The supernatant was stored as the cytoplasmic membrane-enriched cell envelope fraction. The resulting pellet was suspended in 50 mM Tris-HCl buffer (pH 7.8) and used as the outer membrane-enriched cell envelope fraction.
Purification of RCP from P. gingivalis.
Unless stated otherwise, all purification procedures were carried out at 4°C and RCP activity was assayed by the ninhydrin colorimetric method. Cells from a 1-liter culture of P. gingivalis grown to early stationary phase were harvested by centrifugation at 10,000 × g for 40 min, washed three times with phosphate buffered saline (PBS; pH 7.4), and resuspended in a small volume (approximately 100 ml) of PBS. Cells were disintegrated by ultrasonic treatment under the conditions described above. The resulting supernatant was obtained by ultracentrifugation at 100,000 × g for 1 h and was used as the cell extract. The cell extract (103 ml) was dialyzed overnight against 10 mM Tris-HCl buffer (pH 8.2). The dialyzed sample was then applied to a DEAE-Sephacel column (2.6 by 22 cm; Pharmacia) that had been equilibrated with the previous buffer. The enzyme active fractions were eluted with 0.1 M NaCl in the buffer at a flow rate of 1 ml/min. The fractions were collected, concentrated with Centriplus 10 (Amicon Inc., Beverly, Mass.), and then applied to a DEAE-5PW high-performance liquid chromatography (HPLC) column (7.5 mm by 7.5 cm; Tosoh) that had been equilibrated with the buffer. Elution was made in a linear-gradient manner with 0 to 0.5 M NaCl in the buffer at a flow rate 1.0 ml/min. Two major peaks were detected in the eluate with NaCl concentrations of 0.06 to 0.14 M. These enzyme active peaks were collected separately and concentrated. The fraction that had no trypsin-like activity, measured by the method of Erlanger et al. (8) with Nα-benzoyl-dl-arginine-p-nitroanilide as the substrate, was applied to a TSK G2000SWXL gel filtration HPLC column (7.8 mm by 30 cm; Tosoh). The column had been equilibrated with 10 mM Tris-HCl buffer (pH 8.2) containing 0.15 M NaCl. The fractions containing RCP activity were concentrated and run on the same column again.
SDS-PAGE.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed on a slab gel with a concentration gradient of 10 to 20% polyacrylamide including 0.1% SDS in accordance with the method of Laemmli (14). The sample used for SDS-PAGE was pretreated with 0.0625 M Tris-HCl buffer, pH 6.8, including 1% SDS, 5% 2-mercaptoethanol, and 10% glycerol at 100°C for 10 min. Electrophoresis was carried out at a constant current of 40 mA for 1 h. Protein molecular size markers were run simultaneously for calibration. β-Galactosidase (116 kDa), BSA (66 kDa), aldolase (42 kDa), and carbonic anhydrase (30 kDa) were used for gels stained with silver nitrate. The prestained protein molecular size markers maltose-binding protein-β-galactosidase (175 kDa), maltose-binding protein-paramyosin (83 kDa), glutamic dehydrogenase (62 kDa), aldolase (48 kDa), and triose phosphate isomerase (33 kDa) were used for Western blotting gels.
Analysis of N-terminal amino acid sequences.
Purified RCP was first separated by SDS-PAGE. Before electrophoresis, the gels were pretreated with a running buffer containing 0.05 mM reduced glutathione for 2 h. The purified sample was electrophoresed with running buffer containing 0.1 mM sodium thioglycolate. The separated proteins were transferred onto a polyvinylidene difluoride membrane (Biotechnology Systems NEN Research Products, Boston, Mass.) in CAPS buffer (10 mM 3-[cyclohexylamine]1-propanesulfonic acid, 10% methanol, pH 11) by the method of Matsudaira (20). The membrane was rinsed, and proteins were visualized by staining with 0.1% Coomassie brilliant blue R-250. After the membrane was dried, the stained bands were excised and the N-terminal amino acid sequence was analyzed with a pulsed liquid protein sequencer (Hewlett Packard G1005A; Hewlett Packard, Palo Alto, Calif.).
Synthesis of the partial peptide of RCP and production of antibody to the synthetic peptide.
Peptide synthesis and subsequent antiserum production were carried out by Takara Biochemicals (Shiga, Japan). On the basis of the known N-terminal amino acid sequence, the peptide (NAYPTYEAYISMMEEFETKC) was synthesized and purified by reverse-phase HPLC, analyzed by mass spectrometry, and conjugated to keyhole limpet hemocyanin for immunization of a White rabbit. The antibody against the synthetic peptide was prepared as follows. The synthetic peptide (0.75 mg) was injected subcutaneously into a Japan White rabbit in the presence of complete Freund's adjuvant. Subsequent injections were done three times during a 14-day period in a similar manner. The rabbit was bled via the marginal ear vein at day 38 and directly via the heart at day 52, and the serum was collected. The antibody titer was recorded as the dilution of serum that gave enough reactivity with the synthetic peptide by the enzyme-linked immunosorbent assay technique. The antiserum sample, diluted ∼101- to ∼108-fold, was reacted with the synthetic peptide coating a 96-well plate at a concentration of 10 μg/ml and then incubated with horseradish peroxidase-conjugated goat anti rabbit immunoglobulin G (IgG). The color reaction was developed with 2,2′-azino-di-[3-ethyl-benzthiazoline sulfonate] and hydrogen peroxide. The antibody sample diluted 104-fold gave an optical density (OD) at 405 nm of 0.24.
Western blotting.
Following SDS-PAGE, proteins were transferred onto a 0.2-μm-pore-size nitrocellulose membrane that was then incubated with 0.5% BSA-PBS containing 0.05% Tween 20 (PBST). The membrane was rinsed in antibody against the synthetic peptide (1:1,000 dilution) at room temperature for 1 h and washed three times with 0.1% BSA-PBST. The membrane was then incubated with horseradish peroxidase-conjugated goat anti rabbit IgG (1:3,000 dilution; Bio-Rad Laboratories, Richmond, Calif.). The reactive bands were developed with 4-chloro-1-naphthol and hydrogen peroxide as the substrate in accordance with the procedure described in the Bio-Rad technical bulletin supplied with the assay kit.
Localization of RCP in P. gingivalis by immunogold labeling.
Bacterial cells grown to stationary phase were washed three times with PBS and fixed with 4% paraformaldehyde at 4°C for 3 h. The fixed cells were washed four times with PBS and used for postembedding immunogold labeling. The cell pellets were dehydrated through graded ethanol (50 to 90%) and LR-Gold series (London Resin Co., Ltd.) and embedded in LR-Gold. The plastic was allowed to polymerize at −20°C under UV light for 48 h. Thin sections were prepared on an ultramicrotome (2088 Ultramicrotome V; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom), transferred to nickel grids, and incubated with PBS containing 20% normal goat serum and 1% BSA at room temperature for 30 min. The specimens were then reacted with anti-RCP serum (1:10 dilution with 1% BSA in PBS) or rabbit preimmune serum (used as a negative control) at 4°C overnight. The grids were washed three times with PBS containing 1% BSA and reacted with goat anti rabbit IgG conjugated to 10-nm gold particles (1:50 dilution with 1% BSA in PBS; CHEMICON International, Inc.) for 1 h. The reaction was stopped by washing the grids, at least three times for 1 min each time, with 1% BSA in PBST. All specimens were stained with uranyl acetate and lead citrate and observed with a transmission electron microscope (Hitachi H-800) operating at 70 kV.
Analysis of RCP sequence.
The database containing the P. gingivalis W83 genome, available from The Institute for Genomic Research (TIGR), was searched for the presence of nucleotide sequences corresponding to the determined RCP amino acid sequence (YEWNAYPTYEA…) with the TBLASTN algorithm. A homologous sequence, which includes all of the sequence determined, was retrieved from the TIGR database. The position of the RCP gene was localized by using the GENETYX (Software Development Co., Ltd.) open reading frame (ORF) finder. The deduced amino acid sequence, obtained by conceptual translation of the entire ORF, was further used for homology screening by use of the National Center for Biotechnology Information BLAST search tool against the GenBank, EMBL, DDBJ, and PDB databases.
RESULTS
Presence of RCP in putative periodontopathogenic bacteria.
RCP activity in several putative periodontopathogenic bacteria was determined by using cell suspensions in PBS. P. gingivalis, A. actinomycetemcomitans, B. forsythus, F. nucleatum P. intermedia, and T. denticola were used for this experiment. The activity was found in all of the strains of P. gingivalis tested (eight strains shown in Materials and Methods) and in A. actinomycetemcomitans, B. forsythus, and T. denticola. However, F. nucleatum and P. intermedia displayed no detectable activity.
Subcellular distribution of RCP in P. gingivalis.
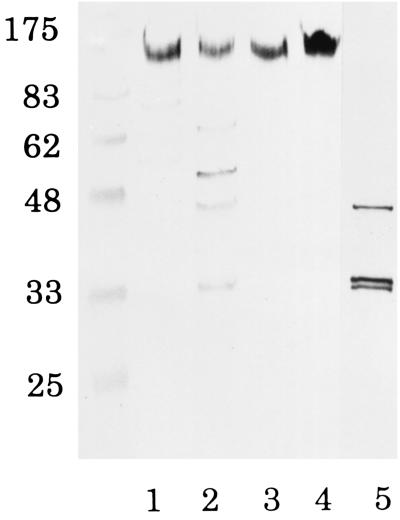
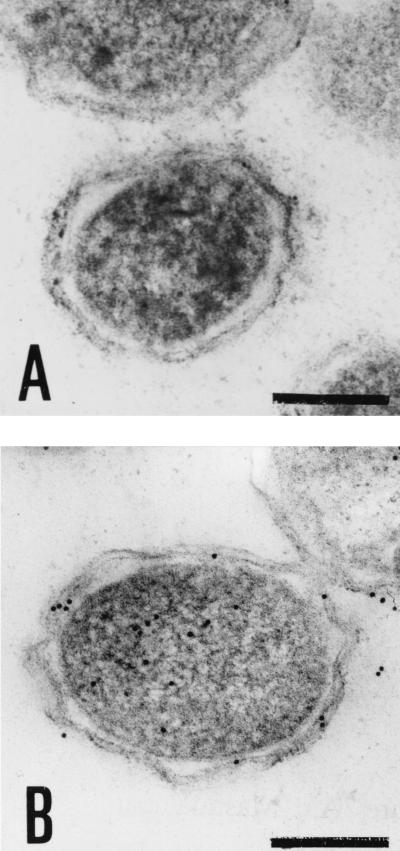
The activities of malate dehydrogenase and alkaline phosphatase were measured to assess the relative purity of the cytoplasm and periplasm as control markers for these fractions by the methods of Ochoa (22) and Yamashita et al. (36), respectively. Relatively higher alkaline phosphatase activity was found in the periplasm, while malate dehydrogenase activity was much higher in the cytoplasm. RCP activity was observed mostly in the cytoplasm and in the outer membrane-enriched cell envelope fraction (Table 1). When cell fractions were analyzed by Western blotting using the antibody against the N-terminal amino acid sequence of purified RCP, one diffuse, immunoreactive band at 120 to 160 kDa common to all of the fractions (periplasm, cytoplasm, and membrane) was found. The cytoplasmic fraction showed five more immunoreactive bands (62, 49, 42, 33, and 32 kDa) (Fig. 1). The intensity of this main band in the cytoplasm (lane 2) looked to be weak in contrast to the RCP activity shown in Table 1. Although no quantitative assay was performed, the apparent total intensities of these five bands in the cytoplasmic fraction looked to be strong enough. Three of the bands (42, 33, and 32 kDa) were consistent with those of RCP purified from the cytoplasmic fraction. Similarly, cytoplasmic fractions from all of the strains of P. gingivalis tested (ATCC 33277, W50, W83, hara-1, and clinical isolates) showed all of these bands (data not shown). A number of immunogold-labeled colloid particles were detected both inside and on the surface structure of cells, as shown by electron microscopy of thin sections (Fig. 2). These findings suggest that RCP is widely distributed in P. gingivalis but occurs mainly in both the cytoplasm and the surface-associated compartments.
TABLE 1.
Subcellular distribution of RCP in P. gingivalis cellsa
| Cell compartment | Total units of activity (%) | Sp actb |
|---|---|---|
| Periplasm | 0.28 (6.2) | 6.5 |
| Cytoplasm | 1.94 (43.0) | 37.4 |
| Cytoplasmic membrane-enriched cell envelope | 0.74 (16.5) | 13.6 |
| Outer membrane-enriched cell envelope | 1.54 (34.2) | 38.7 |
P. gingivalis was cultured in 400 ml of brain heart infusion broth to the log phase of growth.
Specific activity is expressed as units per milligram of protein.
FIG. 1.
Western blotting analysis of cell fractions of P. gingivalis. Western blotting was performed with an antibody against RCP (1:1,000 dilution). Following incubation with the antibody, the nitrocellulose membrane was reacted with a goat anti-rabbit antibody conjugated to horseradish peroxidase (1:3,000 dilution). Lanes: 1, periplasm (30 μg); 2, cytoplasm (30 μg); 3, cytoplasmic membrane-enriched cell envelope (30 μg); 4, outer membrane-enriched cell envelope (30 μg); 5, purified RCP (14 μg). The values on the left are molecular sizes in kilodaltons.
FIG. 2.
Immunogold localization of P. gingivalis RCP. Thin-section electron photomicrographs of P. gingivalis cells. P. gingivalis cells were incubated with preimmune rabbit serum (A) or anti-RCP serum (B), followed by anti-rabbit IgG conjugated to 10-nm gold particles. Note that some of the immunogold-labeled colloid was observed in both the cytoplasm and the surface-associated compartments of P. gingivalis cells. Bars, 0.25 μm.
Isolation and purification of RCP.
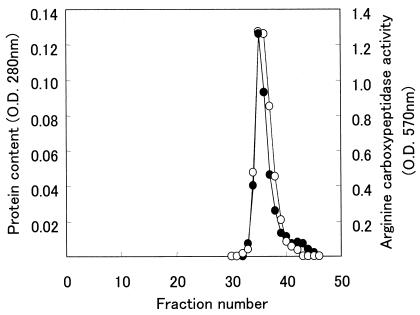
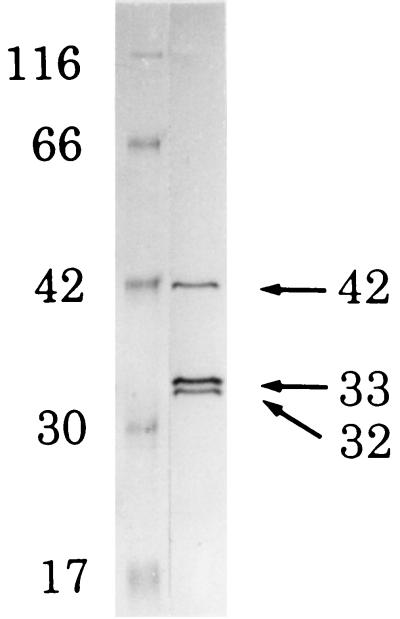
The procedure used for RCP purification from the cytoplasm of P. gingivalis and determination of its activity are summarized in Table 2. By HPLC on DEAE-5PW, two major peaks of RCP were found, one of which was contaminated with a trypsin-like enzyme (data not shown). The other peak was free of trypsin-like activity and was further purified by repeated filtrations on TSK G2000SWXL gel. The fraction containing RCP activity was eluted as a single, symmetric peak (Fig. 3). This purified RCP was further separated by SDS-PAGE into three bands (42, 33, and 32 kDa, respectively; Fig. 4).
TABLE 2.
Purification of RCP from P. gingivalis
| Step | Total protein (mg) | Total activity (U) | Vol (ml) | Sp acta | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Cell extraction | 319.3 | 12.2 | 103.0 | 0.04 | 100 | 1.0 |
| DEAE-Sephacel column chromatography | 19.4 | 9.7 | 45.0 | 0.5 | 80 | 12.5 |
| DEAE-5PW HPLC | 1.0 | 2.0 | 4.8 | 2.0 | 16 | 50.0 |
| 1st TSK G2000SWXL HPLC | 0.5 | 1.2 | 3.0 | 2.4 | 10 | 60.0 |
| 2nd TSK G2000SWXL HPLC | 0.2 | 0.7 | 1.2 | 3.5 | 6 | 87.5 |
Specific activity is expressed as units per milligram of protein.
FIG. 3.
Second gel filtration pattern of the RCP through a TSK G2000SWXL gel HPLC column. One of the enzyme active fractions from the HPLC on DEAE-5PW that contained no trypsin-like enzyme was subjected to gel filtration on an HPLC column packed with TSK G2000SWXL gel that had been equilibrated with 10 mM Tris-HCl buffer (pH 8.2) containing 0.15 M NaCl. The enzyme active fraction was collected, concentrated, and gel filtered again through the same HPLC column. By this repeated gel filtration, the fraction containing RCP activity was eluted as a single, symmetric peak at a position corresponding to an estimated molecular mass of 35 kDa. Symbols: •, protein content; ○, RCP activity
FIG. 4.
SDS-PAGE pattern of the purified RCP from the cytoplasm of P. gingivalis. Lanes: 1, molecular size markers (β-galactosidase [116 kDa], BSA [66 kDa], aldolase [42 kDa], carbonic anhydrase [30 kDa], and myoglobin [17 kDa]); 2, purified enzyme (7 μg of protein was applied). The values shown are molecular sizes in kilodaltons.
Properties of RCP from P. gingivalis. (i) Effect of pH on enzyme activity.
The pH dependence of the purified RCP was determined with hippuryl-arginine as a substrate in the following buffers: 0.1 M sodium acetate buffer (pHs 4.0 to 6.0), 50 mM Tris-acetate buffer (pHs 6.0 to 9.5), and 10 mM Good's buffer (pHs 9.0 to 11.0). No significant activity was observed at pHs 4.0, 5.0, and 11.0. At pHs 6.0, 9.0, and 10.0 in Tris-acetate buffer, the relative activities were 80, 80, and 60%, respectively. The enzyme showed maximum activity between pHs 7.0 and 8.0 and was active between pHs 6.0 and 10.0 in Tris-acetate buffer.
(ii) Effects of inhibitors and metal ions on RCP.
Protease inhibitors were used at a final concentration of 1 or 0.1 mM in the reaction mixture. The activity with hippuryl-arginine in the absence of inhibitors under the conditions specified was regarded as 100%. Serine, cysteine, and aspartic protease inhibitors did not inhibit the activity of RCP from P. gingivalis, whereas the metalloprotease inhibitors such as o-phenanthroline and EGTA showed strong inhibitory activity. MGTA, a specific zinc-binding enzyme inhibitor, inhibited RCP activity remarkably (Table 3). The effects of metal ions on RCP activity were also tested. The enzyme's activity was inhibited 50, 31, and 54% by Cu2+, Zn2+, and Cd2+, respectively, but activated 1.5-fold by Co2+. Ca2+ and Mg2+ had no effect.
TABLE 3.
Inhibition of RCP
| Addition | Final concn (mM) | Relative activitya (%) |
|---|---|---|
| None | 100 | |
| Serine protease inhibitors | ||
| TLCK | 0.1 | 100 |
| 1.0 | 75 | |
| Phenylmethylsulfonyl fluoride | 0.1 | 100 |
| 1.0 | 83 | |
| Diisopropyl fluorophosphate | 0.1 | 100 |
| 1.0 | 67 | |
| Cysteine protease inhibitors | ||
| N-ethylmaleimide | 0.1 | 125 |
| 1.0 | 133 | |
| E64 | 0.1 | 117 |
| 1.0 | 92 | |
| Leupeptin | 100b | 100 |
| Aspartic protease inhibitors | ||
| p-Bromophenacryl bromide | 0.1 | 80 |
| 1.0 | 120 | |
| Pepstatin A | 0.1 | 90 |
| 1.0 | 60 | |
| Metalloprotease and metallocarboxypeptidase inhibitors | ||
| EDTA | 0.1 | 108 |
| 1.0 | 67 | |
| EGTA | 0.1 | 75 |
| 1.0 | 42 | |
| o-Phenanthroline | 0.1 | 83 |
| 1.0 | 25 | |
| MGTA | 0.1 | 17 |
| 1.0 | 17 |
100% relative activity was defined as as activity with no addition.
In micrograms per milliliters.
(iii) Substrate specificity of RCP from P. gingivalis on several peptides.
The ability of the purified enzyme to release amino acid from several hippuryl-amino acid compounds other than hippuryl-arginine was examined to determine the specificity of RCP's activity. The reaction was performed at a final concentration of 2 mM in the reaction mixture, and the purified enzyme only released lysine and/or arginine from the hippuryl-amino acid compounds tested (date not shown). The Km values of hippuryl-arginine and hippuryl-lysine were 0.67 and 0.35 mM, respectively.
The hydrolytic specificity of the purified enzyme was determined by using several peptides that contain an arginine and/or a lysine residue(s) at various positions. As shown in Table 4, the enzyme only reacted with peptides having arginine or lysine at the C terminus. However, the enzyme could not cleave peptides that had the arginine residue at the C terminus adjacent to proline. No activity was found in peptides that contained an arginine or lysine residue(s) inside or at the N terminus.
TABLE 4.
Release of arginine or lysine from synthetic peptides by RCP
| Synthetic peptide (2 mM)a | Released arginine or lysineb |
|---|---|
| Exopeptidase substrates | |
| Bz-Gly-Arg (hippuryl-arginine) | 3.06 |
| Gly-Phe-Tyr-Arg | 1.28 |
| Gly-Gly-Tyr-Arg | 1.42 |
| Thr-Lys-Pro-Argc | NDd |
| Ala-Pro-Gly-Pro-Arg | ND |
| Tyr-Ile-Gly-Ser-Arge | 5.61 |
| His-Leu-Gly-Leu-Ala-Argf | 8.36 |
| Ala-Ser-His-Leu-Gly-Leu-Ala-Argf | 7.19 |
| Arg-Pro-Pro-Gly-Phe-Ser-Pro-Phe-Argg | 2.33 |
| Bz-Gly-Lys (hippuryl-lysine) | 9.44 |
| Thr-Pro-Arg-Lysh | 8.06 |
| Val-His-Leu-Thr-Pro-Val-Glu-Lys | 4.47 |
| Tyr-Gly-Gly-Phe-Leu-Arg-Lys-Tyr-Pro-Lys | ND |
| Endopeptidase substratesi | |
| Asp-Arg-Val-Tyr-His-Pro-Phe | ND |
| Gly-Arg-Gly-Asp-Ser | ND |
Amino acids that are supposed to be released by RCP are in boldface.
Micromoles of arginine or lysine released from synthetic peptide per milligram of protein of the purified enzyme under the specified conditions.
Tuftsin.
N.D., not detected.
C-terminal fragment of laminin.
C-terminal fragment of anaphylatoxin C3a.
Bradykinin.
Kentsin.
The amino acid (aspartic acid or glycine) that existed at the N terminus of the peptide adjacent to arginine was not released.
Amino acid sequence of purified RCP from P. gingivalis.
Thirty cycles of Edman degradation were carried out on the three excised bands of the purified RCP. The N-terminal amino acid sequences of the three bands were identical and determined as YEWNAYPTYEAYISMMEEFQTKYPSLXTXS.
RCP sequence analysis.
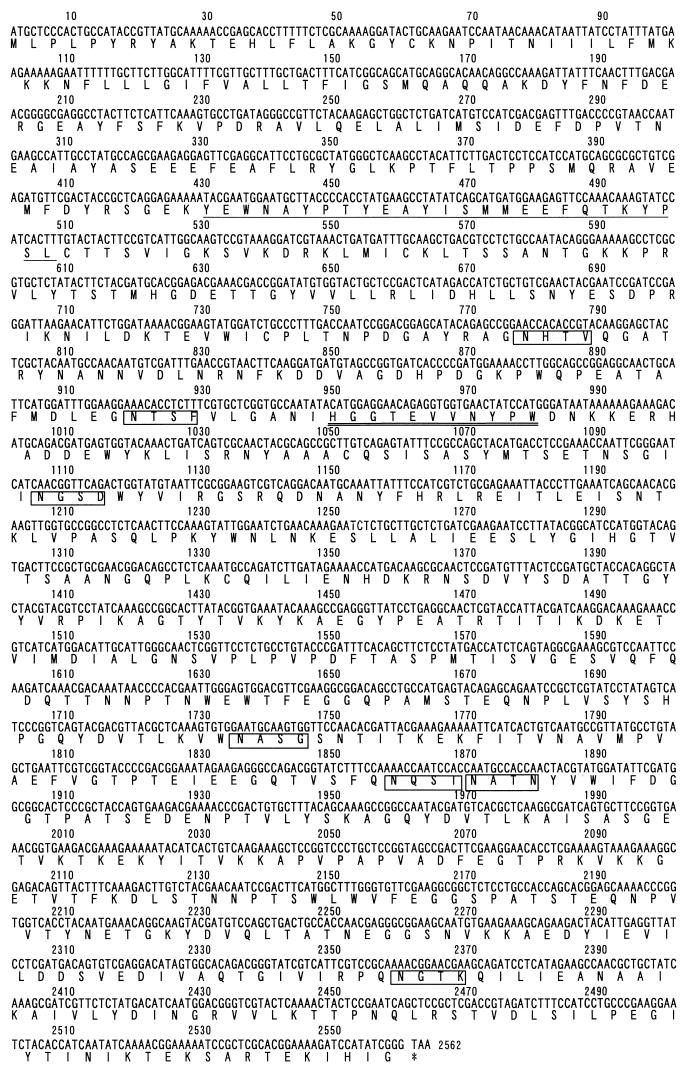
When we searched the P. gingivalis W83 genome for the nucleotide sequence corresponding to the RCP amino acid sequence determined, the exact same sequence was found in an already identified sequence, gnl|TIGR| P. gingivalis_GPG contig Porphyromonas gingivalis W83 unfinished fragment of complete genome. The ORF corresponding to the amino acid sequence of RCP was found and estimated to be 853 amino acids long (Fig. 5) by using GENETYX (Software Development Co., Ltd.).
FIG. 5.
Suspected RCP sequence from the unfinished P. gingivalis W83 genome. The ORF of the DNA sequence and the deduced amino acid sequence are shown. The underlined amino acid sequence was determined by sequencing the N terminus of the isolated protein. The consensus signature sequence of the zinc-binding region conserved in zinc carboxypeptidases is doubly underlined. Potential N-linked glycosylation sites are boxed.
The ORF of the 853-residue RCP protein encoded a protein with consensus sequences for an N-linked glycosylation site (Asn-X-Thr/Ser, where X is any amino acid except Pro) and for a two-zinc-binding-region signature ([PK]-x-[LIVMFY]-x-LIVMFY]-x[4]-H-[STAG]-x-E-x-[LIVM]-[STAG]-x[6]-[LIVMFY], H-[STAG]-X[3]-[LIVMFYW]-P-[FYW]). But one amino acid residue was different from the consensus signature sequence of the former zinc-binding region. A homology search of the amino acid sequences of the deduced RCP and other zinc carboxypeptidases was performed. The ORF showed 33% homology with Aplysia californica (sea hare) carboxypeptidase D (9); 30 and 29% homology with human carboxypeptidases M (34) and N (10), respectively; and 27% homology with rat carboxypeptidase E/H (15, 28). In these sequences, all of the consensus signatures for zinc carboxypeptidase were included. The homology position was found mainly in the amino acid sequence at positions 149 to 491 in the putative sequence of RCP. The homology of bacterial carboxypeptidases with the RCP was lower than that of mammalian zinc carboxypeptidases, i.e., 21 and 26% homology with carboxypeptidase SG from Streptomyces griseus (accession number X65719) and carboxypeptidase T from Thermoactinomyces vulgaris (33), respectively. No consensus signature region for zinc carboxypeptidases was found in the homology position of the bacterial carboxypeptidases.
Effect of MGTA on growth of P. gingivalis.
As RCP activity was inhibited by MGTA, the effect of MGTA on the growth of P. gingivalis in broth was determined by measuring its OD at 660 nm. The growth of this bacterium was affected by the presence of MGTA (46% growth inhibition with 2 mM MGTA and 62% inhibition with 5 mM MGTA). However, addition of free arginine to the broth with MGTA could not reverse growth completely.
DISCUSSION
Many studies have shown that P. gingivalis produces several kinds of protease, but little is known about the carboxypeptidase. We have previously reported that P. gingivalis produces RCP in its culture supernatant and selectively utilizes arginine that has been released in the growth medium by the combined action of arginine aminopeptidase and RCP (17, 19). This fact has suggested the important role of RCP in the growth of this bacterium for obtaining arginine as an essential nutrient, and this might result in the strong virulence of P. gingivalis (19).
In this study, we found that more than 85% of the RCP existed in bacterial cells although the enzyme was also found in the culture supernatant. This meant that arginine might be taken up by P. gingivalis cells as small peptides produced by some kind of protease like Arg/Lys-gingipains. We first demonstrated the localization of RCP in the cell compartment fractions of P. gingivalis. We also showed by immunoelectron microscopy that the enzyme is distributed on both the inside (cytoplasm) and the outside (membrane) of the cells. Since the enzyme is located mostly in the cytoplasm, this fraction was used as the starting material for the purification of RCP. RCP was purified and eluted as a single peak at a molecular size of 35 kDa by gel filtration on TSK G2000SWXL, but when the purified sample was separated by electrophoresis, three bands appeared. It is of interest that the three bands possessed the same N-terminal amino acid sequence. This may indicate that large portions of these proteins are similar and the difference in molecular size revealed by SDS-PAGE may be due to S-S bond cleavage and/or autolysis. The same phenomenon has been reported for gingipains produced by this bacterium (5, 24, 25, 27). The purified RCP was active over a wide range of pHs. Serine and cysteine protease inhibitors did not affect the enzyme, while MGTA and chelating reagents such as o-phenanthroline and EGTA inhibited the enzyme's activity, suggesting that the enzyme is a member of the metallocarboxypeptidase family. MGTA is a specific inhibitor of zinc-containing enzymes that cleaves basic amino acids such as arginine and lysine from the peptide C terminus (26). MGTA also inhibited P. gingivalis growth, suggesting that RCP is needed to produce arginine and that arginine is needed for growth. When we tried to reverse the growth inhibition by adding arginine, the inhibitory effect of MGTA on bacterial growth was not completely reversed by the addition of arginine. This suggests that MGTA has effects on other important systems in P. gingivalis.
The specificity of the enzyme for release of lysine and/or arginine from the C termini of peptides was demonstrated. As the Km values for hippuryl-arginine and hippuryl-lysine were 0.67 and 0.35 mM, respectively, it is suggested that the C-terminal lysine may be released faster than the C-terminal arginine. From this evidence, this enzyme might be termed arginine/lysine carboxypeptidase. However, we prefer to call it RCP because the name has been used historically and fits its biological function in P. gingivalis well.
There are several biologically active peptides that contain arginine and lysine at the carboxyl terminus (32). Release of these amino acids from the carboxyl terminus sometimes results in inactivation of biological activity. Purified RCP actively released arginine from some biologically active substances, such as laminin, anaphylatoxin C3a fragment, bradykinin, and kentsin, suggesting a potential role for RCP in the modification of these peptides. However, if the peptide contains proline adjacent to the arginine, as in tuftsin, hydrolysis by this enzyme could be avoided and biological activity might be protected.
The complex form of this enzyme was revealed by Western blotting analysis using the antibody directed to the recombinant peptide corresponding to the N-terminal amino acid sequence of RCP. The starting material for purification showed several immunoreactive bands consisting of one diffuse band of 120 to 160 kDa and five sharp bands of 62, 49, 42, 33, and 32 kDa, respectively. Three of these (42, 33, and 32 kDa) were consistent with those of RCP. On the other hand, the diffuse band (120 to 160 kDa) commonly existed in all of the cell fractions (periplasm, cytoplasm, and membrane). This fact indicates that the enzyme may have been produced originally as a large-molecular-size complex and that during isolation and purification, the diffuse, large-molecular-size band was cleaved into smaller molecules. On the other hand, it may be that this enzyme is synthesized in multiple forms. Gingipain R, which is produced by this bacterium, has at least three isoforms after posttranslational modification, RgpA (50 kDa), mt-RgpA (∼70 to 90 kDa), and HRgpA (7). RgpA is composed of a catalytic domain, and mt-RgpA is the modified catalytic domain. HRgpA is a heterodimer made of a catalytic domain and a noncatalytic polypeptide chain. Since the RCP ORF encodes a protein with seven consensus sequences for N-linked glycosylation, the observed 120- to 160-kDa band of RCP might be a polyprotein containing a catalytic domain, similar to HgpA or mt-RgpA. As the 42-kDa band of RCP was found in the soluble fractions, such as the culture supernatant (data not shown) and the cytoplasm, this protein might possess a catalytic domain similar to RgpA. The 33- and 32-kDa bands that were detected in the cytoplasm by Western blotting analysis could not be detected when the sample was prepared with a buffer with TLCK, an inhibitor of trypsin-like proteases. These findings suggested that the 33- and 32-kDa proteins appeared because if the degradation of the 42-kDa protein. Further study is required to better define the structure of the gene for RCP and the regulation of its expression. Also, posttranslational modifications of the primary polypeptide need to be studied at the protein level.
Sequence analysis of the protein demonstrated that RCP contains a single ORF of 853 amino acids, including consensus signatures for N-linked glycosylation site (16) and zinc-binding regions (34). These signatures suggest that RCP is a metallocarboxypeptidase composed of glycoproteins. Although data are not shown, the evidence that the 42-kDa band of RCP contains carbohydrate was clearly confirmed by visualization of the transferred membrane from the electrophoresis gel by chemiluminescence. Since the homology position was recognized from position 149 to position 491 of the putative RCP amino acid sequence and the consensus sequence for the zinc-binding region signature was found within this sequence, the RCP could be a zinc carboxypeptidase. However, since these consensus signature sequences have not been found in homologous positions of other bacterial zinc carboxypeptidases, the RCP from P. gingivalis could be a new type of bacterial carboxypeptidase.
The molecular mass calculated from the deduced ORF of RCP was 95,280 Da, but the actual size obtained by gel filtration and SDS-PAGE was different. However, the typical molecular masses of known carboxypeptidases are 40 to 60 kDa (2). This size resembles that of the RCP obtained in this study. Furthermore, when the molecular size was calculated only on the recognized homology position (from position 149 to position 491 of the RCP amino acid sequence), the size was 40,095 Da and was very close to the molecular size of RCP estimated by SDS-PAGE. The calculated molecular mass may be variable, depending upon the initiation codon. Ross et al. (29) have reported the PG21 gene of P. gingivalis strain W50, which encodes an immunoreactive 92-kDa antigen. They also showed that this antigen was reactive with sera from human periodontitis patients, suggesting that this antigen is periodontopathogenic. The deduced ORF found in our study contained exactly the same sequence as the PG21 gene, suggesting that RCP is similar to the 92-kDa antigen protein described by Ross et al. This fact might indicate that RCP is also related to periodontopathogenicity. We have previously found that P. gingivalis utilizes arginine and have demonstrated that this bacterium can obtain energy through arginine catabolism (19). RCP may function to release arginine after an endopeptidase like Arg-gingipain has initially cleaved the peptide precursor to expose arginine at the C terminus. Consequently, in periodontal disease, the bacterium may obtain free arginine from the surrounding host tissues and catabolize the amino acid to obtain energy for its own growth through the arginine deiminase pathway. This underlines the important role that an arginine-releasing peptidase may possess in the disease process.
Acknowledgments
We sincerely thank Denis Mayrand and Daniel Grenier, University of Laval, Quebec, Canada, for helpful discussions and critical reading of the manuscript. We also thank Jan Potempa, Jagiellonian University, Cracow, Poland, for a helpful suggestion. We thank Tsuneko Ono, University of Tokushima, for help in identifying the RCP-encoding gene and Kazumi Ozaki, University of Tokushima, for the localization study using transmission electron microscopy.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Banbila, A., P. Mak, M. Bugno, J. Silberring, A. Dubin, D. Nelson, J. Travis, and J. Potempa. 1999. Prolyl tripeptidyl peptidase from Porphyromonas gingivalis. J. Biol. Chem. 274:9246-9252. [DOI] [PubMed] [Google Scholar]
- 2.Barrett, A. J., N. D. Rawling, and J. F. Woessner (ed.). 1998. Handbook of proteolytic enzymes, p. 1318-1351. Academic Press, London, England.
- 3.Birkedal, H., R. E. Taylar, J. J. Zambor, P. K. Barwa, and M. E. Neiders. 1988. Characterization of collagenolytic activity from strains of Bacteroides gingivalis. J. Periodontal Res. 23:258-264. [DOI] [PubMed] [Google Scholar]
- 4.Braham, P. H., and B. J. Moncla. 1992. Rapid presumptive identification and further characterization of Bacteroides forsythus. J. Clin. Microbiol. 30:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciborowski, P., M. Nishikata, R. D. Allen, and M. S. Lantz. 1994. Purification and characterization of two forms of a high-molecular-weight cysteine proteinase (porphypain) from Porphyromonas gingivalis. J. Bacteriol. 176:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB protease of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erlanger, B. F., N. Kokowsky, and W. Cohen. 1961. The preparation and properties of two new chromogenic substrates of trypsin. Arch. Biochem. Biophys. 95:271-278. [DOI] [PubMed] [Google Scholar]
- 9.Fan, X., Y. Qian, L. D. Fricker, D. B. G. Akalal, and G. T. Nagle. 1999. Cloning and expression of Aplysia carboxypeptidase D, a candidate prohormone processing enzyme. DNA Cell Biol. 18:121-132. [DOI] [PubMed] [Google Scholar]
- 10.Gebhard, W., M. Schube, and M. Eulitz. 1989. cDNA cloning and complete primary structure of the small, active subunit of human carboxypeptidase N (kininase I). Eur. J. Biochem. 178:603-607. [DOI] [PubMed] [Google Scholar]
- 11.Grenier, D. 1994. Characteristics of a protease inhibitor produced by Prevotella intermedia. FEMS Microbiol. Lett. 119:13-18. [DOI] [PubMed] [Google Scholar]
- 12.Hartree, E. F. 1972. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 48:422-427. [DOI] [PubMed] [Google Scholar]
- 13.Hinode, D., H. Hayashi, and R. Nakamura. 1992. Purification and characterization of three types of protease from culture supernatants of Porphyromonas gingivalis. Infect. Immun. 59:3060-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Manser, E., D. Fernandez, L. Loo, P. Y. Goh, C. Monfries, C. Hall, and L. Lim. 1990. Human carboxypeptidase E. Isolation and characterization of the cDNA, sequence conservation, expression and processing in vitro. Biochem. J. 267:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall, R. D. 1972. Glycoproteins. Annu. Rev. Biochem. 41:673-702. [DOI] [PubMed] [Google Scholar]
- 17.Masuda, K., H. Hayashi, M. Yoshioka, and R. Nakamura. 1995. Presence of an arginine carboxypeptidase in culture supernatants of Porphyromonas gingivalis. J. Dent. Res. 74:974. [Google Scholar]
- 18.Masuda, K., M. Yoshioka, D. Hinode, and R. Nakamura. 1999. Characterization of arginine carboxypeptidase from Porphyromonas gingivalis. J. Dent. Res. 78:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda, K., K. Tomita, H. Hayashi, M. Yoshioka, D. Hinode, and R. Nakamura. 2001. Consumption of peptide-derived arginine by a periodontopathogenic bacterium, Porphyromonas gingivalis. Anaerobe 7:209-217. [Google Scholar]
- 20.Matsudaira, P. 1987. Sequence from picomole quantities of protein electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 21.McGraw, W. T., J. Potempa, D. Farley, and J. Travis. 1999. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 67:3248-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochoa, S. 1955. Malic dehydrogenase from pig heart. Methods Enzymol. 1:735-739. [Google Scholar]
- 23.Ohta, K., K. K. Makinen, and W. J. Loesche. 1986. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect. Immun. 53:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto, K., Y. Misumi, T. Kadowaki, M. Yoneda, K. Yamamoto, and Y. Ikehara. 1995. Structural characterization of argingipain, a novel arginine-specific cysteine protease as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch. Biochem. Biophys. 316:917-925. [DOI] [PubMed] [Google Scholar]
- 25.Pike, R., W. McGraw, J. Potempa, and J. Travis. 1994. Lysine-and arginine-specific proteinase from Porphyromonas gingivalis. J. Biol. Chem. 269:406-411. [PubMed] [Google Scholar]
- 26.Plummer, T. H., Jr., and T. J. Ryean. 1981. A potent mercapto bi-product analogue inhibitor for human carboxypeptidase N. Biochem. Biophys. Res. Commun. 98:448-454. [DOI] [PubMed] [Google Scholar]
- 27.Potempa, J., R. Pike, and J. Travis. 1995. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63:1176-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriquez, C., K. A. Brayton, M. J. Brownstein, and J. E. Dixon. 1989. Rat preprocarboxypeptidase H: cloning, characterization, and sequence of the cDNA and regulation of the mRNA by corticotropin-releasing factor. J. Biol. Chem. 264:5988-5995. [PubMed] [Google Scholar]
- 29.Ross, B. C., I. Barr, M. Patterson, C. Agius, L. Rothel, M. Margetts, D. Hocking, and E. Wess. December 1998. Porphyromonas gingivalis polypeptides and nucleic acids. Australian patent (AU 98/01023)-PCT.
- 30.Shibata, K.-I., and T. Watanabe. 1986. Carboxypeptidase activity in human mycoplasmas. J. Bacteriol. 168:1045-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata, K., and T. Watanabe. 1988. Purification and characterization of an arginine-specific carboxypeptidase from Mycoplasma salivarium. J. Bacteriol. 170:1759-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skidgel, R. A. 1988. Basic carboxypeptidase: regulators of peptide hormone activity. Trends Pharmacol. Science 9:299-304. [DOI] [PubMed] [Google Scholar]
- 33.Sumlevitch, S. V., A. L. Osterman, O. V. Galperina, M. V. Matz, O. P. Zagnitko, R. M. Kadyrov, I. A. Tsaplina, N. V. Grishin, G. G. Chestukhina, and V. M. Stepanov. 1991. Molecular cloning and primary structure of Thermoactinomyces vulgarius carboxypeptidase T. FEBS Lett. 291:75-78. [DOI] [PubMed] [Google Scholar]
- 34.Tan, F., S. J. Chan, D. F. Steiner, J. W. Schilling, and R. A. Skidgel. 1989. Molecular cloning and sequencing of the cDNA for human membrane-bound carboxypeptidase M: comparison with carboxypeptidase A, B, H and N. J. Biol. Chem. 264:13165-13170. [PubMed] [Google Scholar]
- 35.Toda, K., M. Otsuka, Y. Ishikawa, M. Sato, Y. Yamamoto, and R. Nakamura. 1984. Thiol-dependent collagenolytic activity in culture media of Bacteroides gingivalis. J. Periodontal Res. 19:372-381. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita, Y., K. Toyoshima, M. Yamazaki, N. Hanada, and T. Takahara. 1990. Purification and characterization of alkaline phosphatase of Bacteroides gingivalis 381. Infect. Immun. 58:2882-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yemm, E. W., and E. C. Cocking. 1954. Estimation of amino acids by ninhydrin. Biochem. J. 58:xii. [PubMed] [Google Scholar]