Abstract
A major obstacle to studying the functions of particular gene products in the mouse-tick infectious cycle of Borrelia burgdorferi has been an inability to knock out genes in pathogenic strains. Here, we investigated conditions for site-directed mutagenesis in B31 MI, the low-passage-number, infectious B. burgdorferi strain whose genome was sequenced. We inactivated several plasmid and chromosomal genes in B31 MI and determined that clones carrying these mutations were not infectious for mice. However, we found extensive heterogeneity among clones and mutants derived from B31 MI based on colony phenotype, growth rate, plasmid content, protein profile, and transformability. Significantly, several B31 MI clones that were not subjected to mutagenesis but that lacked particular plasmids also exhibited defects at various stages in the infectious cycle. Therefore, the high degree of clonal polymorphism within B31 MI complicates the assessment of the contributions of individual genes to the observed phenotypes of the mutants. Our results indicate that B31 MI is not an appropriate strain background for genetic studies in infectious B. burgdorferi, and a well-defined isogenic clone is a prerequisite for targeted mutagenesis. To this end, we derived several wild-type clones from B31 MI that were infectious for mice, and gene inactivation was successful in one of these clones. Due to the instability of the genome with in vitro propagation, careful monitoring of plasmid content of derived mutants and complementation of inactivated genes will be crucial components of genetic studies with this pathogen.
Lyme disease is caused by Borrelia burgdorferi, a spirochete transmitted by ticks of the genus Ixodes and maintained within an enzootic cycle between the tick vector and mammalian hosts, most importantly small rodents (7, 11, 19). The clinical manifestations of this zoonosis can include a multisystem disorder affecting skin and joints and the nervous, lymphoreticular, and cardiovascular systems (39, 40).
The organization of the B. burgdorferi genome is unique among bacteria in that the genome is composed of a linear chromosome and a large number of linear and circular plasmids (8, 14). The complete genome sequence of an infectious B. burgdorferi isolate, the type strain B31, identified 21 linear and circular plasmids (8). In vitro propagation of B. burgdorferi can lead to plasmid loss and concurrent loss of infectivity for mice (3, 21, 22, 30, 33). Although increasing evidence suggests that certain Borrelia plasmids are important for infection in mice (18, 25), this hypothesis has not been experimentally verified, and the roles of most plasmid-encoded genes in the infectious cycle are unknown. A number of plasmid- and chromosomally encoded genes have been inactivated in the high-passage-number, noninfectious clone B31-A (4, 5, 12, 17, 20, 43, 45, 46), but gene inactivation in a low-passage-number, infectious strain background has not been reported.
Here, we investigate conditions for site-directed mutagenesis in B31 MI, the low-passage-number, infectious B. burgdorferi strain whose genome has been sequenced. We chose this strain because the sequence information allows for comparison and monitoring of plasmid content among different clones and after passage of a given clone. Also, the availability of the complete genome sequence greatly facilitates gene inactivation and opens the possibility for future studies to analyze gene expression on a genome-wide basis.
Since genetic studies require comparison of a wild-type clone with isogenic mutants, we evaluated the heterogeneity and genetic transformability of B31 MI and clonal derivatives (outlined in Fig. 1). A primary purpose of these experiments was to identify infectious clones derived from B31 MI that were suitable for genetic studies. In addition, distinct plasmid profiles and in vivo phenotypes of clonal variants and mutants suggest factors influencing vector-host transmission, host immune response, infectivity, and regulation of protein expression of B. burgdorferi during host adaptation.
FIG. 1.
Experimental outline.
The possibility for gene inactivation and complementation in low-passage-number, infectious B. burgdorferi has important implications for future research with this pathogen. It creates a basis for experimentally assessing the functions of specific genes or plasmids within the infectious cycle. However, the genetic instability of B. burgdorferi with in vitro propagation, as well as possible traits of transformation itself, represents potential caveats for using the present genetic system in generating mutants for in vivo studies. Careful and well-controlled experiments are needed to assess the phenotype of specific mutants in the infectious cycle.
MATERIALS AND METHODS
B. burgdorferi strains and growth conditions.
B. burgdorferi B31 (ATCC 35210) was previously described (7). Prior to isolation of DNA for genomic sequencing by Fraser et al. (14), strain B31 was subjected to four successive rounds of infection in mice, after which the strain underwent three consecutive in vitro passages (14). This low-passage-number, infectious B31 was previously denoted B31 MI (8). For experiments in this study, 5-ml cultures were inoculated from frozen stocks of a passage 3 (P3) culture and used directly or were passaged no more than twice (P5). From this uncloned, parental B31 MI isolate, a number of clones were isolated from individual colonies grown in solid Barbour-Stoenner-Kelly (BSK) medium (26, 27). In addition, three clonal mutants derived by allelic exchange from uncloned, parental B31 MI were further characterized. Bacteria were grown at 35°C in BSK-II medium (2) supplemented with 6% rabbit serum, unless otherwise indicated.
Growth and temperature upshift experiments.
Cultures inoculated with B. burgdorferi directly from P3 glycerol stocks and grown at 35°C to mid-exponential phase (5 × 107/ml) were diluted to 1 × 105 to 5 × 105 cells/ml and grown at 23°C. At a cell density of 5 × 107/ml, cultures at 23°C were diluted to 1 × 105 to 5 × 105 cells/ml and shifted back to 35°C. The number of spirochetes in each culture was determined daily by dark-field microscopy with a Petroff-Hausser counting chamber. All experiments were done in triplicate. For each triplicate data set at a given time point, the arithmetic mean and standard deviation were calculated and plotted using Microsoft Excel software (Microsoft Corporation, Redmond, Wash.). Similarly, doubling times in exponential growth phase were calculated as arithmetic means for each triplicate culture. Subsequent single-factor analysis of variance with the Microsoft Excel Analysis ToolPak tested the hypothesis that means of doubling times from different cultures were equal.
Construction of oppAII::kan.
Plasmid pOK25.B2 contains a 6.6-kb fragment of the chromosomal opp locus including oppAI (BB328), oppAII (BB329), oppAIII (BB330), and part of oppB (BB332) (5, 14). The flaBp-kan cassette was amplified from pTAkanA (4) with primers PflaB-5′+KpnI (5′-GGTACCTGTCTGTCGCCTCTTGTGGCTTCCGG) and pOK.7+MluI (5′-ACGCGTGGCGAATGAGCTTGCGCCGTCCC) and cloned into KpnI-MluI-digested pOK25.B2, resulting in a deletion of 1,036 nucleotides from oppAII, the oppAII-oppAI intervening sequence, and 47 nucleotides from the 3′ end of oppAI.
Construction of rpoS::kan.
The inactivation of rpoS in high-passage-number, noninfectious B31-A with the mutated form of gyrB that encodes coumermycin resistance as a selectable marker has been described previously (12). We used the kanamycin resistance cassette (Kanr) (4) to create an rpoS mutant in low-passage-number B. burgdorferi. The 4.2-kb rpoS-containing chromosomal fragment cloned into pOK12 was digested with BbsI as described previously (12) and ligated with flgBp-kan that had been excised from pTAkanG (4) by EcoRI digestion, resulting in the disruption of rpoS (rpoS::kan).
Construction of a plasmid for chbC inactivation by insertion-deletion.
The plasmid used for inactivation of the chbC gene was made as previously described (45), except that an flgB promoter-aacC1 cassette conferring gentamicin resistance Gmr (unpublished data) was used instead of the mutated form of gyrB that encodes coumermycin resistance to replace part of the gene. Briefly, the flgBp-aacC1 fusion was amplified from a pCR2.1 clone using primers flgBp-5′+NcoI (5′-CGCCATGGCTAATACCCGAGCTTCAAGGA) and aacC1-3′+NheI (5′-GCTAGCCGATCTCGGCTTGAACG). The amplified fragment was cloned into pCR2.1-TOPO, and a clone with the insert oriented so as to be released by NcoI-KpnI digestion was selected. This insert was recloned as described previously (45) into pOK12 carrying the chbC gene and flanking sequences, resulting in plasmid pKK82 with insertion-deletion of chbC (chbC::aaC1).
The constructs used for gene inactivations were confirmed by DNA sequencing with an Applied Biosystems 377 automated sequencer (Foster City, Calif.). Purification of plasmid DNA from Escherichia coli, restriction digestions, and ligations were done as described previously (12).
Preparation of electrocompetent B. burgdorferi and electroporation conditions.
Low-passage-number B. burgdorferi isolates were grown in 100 ml of BSK-II medium and harvested at cell densities of 5 × 107 to 9 × 107/ml, corresponding to mid- to late exponential phase. In some experiments, borrelial cultures were incubated at 37 or 42°C prior to the preparation of electrocompetent cells. Cells were concentrated 500- to 1,000-fold by a series of three washes with EPS (0.27 M sucrose, 15% [vol/vol] glycerol) at room temperature. Each washing step included centrifugation at 4,000 × g for 10 min followed by gentle vortexing to achieve resuspension of the cells in EPS. For electroporation, approximately 5 × 109 to 1 × 1010 freshly prepared electrocompetent cells in 50 to 100 μl of EPS were transferred to an 0.2-cm electroporation cuvette after the addition of 20 to 70 μg of DNA and transformed with a pulse generator (Bio-Rad, Hercules, Calif.) set at 2.5 kV, 25 μF, and 200 Ω (32).
TSS transformation.
A cell concentrate (109 to 1010 cells) prepared from 100 ml of a mid-exponential-phase culture was added to 200 μl of freshly prepared modified transformation and storage solution (TSS) (9) containing BSK-II medium instead of the L broth component (10% polyethylene glycol 3350, 5% dimethyl sulfoxide, 20 mM magnesium sulfate, and BSK-II medium to a final volume). After gentle vortexing, 20 to 70 μg of DNA was added and the mixture was incubated for 5 to 15 min at room temperature. The entire transformation volume was then transferred to 5 ml of fresh BSK-II medium and incubated overnight at 35°C. Concentration of cells included either a series of washes in EPS or HN buffer (100 mM sodium chloride, 20 mM HEPES) or a one-step centrifugation and direct resuspension in modified TSS.
Selection and screening of transformants.
After overnight incubation, all cells from the 5-ml cultures were plated in solid BSK medium (27). Cells were concentrated approximately two- to fourfold by centrifugation and resuspension in fresh BSK-II medium (1.5 to 3 ml), and 200 to 400 μl of this concentrate was used per plate. Depending on the antibiotic resistance marker, plates with solid BSK medium contained kanamycin (400 μg/ml) or gentamicin (40 μg/ml). Screening for rpoS mutants by PCR was done as previously described with primers rpoS-F1 and rpoS-B1 (12). Primers oppA0.61 (5′-GAGGGAAGACCAACAACAGCAG) and oppAII-3′sp. (5′-CTGCAATATCATGATAACCC) were used to screen for oppAII::kan mutants. Mutants at the chbC locus were identified with primers celB-F (5′-CAGAGTCTGTTCCGCCTGCTG) and celB-R (5′-AAGTAGAGCCTTATCATACGC) flanking the insertion site of the gentamicin resistance cassette. The transformation frequency was the number of mutants recovered relative to the total number of CFU in the transformed cultures.
Determination of plasmid content.
For plasmid profiles, borrelial plasmid DNA was obtained with the Qiagen Midi plasmid kit (Qiagen, Chatsworth, Calif.) or the Wizard Genomic DNA purification kit (Promega, Madison, Wis.) and separated by electrophoresis through an 0.3% agarose gel as previously described (42). Plasmid content was determined by PCR (50 ng of borrelial DNA template in 20-μl reaction mixtures) with 29 unique primer pairs spanning all or most of an open reading frame (ORF) from each plasmid (Table 1). Sequence information and ORF designations were obtained from The Institute for Genomic Research website (http://www.tigr.org/tdb/mdb/bbdb/bbdb.html). Due to the extensive similarity between lp56 and the multiple cp32s, primers were designed to specifically amplify ORFs from individual cp32s or lp56 that would not result in cross-amplification of paralogous ORFs (two sets of primers to different ORFs on each cp32 and lp56 were generated per plasmid). Sequence information for each primer pair is available upon request from Darrin R. Akins.
TABLE 1.
Oligonucleotides used to determine plasmid contents
| Plasmid | Primer | Primer coordinates | Corresponding gene (coordinates) |
|---|---|---|---|
| lp54 | BBA60-5′ | 40981-40957 | BBA60, surface lipoprotein P27 (40981-40151) |
| BBA60-3′ | 40151-40175 | ||
| cp26 | BBB03-5′ | 2186-2162 | BBB03, hypothetical protein (2186-840) |
| BBB03-3′ | 840-864 | ||
| cp9 | BBC10-5′ | 6808-6784 | BBC10, Rev protein (rev) (6808-6284) |
| BBC10-3′ | 6284-6308 | ||
| 1p17 | BBD11-5′ | 6681-6705 | BBD11, conserved hypothetical protein (6681-7631) |
| BBD11-3′ | 7631-7607 | ||
| 1p25 | BBE17-5′ | 10709-10685 | BBE17, hypothetical protein (10709-10203) |
| BBE17-3′ | 10203-10227 | ||
| lp28-1 | BBF18-5′ | 9561-9585 | BBF18, putative transposase-like protein, authentic frameshift (9561-10049) |
| BBF18-3′ | 10049-10025 | ||
| lp28-2 | BBG13-5′ | 12355-12331 | BBG13, hypothetical protein (12355-11504) |
| BBG13-3′ | 11504-11528 | ||
| lp28-3 | BBH18-5′ | 12105-12129 | BBH18, hypothetical protein (12105-13217) |
| BBH18-3′ | 13217-13193 | ||
| lp28-4 | BBI28-5′ | 17874-17850 | BBI28, hypothetical protein (17874-17305) |
| BBI28-3′ | 17305-17329 | ||
| lp38 | BBJ23-5′ | 16505-16529 | BBJ23, hypothetical protein (16505-17326) |
| BBJ23-3′ | 17326-17302 | ||
| lp36 | BBK23-5′ | 15760-15736 | BBK23, hypothetical protein (15760-14837) |
| BBK23-3′ | 14837-14861 | ||
| cp32-8 | BBL39-5′ | 26370-26394 | BBL39, ErpA protein (26370-26900) |
| BBL39-3′ | 26900-26876 | ||
| BBL40-5′ | 26931-26955 | BBL40, ErpB2 protein (26931-28064) | |
| BBL40-3′ | 28064-28040 | ||
| cp32-6 | BBM32-5′ | 20732-20756 | BBM32, plasmid partition protein, putative (20721-21467) |
| BBM32-3′ | 21417-21393 | ||
| BBM38-5′ | 26380-26404 | BBM38, ErpK protein (26245-27012) | |
| BBM38-3′ | 26887-26863 | ||
| cp32-9 | BBN28-5′ | 17302-17326 | BBN28, lipoprotein (17302-17727) |
| BBN28-3′ | 17596-17572 | ||
| BBN32-5′ | 20828-20852 | BBN32, plasmid partition protein, putative (20817-21569) | |
| BBN32-3′ | 21449-21425 | ||
| cp32-7 | BBO32-5′ | 20777-20801 | BBO32, plasmid partition protein, putative (20766-21512) |
| BBO32-3′ | 21382-21358 | ||
| BBO40-5′ | 27123-27147 | BBO40, ErpM protein (26893-27981) | |
| BBO40-3′ | 27615-27591 | ||
| cp32-1 | BBP32-5′ | 20777-20801 | BBP32, plasmid partition protein, putative (20766-21503) |
| BBP32-3′ | 21440-21416 | ||
| BBP38-5′ | 26235-26259 | BBP38, ErpA protein (26247-26765) | |
| BBP38-3′ | 26765-26741 | ||
| lp56 | BBQ05-5′ | 2744-2768 | BBQ05, antigen, P35, putative (2744-3493) |
| BBQ05-3′ | 3493-3469 | ||
| BBQ08-5′ | 5814-5790 | BBQ08, plasmid partition protein, putative (5830-5072) | |
| BBQ08-3′ | 5185-5209 | ||
| cp32-4 | BBR41-5′ | 26109-26133 | BBR41, conserved hypothetical protein (26077-26817) |
| BBR41-3′ | 26707-26683 | ||
| BBR42-5′ | 26922-26946 | BBR42, outer surface protein F (26853-27524) | |
| BBR42-3′ | 27370-27346 | ||
| cp32-3 | BBS35-5′ | 21268-21292 | BBS35, plasmid partition protein, putative (21257-21994) |
| BBS35-3′ | 21806-21782 | ||
| BBS41-5′ | 26743-26767 | BBS41, outer surface protein G (26708-27295) | |
| BBS41-3′ | 27199-27175 | ||
| lp5 | BBT03-5′ | 1208-1232 | BBT03, hypothetical protein (1208-1573) |
| BBT03-3′ | 1573-1549 | ||
| lp21 | BBU04-5′ | 1493-1517 | BBU04, conserved hypothetical protein (1486-2625) |
| BBU04-3′ | 2578-2554 |
PCR conditions included an initial denaturation step at 94°C (5 min) followed by 30 cycles of 94°C for 30 s, 50 or 55°C for 30 s, and 72°C for 1 min and a final extension at 72°C for 7 min.
For Southern blot analysis, total borrelial DNA extracted from B31 MI and clonal derivatives was separated through 0.3% agarose gels by electrophoresis. Equal amounts of borrelial DNAs (500 ng) were added to each lane, and equivalent loadings were confirmed visually by ethidium bromide staining. Blotting and hybridization were performed as previously described (29) using [α-32P]dATP-labeled probes for lp25 (BBE17), lp28-1 (BBF18), or lp28-4 (BBI28). Probe fragments were generated by PCR with plasmid-specific primer pairs (Table 1).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis.
B. burgdorferi whole-cell lysates derived from mid-exponential-phase cultures (5 × 107cells/ml) and E. coli lysates expressing B. burgdorferi protein P39 (BmpA) from plasmid pSPR33 were prepared as previously described (38, 41).
Equivalent amounts of cells were resuspended, boiled in reducing Laemmli sample buffer (31), and separated through 12.5% polyacrylamide gels in a Hoefer SE600 gel apparatus (Hoefer Scientific Instruments, San Francisco, Calif.) or a Bio-Rad Minicell system. Proteins were visualized with the Silver Stain Plus kit (Bio-Rad). Alternatively, proteins were transferred to nitrocellulose membranes (Bio-Rad) and immunoblotted as previously described (43).
Antisera.
Rabbit polyclonal antiserum to OspC (37) and H4610, a monoclonal antibody that recognizes the amino-terminal segment of OspB (28), were incubated at 1:500 and 1:50 dilutions, respectively. A rabbit polyclonal antiserum against B. burgdorferi Sh-2-82 for indirect fluorescent antibody (IFA) staining and mouse sera from the infection experiments were used at 1:100 dilutions.
Experimental mouse-tick cycle with B31 MI and its clonal derivatives.
White-footed mice (Peromyscus leucopus) were from a colony maintained at Rocky Mountain Laboratories, Hamilton, Mont., and were free of B. burgdorferi prior to inoculation. Nonclonal B31 MI (P5) and the clonal strains A1 (P3) and B1 (P3) were tested in a mouse-tick cycle. Mice were inoculated with 5 × 103 spirochetes by intraperitoneal (4 × 103 spirochetes) and subcutaneous (1 × 103 spirochetes) injection as previously described (34). At 3 weeks postinoculation, blood was obtained from the retro-orbital sinus for serologic analysis. At 5 weeks postinoculation, unfed Ixodes scapularis larvae were fed to repletion on mice injected with B31 MI, A1, or B1 (two mice per strain, approximately 20 to 30 larvae per mouse). A subset of engorged larvae was analyzed soon after feeding for the presence of spirochetes by IFA as previously described (36). The remaining engorged larvae were kept in a humidified jar at 25°C until they molted to the nymphal stage. The nymphs were fed to repletion on uninfected mice (approximately 15 to 20 nymphs per mouse) and subsequently analyzed for the presence of spirochetes by IFA. Four weeks after tick feeding, all mice were bled from the retro-orbital sinus and sacrificed, and tissues (bladder, ankle joint synovia, and ear skin) were cultured for the presence of spirochetes as previously described (33, 34).
In a subsequent experiment, mice were inoculated with 104 spirochetes of B31 MI clones A1, A3, and C1, as well as mutants MI-ΔoppAII7.3, MI-ΔoppAII44, and MI-ΔchbC37 by intraperitoneal (8 × 103 spirochetes) and subcutaneous (2 × 103 spirochetes) injection. At 3 weeks postinoculation, animals were bled from the retro-orbital sinus and sacrificed to obtain organs for culture isolation of spirochetes.
Analysis of host adaptation in the dialysis chamber model.
To obtain B. burgdorferi in a mammalian host-adapted state, spirochetes were cultivated in BSK-H medium at 23°C to mid-exponential phase. Organisms (103/ml) were added to dialysis membrane chambers (DMCs; 8-kDa cutoff) containing 8 ml of BSK-H medium and subsequently implanted into the peritoneal cavities of Sprague-Dawley rats. DMCs were harvested 14 days postimplantation as previously described (1).
RESULTS
Clonal analysis of B. burgdorferi B31 MI.
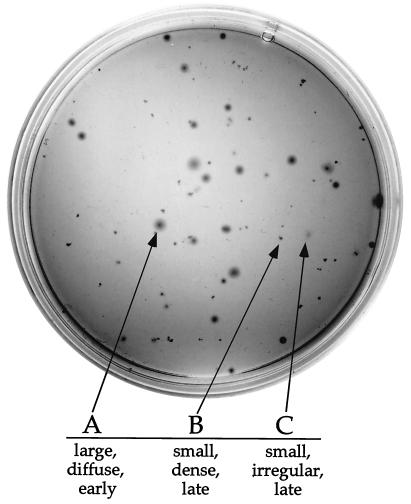
Clonal derivatives of B31 MI (P3) were obtained from individual colonies in solid BSK medium (Fig. 2). The colony morphology and time of colony appearance of B31 MI were heterogeneous (Fig. 2). Some colonies appeared earlier (5 to 6 days after plating), were regularly shaped, were relatively large, and had a dense center with a diffuse border. Three of the clones with this colony phenotype were further characterized and designated A clones. Other colonies appeared later (10 or more days after plating) and were significantly smaller. Of those, some were dense and regularly shaped but lacked a diffuse border (B clones). A few of the smaller colonies were irregular in shape and lacked the dense center (C clones). The colony phenotypes of A, B, and C clones were reproducible when bacteria were grown in liquid BSK-II medium and subsequently plated again in solid BSK medium (data not shown).
FIG. 2.
Heterogeneous colony morphology of parental B31 MI (P3) with representative examples of A, B, and C clones (arrows).
B31 MI-derived clones had characteristic differences in protein and plasmid profiles that coincided with their distinct colony morphologies. The A clones lacked a 34-kDa protein that was present in the B and C clones, as well as in the parental B31 MI (Fig. 3A and data not shown). OspB monoclonal antibody H4610 reacted with the 34-kDa protein, confirming the presence of full-length OspB in B31 MI clones B and C (Fig. 3B and Table 2). In contrast, reactivity of this antibody with a band of approximately 18 kDa in A clones was consistent with truncated OspB (Fig. 3B and Table 2). This phenotype has been described previously (6, 10, 13, 15, 30). The parental B31 MI strain contained both full-length and truncated OspB, as would be expected of a heterogeneous population (Fig. 3B and Table 2).
FIG. 3.
Protein profiles, OspB immunoblotting, and plasmid profiles of B31 MI and representative clonal derivatives. (A) SDS-PAGE of whole-cell lysates stained with silver. A 34-kDa band corresponding to OspB is present in B31 MI and clones B1 and C1 (arrow) but absent in clone A1 and mutant MI-ΔoppAII-7.3. Each lane contains borrelial lysate equivalent to 5 × 107 cells. Molecular masses are indicated on the left. (B) Immunoblotting of whole-cell lysates probed with H4610, a monoclonal antibody that recognizes the amino-terminal segment of OspB. Closed arrow, full-length OspB; open arrow, truncated OspB. Each lane contains borrelial lysates equivalent to 8 × 107 cells. Molecular masses are indicated on the left. (C) Plasmid profiles of B31 MI and clonal derivatives. Total plasmid DNA was separated by electrophoresis through an 0.3% agarose gel and stained with ethidium bromide. Molecular sizes are indicated on the left. Supercoiled (S), linear (L), and nicked (N) forms of cp9 are indicated by arrows on the right.
TABLE 2.
OspB phenotype, plasmid content, transformability, and infectivity of B31 MI and clonal derivatives
| Strain | OspB phenotype | Plasmid(s) lost | Transformation result
|
Infectivity in micea
|
||
|---|---|---|---|---|---|---|
| Shuttle vector | Allelic exchangeb | No. of mouse sera reactive with P39/total no. | No. of culture-positive mice/total no.c | |||
| B31 MI | Heterogenous | None | + | + | 3/3 | 3/3 |
| A1 | Truncated | cp9, lp28-1 | + | + | 3/3, 3/3 | 0/3, 0/3 |
| A2 | Truncated | cp9, lp25d | NAe | NA | NA | NA |
| A3 | Truncated | cp9 | + | + | 3/3 | 3/3 |
| B1 | Full length | lp28-4 | + | − | 3/3 | 3/3 |
| B2 | Full length | lp21, lp25, lp28-1/-4 | NA | NA | NA | NA |
| B3 | Full length | lp28-4 | NA | NA | NA | NA |
| C1 | Full length | lp5, lp28-4, cp32-3/-9 | + | − | 3/3 | 3/3 |
| C2 | Full length | lp5, lp25, lp28-1/-4, cp32-3/-9 | NA | NA | NA | NA |
| MI-ΔoppAII-7.3 | Truncated | lp5, cp9, lp25 | NA | + | 0/3 | 0/3 |
| MI-ΔoppAII-44 | Truncated | lp5, cp9 | NA | + | 0/3 | 0/3 |
| MI-ΔchbC-37 | Truncated | lp5, cp9 | NA | + | 3/3 | 0/3 |
After needle inoculation.
Electroporation or TSS transformation with allelic-exchange constructs oppAII::kan, chbC::gent, or rpoS::kan as described in the text; transformation frequencies were between 10−9 and 10−8 and independent of the construct or transformation method.
Growth of spirochetes in cultures of skin, joints, and bladder in BSK-II medium. Cultures were considered negative when no spirochetes could be detected by 6 weeks after tissue inoculation.
PCR with the lp25-specific primer pair resulted in a faint product of the expected size. However, Southern blot analysis with an lp25-specific probe did not result in a detectable band (Fig. 4).
NA, not addressed.
The A clones lacked the 9-kb circular plasmid (cp9) as seen by agarose gel electrophoresis of total plasmid DNA (Fig. 3C and data not shown). This plasmid was present in the parental B31 MI, as well as in B and C clones (Fig. 3C and data not shown). Analysis of the total plasmid content of uncloned B31 MI by PCR using plasmid-specific primers (Table 1) confirmed the presence of all 21 plasmids previously described for B31 MI (Table 2) (8). However, none of the clones had the entire plasmid set, and different plasmids were lacking among those clones (Table 2). PCR analysis was consistent with plasmid profiles obtained by agarose gel electrophoresis (Table 2 and Fig. 3). The presence or absence of plasmids lp25, lp28-1, and lp28-4 in B31 MI derivatives was confirmed by Southern blot analysis (Fig. 4).
FIG. 4.
Southern blot analysis of genomic DNA from B31 MI and clonal derivatives. DNAs (500 ng/lane) were separated by electrophoresis and hybridized with probes specific for lp25 (A), lp28-1 (B), or lp28-4 (C). Arrows to the left of each panel indicate the positions of the plasmids. Probes were from BBE17 (lp25), BBF18 (lp28-1), and BBI28 (lp28-4) and were generated with the same primers used to assess plasmid content (Table 1).
In summary, individual clones derived from uncloned, low-passage-number B31 MI (P3) differed in colony morphology, OspB phenotype, and plasmid content. These results indicate that low-passage-number B31 MI is heterogeneous, despite its recent reisolation from an infected mouse.
Growth at 23 and 35°C.
We determined growth curves and protein profiles of clonal variants of B31 MI grown at 23 and 35°C. Two growth phenotypes were observed among the B31 MI-derived clones. At 23°C, A clones had a doubling time ranging from 28.2 to 30.6 h, whereas the doubling times of B and C clones were between 33.9 and 43.4 h (data not shown). At 35°C, A clones doubled approximately every 5 h and B and C clones doubled every 8 h (Fig. 5). Doubling times at 35°C were not statistically different among A clones or within the group of B and C clones (P = 0.22 and P = 0.12, respectively, single-factor analysis of variance). In contrast, the difference in doubling time between A clones and the group of B and C clones was highly significant (P < 0.0001) (Fig. 5). The faster growth of A clones in liquid medium than of B and C clones was consistent with the earlier appearance of A colonies in solid BSK medium. A, B, and C clones all had higher OspC levels after temperature upshift (data not shown). Parental B31 MI grew similarly to A clones with doubling times of 25.9 and 5 h at 23 and 35°C, respectively (data not shown).
FIG. 5.
Growth curves in liquid BSK-II medium of B31 MI and clonal derivatives at 35°C. Each data point represents the arithmetic mean of the number of spirochetes determined in triplicate. Error bars indicate the standard deviation for each triplicate set. Doubling times and standard deviations are indicated to the right. B31 MI and derivatives with faster growth (F) are in the upper panel of the key, and derivatives with slower growth (S) are in the lower panel.
Experimental mouse-tick cycle with B31 MI and its clonal derivatives.
The uncloned B31 MI and two of its clonal derivatives, A1 and B1, were tested for mouse infectivity and tick transmission. Three uninfected mice per strain were inoculated by needle injection with 5 × 103 spirochetes. Three weeks postinoculation, the mouse sera were tested for evidence of B. burgdorferi infection by serologic conversion to the B. burgdorferi protein P39 (Table 2 and Fig. 6A). Previous studies by Simpson et al. demonstrated that seroconversion to P39 after needle inoculation of B. burgdorferi is a reliable marker of active infection in mice (38). All mouse sera were reactive with P39 (Table 2 and Fig. 6A), suggesting that infections with B. burgdorferi had been established. Five weeks postinoculation, I. scapularis larvae were fed to repletion on all three mice inoculated with A1 or B1 and one of the three mice inoculated with B31 MI. When a subset of fed larvae was examined by IFA, none of the larvae (zero of five) fed on mice inoculated with A1 contained detectable spirochetes, despite the positive serology. In contrast, spirochetes were detected in larvae fed on mice inoculated with B31 MI (4 of 5) or B1 (7 of 10) (data not shown). Consistent with these findings, no spirochetes were reisolated from mice inoculated with A1, whereas cultures of all three mice inoculated with B31 MI or B1 contained detectable spirochetes within 6 days (Table 2).
FIG. 6.
Immunoblot of mouse sera probed against recombinant P39. (A) Sera from mice needle inoculated with 5 × 103 spirochetes. (B) Sera of mice collected 4 weeks after being fed upon by B31 MI- or B1-infected nymphs. Arrows indicate reactivity with p39.
The majority of I. scapularis larvae (67%) molted to the nymphal stage after feeding to repletion. Nymphs that molted from larvae fed on mice infected with B31 MI and B1 were allowed to feed on uninfected mice (approximately 15 to 20 ticks per mouse, two mice per strain). Four weeks later, sera and tissues (bladder, ankle joint synovia, and ear skin) were obtained from each mouse. Spirochetes were recovered from both mice on which B31 MI-infected nymphs had fed, and sera from the mice recognized P39 (Table 3 and Fig. 6B). However, no spirochetes were recovered from either of the two mice used to feed B1-infected nymphs (Table 3). A second set of approximately 20 nymphs, molted from larvae that had fed on B1-infected mice, was allowed to feed to repletion on a third uninfected mouse. Clone B1 could not be reisolated from this mouse 4 weeks postrepletion (Table 3). None of the three mice showed seroreactivity with P39 (Fig. 6B). In contrast, the infection rate of nymphal midguts with B1 determined by IFA assay in a subset of fed nymphs was 71.4% (five of seven nymphs positive) (Table 3).
TABLE 3.
Serology and reisolation from mice after feeding infected I. scapularis nymphs
| Strain | No. of nymphs/mouse | % of infected nymphsa | No. of culture-positive miceb
|
No. of mouse sera reactive with P39 | ||
|---|---|---|---|---|---|---|
| Bladder | Skin | Joints | ||||
| B31 MI | 15 | 50 | 2/2 | 2/2 | 2/2 | 2/2 |
| B1 | 20 | 71.4 | 0/3 | 0/3 | 0/3 | 0/3 |
The percentage of infected nymphs was determined by immunofluorescence microscopy of tick midguts with polyclonal rabbit serum against B. burgdorferi.
Growth of spirochetes from cultures of skin, joints, and bladder in BSK-II medium. Cultures were considered negative when no spirochetes could be detected by 6 weeks after tissue inoculation.
Infectivity of additional B31 MI-derived clones after needle inoculation into mice.
Clones A1, A3, and C1 were tested for their ability to establish an infection in mice after needle inoculation. A1 was included to verify our previous inability to recover this clone from mice after needle inoculation (see above). A3 was tested for infectivity because it lacked only cp9 and was a potential candidate for genetic manipulation (see below). Spirochetes were cultured from all mice inoculated with A3 and C1 (Table 2). Consistent with previous results (see above), no spirochetes were recovered from any of the three mice inoculated with A1 (Table 2). Sera collected from mice 4 weeks postinoculation with either A1, A3, or C1 contained antibody to P39 (Table 2).
Growth and host adaptation of B31 MI-derived clonal variants in the dialysis chamber model.
We analyzed the phenotypes of clones A1, B1, and C1 in the dialysis chamber model. In this system, spirochetes are grown in BSK medium-containing DMCs implanted into peritoneal cavities of rats. A molecular mass cutoff of 8 kDa allows for some exchange with the host environment across the dialysis membrane. Previous work has shown that regulation of protein expression in B. burgdorferi during growth in DMCs simulates patterns seen during adaptation to the mammalian host environment, allowing direct analysis of B. burgdorferi without in vitro culture (1, 16).
Clone A1, which previously was not recovered from mice (see above), as well as clones B1 and C1, survived in the chambers, and the three clones grew similarly to approximately 107 spirochetes/ml within 14 days in the DMC (data not shown). All three clones grown in DMCs showed protein synthesis patterns typical of host adaptation, such as a reduced amount of OspA and an increased amount of OspC (Fig. 7). However, A1 exhibited a more prominent adaptive response than did B1 and C1, with OspA expression decreasing to nondetectable levels (Fig. 7).
FIG. 7.
Whole-cell lysates of clones A1, B1, and C1 separated by SDS-PAGE and stained with silver. The first two lanes of each panel show lysates derived from cultures grown in vitro at 23°C and after temperature upshift to 35°C, respectively. The third lane of each panel shows lysates derived from spirochetes grown in DMCs. Closed arrow, OspA; open arrow, OspC. Molecular masses are indicated on the left.
Electrotransformation conditions allowing for recovery of mutants in B31 MI.
We developed electrotransformation conditions allowing gene inactivation in low-passage-number B31 MI. Electroporation with various gene inactivation constructs (oppAII::kan, rpoS::kan, and chbC::gent) resulted in mutants at frequencies between 10−8 and 10−9 (Table 2). Factors that appeared to be important for successful recovery of mutants in B31 MI were extremely high numbers of competent spirochetes (approximately 5 × 109 to 1 × 1010 cells per transformation) and the use of freshly prepared electrocompetent cells, as well as growth phase and cell density. Spirochetes in stationary phase were difficult to resuspend during washes, requiring more rigorous vortexing and resulting in decreased survival after electroporation. However, spirochetes in early exponential phase had decreased survival rates after electroporation. Therefore, the best results were achieved with cultures in mid- to late exponential phase, corresponding to a cell density of approximately 5 × 107 to 1 × 108/ml.
Incubation of B31 MI at elevated temperatures (37 or 42°C) prior to the preparation of electrocompetent cells and/or after electroporation did not have a significant effect on transformation frequencies, nor did growth at a different pH (data not shown). Exposure of spirochetes to temperatures higher than 45°C resulted in increased cell death, as determined by the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Wilsonville, Oreg.) (data not shown).
TSS transformation as an alternative method to transform B. burgdorferi.
We adapted a previously described method that uses polyethylene glycol and dimethyl sulfoxide (9) to transform B. burgdorferi. With a modified TSS (9) containing BSK-II medium, TSS transformation yielded B. burgdorferi mutants with transformation frequencies similar to those obtained with electroporation. No significant differences in transformation frequencies were observed when the time during which the cells were incubated with DNA in modified TSS varied from 5 min to 15 min or when washing steps prior to TSS incubation were omitted (data not shown).
TSS transformation involves a milder treatment than does electroporation (44). This results in a better survival rate for TSS-transformed spirochetes, with up to 10-fold more bacteria following transformation by TSS than after electroporation, under otherwise identical conditions (data not shown). Although this did not alter overall transformation frequency, TSS transformation required fewer bacteria per experiment, allowing for transformation of cultures earlier in exponential phase (<5 × 107/ml).
Targeted mutagenesis in low-passage-number B31 MI and its clonal derivatives.
We compared parental B31 MI with some of its clonal derivatives for the ability to inactivate genes by allelic exchange. The clones were transformed by either electroporation or TSS transformation with suicide plasmids that contained oppAII::kan, rpoS::kan, or chbC::aacC1 allelic-exchange constructs. These constructs were chosen for transformation experiments in low-passage-number B31 MI on the basis of previous work, in which mutations in oppAII, rpoS, and chbC were obtained with reproducible and comparable transformation frequencies in high-passage-number B31-A (12, 45). These genes may be important for nutrient acquisition or adaptation during the infectious cycle, but our previous data indicate that they are not required for in vitro growth.
Transformation frequencies in the parental B31 MI were between 1.5 × 10−9 and 4 × 10−8 with allelic-exchange constructs oppAII::kan, chbC::aacC1, and rpoS::kan. In clones A1 and A3, transformation frequencies with the same constructs ranged from 1 × 10−9 to 3.2 × 10−8. In contrast, several attempts to obtain mutants in B1 or C1 were unsuccessful. However, like the parental B31 MI and its clonal derivatives A1 and A3, B and C clones could be transformed with the shuttle vector pBSV2 (Table 2). Even without an efficient method for gene inactivation, shuttle vector transformation can be used to investigate the defect in transmission of B1.
In summary, by using either of the described transformation methods and different plasmid constructs, mutants could be recovered from the clones A1 and A3 with frequencies similar to those for the parental, uncloned B31 MI strain. However, only one of these clones, A3, was reisolated from mice.
Clonal analysis and infectivity of B31 MI mutants.
Mutants created in B31 MI had colony phenotypes similar to those of A clones when grown in solid BSK medium and comparable growth rates when grown in liquid medium (Table 2 and Fig. 5). Also, protein and plasmid profiles of these and other B31 MI mutants studied to date were similar to those for the A clones, which have a truncated OspB (Fig. 3A and B) and lack cp9 (Fig. 3C).
Three mutants derived from the uncloned, parental B31 MI were tested for infectivity in mice. Three mice per mutant strain were injected with 104 spirochetes and sacrificed 4 weeks postinoculation for reisolation of spirochetes (bladder, ankle joint synovia, and ear skin) and serology. None of the mutants could be reisolated from mice (Table 2), but the serologic response varied among the mutants. The chbC mutant elicited a seroconversion to P39 in all three mice, whereas no reactivity to P39 was detected in the two oppAII mutants (Table 2). Thus, the phenotype of the chbC mutant in the mouse model was similar to that for clone A1, which could not be recovered from needle-inoculated mice but elicited a seroconversion to P39.
The two oppAII mutants were tested for growth in DMCs. Neither grew well in DMCs, reaching 10- to 50-fold-lower cell densities than those for the A1, B1, and C1 clones after 14 days, the time at which the chambers were removed from the peritoneal cavities (data not shown). Due to the low spirochetal loads, no protein analysis was performed with the recovered mutants.
DISCUSSION
We have analyzed the in vitro and in vivo phenotypes of B. burgdorferi clones derived from the nonclonal, low-passage-number (P3), infectious strain B31 MI and determined conditions for site-directed mutagenesis in this strain background. We found that B31 MI (P3), the source strain for the B. burgdorferi genomic sequence, is a heterogeneous mixture of clonal variants with differences in growth phenotype, plasmid content, and protein profile.
The clonal polymorphism observed in low-passage-number B31 MI has important implications for genetic transformation of infectious B. burgdorferi and subsequent studies in vivo. Although parental B31 MI was both infectious in the mouse-tick cycle and transformable by allelic exchange, only a subset (A3, B1, and C1) of the clones derived from B31 MI could be recovered from mice after needle inoculation and gene inactivation was successful in only one (A3) of these clones. Because allelic exchange at a targeted locus is an extremely rare event in low-passage-number B. burgdorferi (44), it requires a very large number of spirochetes. Thus, in a heterogenous strain such as B31 MI, phenotypes of mutants may result from other genetic differences between clonal variants (e.g., loss of lp25 in MI-ΔoppAII-7.3) rather than from the introduced mutation itself. Consequently, uncloned B31 MI is not an appropriate strain background for genetic studies with infectious B. burgdorferi, given the observed degree of clonal polymorphism.
In contrast, B31 MI derivative A3 is clonal, has a full complement of plasmids except cp9, and is infectious in mice and transformable by allelic exchange, making it suitable for genetic studies in vivo. An important question to answer by using such a clone will be whether mutations in oppAII, chbC, and rpoS result in loss of infectivity, as suggested by the present experiments. These genes are predicted to be important in the infectious cycle because of their implied roles in nutrient acquisition, host adaptation, and regulation of gene expression. Gene inactivation in the infectious clone A3 should allow us to verify the in vivo functions of these genes. Complementation of specific mutations is now feasible through introduction of a wild-type copy of the inactivated gene on a shuttle vector (42) and represents an important component of such studies.
Mutants could be recovered from A clones (A1 and A3) but not from B or C clones, and mutants derived from uncloned B31 MI (MI-ΔoppAII-7.3, MI-ΔoppAII-44, and MI-ΔchbC-37) resembled A clones with respect to growth phenotype and protein profiles. This suggests that transformability represents a phenotype that varies within the population of B31 MI. The genetic basis for the different transformation frequencies among clones has not been defined. However, our results suggest that transformability is related to certain clonal phenotypes (A versus B or C clones) but not necessarily to infectivity (A1 versus A3). Whether the truncation of OspB or lack of cp9 seen in clones A1 and A3 is coincidental with or a prerequisite for transformation remains to be addressed in further studies.
Clones or mutants with truncated OspB and no cp9 also had significantly shorter in vitro doubling times. These commonly observed in vitro phenotypes (6, 10, 13, 15, 30) could result in outgrowth of such clones within a heterogenous population. Such outgrowth may explain the rapid loss of plasmids and infectivity from the population during in vitro passage of uncloned B. burgdorferi isolates. This explanation is consistent with our observation that the population of uncloned B31 MI contains all plasmids, yet all 11 clonal derivatives obtained before mouse passage are lacking one or more plasmids. Similarly, although we have obtained A3 transformants that retain a full complement of plasmids (except cp9), we have also noted loss of plasmids in other A3 transformants (data not shown). These results emphasize that starting with a clonal population does not guarantee that subsequent derivatives will retain the original plasmid complement. In contrast, clonal analysis of reisolates from mice injected with the parental B31 MI indicated that all clones had a full complement of plasmids (with the exception of cp9 in some clones [data not shown]), suggesting a selective advantage for those clones in vivo but not in vitro.
Our inability to recover clone A1, which lacks lp28-1, after needle inoculation of mice supports previous reports correlating the loss of this plasmid with reduced infectivity in mice (Table 2) (18, 25, 47). The similarity between A1 and the infectious clone A3 also points to the importance of lp28-1. Both were derived from the same parental B31 MI culture and are similar in colony morphology, growth phenotype, and protein and plasmid profiles, other than the presence or absence of lp28-1. The seroconversion to P39 seen for mice after needle injection of A1 is indistinguishable from an infection with the parental B31 MI or infectious clone A3, B1, or C1. Therefore, A1 may be able to initiate an infection, but spirochetes may subsequently be cleared or reduced in number by the host immune response.
Lack of the vlsE locus located on lp28-1 in clone A1 could play a role in clearance of spirochetes by restricting antigenic variation of A1 during host infection (47, 48). This hypothesis is supported by our results with peritoneal dialysis chambers, which allow B. burgdorferi to grow in an immunoprivileged site in the rat, with the dialysis chambers functioning as barriers against host immune cells and antibodies. In this model, A1 grew to normal densities and showed the adaptive gene expression pattern expected in a mammalian host environment (decreased expression of OspA and increased expression of OspC).
Clone B1 was infectious in mice after needle inoculation but appeared to be defective in transmission to mice by infected ticks. This clone could not be reisolated from mice on which B1-infected I. scapularis nymphs had fed to repletion. The infection rate with B1 in those nymphs was comparable to the infection rate in the larvae from which they had molted. Similar numbers were obtained for the parental B31 MI strain, which, in contrast to B1, was transmitted to mice by infected nymphal ticks. The seroconversion to P39 seen for mice infected with B1 by needle inoculation, but absent in mice on which B1-infected nymphs had fed, suggests a block in transmission of B1 from ticks.
Whether the incomplete down-regulation of OspA observed for B1 during growth in DMCs plays a role in the transmission defect remains to be addressed. As shown previously, the number of spirochetes expressing OspA decreases after ticks feed on blood (35, 36), and recent studies suggest that OspA may be involved in tick midgut adhesion (23, 24, 35). Therefore, a defect in OspA down-regulation could compromise the ability of spirochetes to disseminate from the midgut and result in a decreased transmission efficiency.
Our data suggest that lp28-4, which is the only plasmid missing in B1, may be required for transmission from the tick vector. Lack of lp28-4 has previously been associated with modest attenuation in mice after needle inoculation (18), but the role of this plasmid and any other in tick transmission has not been investigated. Further research is needed to confirm the relationship between loss of lp28-4 and the transmission defect observed for B1 or to determine where at the vector-host interface an interruption of B1 transmission takes place.
In summary, we derived clonal variants from B. burgdorferi B31 MI with distinct in vitro characteristics and different in vivo profiles. We demonstrated transformation with a shuttle vector and gene inactivation by allelic exchange in the low-passage-number B31 MI and in various clonal derivatives, some of which were infectious in mice. Our results indicate that a well-defined, clonal wild-type strain, such as A3, is a prerequisite for targeted mutagenesis of infectious B. burgdorferi. Only careful monitoring of plasmid content and infectivity, combined with complementation, will allow the interpretation of mutant phenotypes.
Acknowledgments
We thank Paul Policastro for assistance with ticks; P. Scott Hefty for help with primer design; Dan Hogan for sequencing; Cynthia Gonzales for DMC cultivation; Stanley Falkow, Frank Gherardini, and James Musser for helpful comments on the manuscript; James Carroll for assistance with growth of B. burgdorferi at different pHs; Anita Mora and Gary Hettrick for graphic support; and Penny Gaddy-Rhodes for secretarial assistance.
Justin D. Radolf is supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant AI-29735). Darrin R. Akins is supported by the American Heart Association (grant 0030130N) and the National Center for Research Resources, National Institutes of Health (grant RR-15564).
Editor: D. L. Burns
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, A. G. 1988. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J. Clin. Microbiol. 26:475-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bono, J. L., K. Tilly, B. Stevenson, D. Hogan, and P. Rosa. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 6.Bundoc, V. G., and A. G. Barbour. 1989. Clonal polymorphisms of outer membrane protein OspB of Borrelia burgdorferi. Infect. Immun. 57:2733-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 9.Chung, C. T., S. L. Niemla, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, J. L., R. C. Rogers, P. A. Rosa, and J. L. Benach. 1994. Variations in the ospB gene of Borrelia burgdorferi result in differences in monoclonal antibody reactivity and in production of escape variants. Infect. Immun. 62:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donahue, J. G., J. Piesman, and A. Spielman. 1987. Reservoir competence of white-footed mice for Lyme disease spirochetes. Am. J. Trop. Med. Hyg. 36:92-96. [DOI] [PubMed] [Google Scholar]
- 12.Elias, A. F., J. L. Bono, J. A. Carroll, P. Stewart, K. Tilly, and P. Rosa. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 182:2909-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fikrig, E., H. Tao, F. S. Kantor, S. W. Barthold, and R. A. Flavell. 1993. Evasion of protective immunity by Borrelia burgdorferi by truncation of outer surface protein B. Proc. Natl. Acad. Sci. USA 90:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 15.Golde, W. T., and M. C. Dolan. 1995. Variation in antigenicity and infectivity of derivatives of Borrelia burgdorferi, strain B31, maintained in the natural, zoonotic cycle compared with maintenance in culture. Infect. Immun. 63:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight, S. W., B. J. Kimmel, C. H. Eggers, and D. S. Samuels. 2000. Disruption of the Borrelia burgdorferi gac gene, encoding the naturally synthesized GyrA C-terminal domain. J. Bacteriol. 182:2048-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36:587-609. [DOI] [PubMed] [Google Scholar]
- 20.Motaleb, M. A., L. Corum, J. L. Bono, A. F. Elias, P. Rosa, D. S. Samuels, and N. W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 97:10899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norris, S. J., C. J. Carter, J. K. Howell, and A. G. Barbour. 1992. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect. Immun. 60:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norris, S. J., J. K. Howell, S. A. Garza, M. S. Ferdows, and A. G. Barbour. 1995. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect. Immun. 63:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal, U., R. R. Montgomery, D. Lusitani, P. Voet, V. Weynants, S. E. Malawista, Y. Lobet, and E. Fikrig. 2001. Inhibition of Borrelia burgdorferi-tick interactions in vivo by outer surface protein A antibody. J. Immunol. 166:7398-7403. [DOI] [PubMed] [Google Scholar]
- 25.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosa, P., D. S. Samuels, D. Hogan, B. Stevenson, S. Casjens, and K. Tilly. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J. Bacteriol. 178:5946-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosa, P. A., and D. Hogan. 1992. Colony formation by Borrelia burgdorferi in solid medium: clonal analysis of osp locus variants, p. 95-103. In U. G. Munderloh and T. J. Kurtti (ed.), First International Conference on Tick Borne Pathogens at the Host-Vector Interface: an agenda for research. University of Minnesota, St. Paul.
- 28.Rosa, P. A., T. Schwan, and D. Hogan. 1992. Recombination between genes encoding major outer surface proteins A and B of Borrelia burgdorferi. Mol. Microbiol. 6:3031-3040. [DOI] [PubMed] [Google Scholar]
- 29.Rosa, P. A., and T. G. Schwan. 1989. A specific and sensitive assay for the Lyme disease spirochete Borrelia burgdorferi using the polymerase chain reaction. J. Infect. Dis. 160:1018-1029. [DOI] [PubMed] [Google Scholar]
- 30.Sadziene, A., A. G. Barbour, P. A. Rosa, and D. D. Thomas. 1993. An OspB mutant of Borrelia burgdorferi has reduced invasiveness in vitro and reduced infectivity in vivo. Infect. Immun. 61:3590-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwan, T. G., W. Burgdorfer, and C. F. Garon. 1988. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect. Immun. 56:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwan, T. G., W. Burgdorfer, M. E. Schrumpf, and R. H. Karstens. 1988. The urinary bladder, a consistent source of Borrelia burgdorferi in experimentally infected white-footed mice (Peromyscus leucopus). J. Clin. Microbiol. 26:893-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwan, T. G., M. E. Schrumpf, R. H. Karstens, J. R. Clover, J. Wong, M. Daugherty, M. Struthers, and P. A. Rosa. 1993. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J. Clin. Microbiol. 31:3096-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson, W. J., W. Burgdorfer, M. E. Schrumpf, R. H. Karstens, and T. G. Schwan. 1991. Antibody to a 39-kilodalton Borrelia burgdorferi antigen (P39) as a marker for infection in experimentally and naturally inoculated animals. J. Clin. Microbiol. 29:236-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steere, A. C. 1989. Lyme disease. N. Engl. J. Med. 321:586-596. [DOI] [PubMed] [Google Scholar]
- 40.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in tick-bite-infected mammals, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 43.Tilly, K., S. Casjens, B. Stevenson, J. L. Bono, D. S. Samuels, D. Hogan, and P. Rosa. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol. 25:361-373. [DOI] [PubMed] [Google Scholar]
- 44.Tilly, K., A. F. Elias, J. L. Bono, P. Stewart, and P. Rosa. 2000. DNA exchange and insertional inactivation in spirochetes. J. Mol. Microbiol. Biotechnol. 2:433-442. [PubMed] [Google Scholar]
- 45.Tilly, K., A. F. Elias, J. Errett, E. Fischer, R. Iyer, I. Schwartz, J. L. Bono, and P. Rosa. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tilly, K., L. Lubke, and P. Rosa. 1998. Characterization of circular plasmid dimers in Borrelia burgdorferi. J. Bacteriol. 180:5676-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, J.-R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:1-20. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, J. -R., and S. J. Norris. 1998. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect. Immun. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]