Abstract
Three enterotoxins from the Aeromonas hydrophila diarrheal isolate SSU have been molecularly characterized in our laboratory. One of these enterotoxins is cytotoxic in nature, whereas the other two are cytotonic enterotoxins, one of them heat labile and the other heat stable. Earlier, by developing an isogenic mutant, we demonstrated the role of a cytotoxic enterotoxin in causing systemic infection in mice. In the present study, we evaluated the role of these three enterotoxins in evoking diarrhea in a murine model by developing various combinations of enterotoxin gene-deficient mutants by marker-exchange mutagenesis. A total of six isogenic mutants were prepared in a cytotoxic enterotoxin gene (act)-positive or -negative background strain of A. hydrophila. We developed two single knockouts with truncation in either the heat-labile (alt) or the heat-stable (ast) cytotonic enterotoxin gene; three double knockouts with truncations of genes encoding (i) alt and ast, (ii) act and alt, and (iii) act and ast genes; and a triple-knockout mutant with truncation in all three genes, act, alt, and ast. The identity of these isogenic mutants developed by double-crossover homologous recombination was confirmed by Southern blot analysis. Northern and Western blot analyses revealed that the expression of different enterotoxin genes in the mutants was correspondingly abrogated. We tested the biological activity of these mutants in a diet-restricted and antibiotic-treated mouse model with a ligated ileal loop assay. Our data indicated that all of these mutants had significantly reduced capacity to evoke fluid secretion compared to that of wild-type A. hydrophila; the triple-knockout mutant failed to induce any detectable level of fluid secretion. The biological activity of selected A. hydrophila mutants was restored after complementation. Taken together, we have established a role for three enterotoxins in A. hydrophila-induced gastroenteritis in a mouse model with the greatest contribution from the cytotoxic enterotoxin Act, followed by the Alt and Ast cytotonic enterotoxins.
Among various Aeromonas species, A. hydrophila is most commonly involved in causing human infections such as septicemia and gastroenteritis (16). Isolation of A. hydrophila from water and food sources, as well as the increasing resistance of this organism to antibiotics and chlorination in water, presents a significant threat to public health (2, 4, 10, 11, 16, 26, 27, 30, 34, 48). Although Aeromonas-induced gastroenteritis is most common in young children, the organism is being isolated lately with high frequency from patients with traveler's diarrhea (14, 48).
The pathogenesis of A. hydrophila infection is complex and multifactorial, with the involvement of a number of virulence factors (1, 5). After initial colonization of the epithelial cells through type IV pili (8, 30, 31, 32), A. hydrophila may cause diarrhea by producing enterotoxins (14, 33). Asao et al. (6) first purified a 49- to 52-kDa β-hemolysin to homogeneity from a species of Aeromonas that induced fluid secretion in an animal model. Subsequently, a β-hemolysin-related aerolysin from A. bestiarum and A. trota (12, 29) and a cytotoxic enterotoxin (Act, containing 493 amino acid residues) from the A. hydrophila diarrheal isolate SSU were molecularly characterized (17). Act and aerolysin are pore-forming toxins, and we have demonstrated previously that Act has hemolytic, cytotoxic, and enterotoxic activities (17, 47). Recently, we have shown that Act activates proinflammatory cytokine and eicosanoid cascades in macrophages and a rat intestinal epithelial cell line (IEC-6), leading to tissue damage and a fluid secretory response (22; unpublished data).
In addition to Act, we have cloned two cytotonic enterotoxin genes in Escherichia coli from the genomic library of A. hydrophila SSU (21). Unlike Act, these cytotonic enterotoxins did not cause degeneration of crypts and villi of the small intestine (15, 20, 21). The cell lysates from E. coli clones harboring cytotonic enterotoxin genes caused Chinese hamster ovary (CHO) cells to elongate and to produce cyclic AMP, which are typical enterotoxic responses (21). One of these cytotonic enterotoxins was heat labile at 56°C and was referred to as Alt, while the other was heat stable at the same temperature and was designated Ast (20, 21). An Ast-related cytotonic enterotoxin gene was first cloned from A. hydrophila (reclassified as A. trota) by Chakraborty et al. (13); however, it was not further characterized. These investigators demonstrated that cell lysates from their E. coli clone caused fluid secretion in the rabbit ligated ileal loop and suckling mouse assays (13).
We showed previously that the purified native Alt from A. hydrophila SSU exhibited a size of 44 kDa, elongated CHO cells, and evoked fluid secretion in the rabbit ligated ileal loop model (15, 21). More detailed characterization of the recombinant Alt from E. coli revealed that it consisted of a single polypeptide chain with 368 amino acid residues (20) and that purified Alt elevated cyclic AMP and prostaglandin E2 levels in CHO and rat intestinal epithelial cells (20, 21). Our recent DNA sequence analysis of the ast gene, which is presented in this paper, revealed that it was encoded by a 1,911-bp open reading frame (ORF), contained 636 amino acid residues, and had a predicted molecular mass of 71 kDa with an isoelectric point of 6.9 based on computer analysis. Both Alt and Ast represent novel molecules with no significant homology to known bacterial enterotoxins (15, 20, 21).
The molecular characterization of these three enterotoxins (Act, Alt, and Ast) (15, 17, 20, 21) will now allow us to define their individual contributions in evoking diarrhea and a possible interaction among these enterotoxins during A. hydrophila infections. In this report, we have generated various enterotoxin gene-deficient mutants of A. hydrophila to precisely evaluate their role in secretory diarrhea.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The sources of A. hydrophila and E. coli strains, as well as the plasmids used in this study, are listed in Table 1. Briefly, the pBlue alt recombinant plasmid contained a 4.0-kb SalI DNA fragment from the chromosome of A. hydrophila SSU that harbored the alt gene in pBluescript (Stratagene, La Jolla, Calif.) and was generated from the original pSL24 recombinant plasmid (21). The pBlue ast recombinant plasmid contained a 4.8-kb SalI/BamHI A. hydrophila SSU chromosomal DNA fragment in pBluescript and harbored the ast gene (21). This plasmid was generated from the original clone pSBS32, which contained a 6.0-kb SalI DNA fragment harboring the ast gene (21). The suicide vector pJQ200SK contained a P15A origin of replication (ori), a levansucrase gene (sacB) from Bacillus subtilis, and a gentamicin resistance (Gm) gene (28, 41). Suicide vectors pDMS197 and pRE112 had a conditional R6K ori and a sacB gene and also a tetracycline resistance (Tc) gene and a chloramphenicol resistance (Cm) gene, respectively (24). An act isogenic mutant of A. hydrophila SSU (designated SSUΔact) was previously generated in our laboratory via homologous recombination using suicide vector pJQ200SK (47).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Sourcea or reference |
|---|---|---|
| A. hydrophila SSU | CDC, Atlanta, Ga | |
| SSU-R | Rifr | Laboratory stock |
| SSUΔact | act isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr; sucrose resistance | 47 |
| SSUΔalt | alt isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr; sucrose resistance | This study |
| SSUΔast | ast isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr; sucrose resistance | This study |
| SSUΔalt,ast | alt and ast gene double-knockout isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr Smr Spr; sucrose resistance | This study |
| SSUΔact,ast | act and ast gene double-knockout isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr Smr Spr; sucrose resistance | This study |
| SSUΔact,alt | act and alt gene double-knockout isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr Tcr; sucrose resistance | This study |
| SSUΔact,alt,ast | act, alt, and ast gene triple-knockout isogenic mutant of A. hydrophila SSU-R generated by double crossover; Rifr Kmr Smr Spr Tcr; sucrose resistance | This study |
| E. coli | ||
| HB101 | recA13 hsdS20 supE44 | Promega |
| DH5α | recA gyrA | Laboratory stock |
| SM10 | Kmr λpir | 35 |
| S17-1 | Smr; trimethoprim resistance; λpir | 28 |
| HMS174(DE3) | Rifr | Novagen |
| Plasmids | ||
| pRK2013 | Helper plasmid, Km | ATCC, Manassas, Va. |
| pBR322 | Ap Tc | Amersham |
| pBluescript-SK | Ap | Stratagene |
| pUC-4K | Contains a 1.2-kb Km gene cassette | Amersham |
| pHP45Ω | Contains a 2.0-kb Sm/Sp gene cassette | 40 |
| pJQ200SK | Suicide vector; P15A ori sacB Gm | 28, 41 |
| pDMS197 | Suicide vector; R6K ori sacB Tc | 24 |
| pRE112 | Suicide vector; R6K ori sacB Cm | 24 |
| pSL24 | Contains a 4.0-kb SalI DNA fragment harboring the alt gene in pT7-6 vector; Ap | 21 |
| pSBS32 | Contains a 6.0-kb SalI DNA fragment harboring the ast gene in pBluescript-SK vector; Ap | 21 |
| pBlue alt | pBluescript recombinant plasmid with a 4.0-kb SalI DNA fragment from plasmid pSL24 containing the alt gene | This study |
| pBlue ast | pBluescript recombinant plasmid with a 4.6-kb SalI/BamHI DNA fragment from plasmid pSBS32 containing the ast gene | This study |
| pB ast-Sm/Sp | ast gene in plasmid pBlue ast truncated at the SmaI site with a Sm-Sp gene cassette | This study |
| pB ast-Km | ast gene in plasmid pBlue ast truncated at the SmaI site with a Km gene cassette | This study |
| pB alt-Km | alt gene in plasmid pBlue alt truncated at the BglII site with a Km gene cassette | This study |
| pB alt-Tc | alt gene in plasmid pBlue alt truncated at the BglII site with a Tc gene cassette | This study |
| pJQ alt-Km | Vector pJQ200SK containing a Km gene cassette-truncated alt gene with its flanking sequences for generating mutant SSUΔalt | This study |
| pJQ ast-Km | Vector pJQ200SK containing a Km gene cassette-truncated ast gene with its flanking sequences for generating mutant SSUΔast | This study |
| pDMS ast-Sm/Sp | Vector pDMS197 containing a Sm-Sp gene cassette-truncated ast gene with its flanking sequences for generating SSUΔalt,ast and SSUΔact,ast mutants | This study |
| pRE alt-Tc | Vector pER112 containing a Tc gene cassette-truncated alt gene with its flanking sequences for generating SSUΔact,alt and SSUΔact,alt,ast mutants | This study |
| pBR alt | A. hydrophila alt gene with its putative promoter region, cloned in pBR322 at the EcoRI site | This study |
| pBR ast | A. hydrophila ast gene with its putative promoter region, cloned in pBR322 at the EcoRI/PstI site | This study |
| pBR act | A. hydrophila act gene with its putative promoter region, cloned in pBR322 at the EcoRI/PstI site | This study |
| pGP1-2 | Contains a T7 RNA polymerase gene which is regulated by a thermally inactivated repressor, cI857; Km | Laboratory stock |
| pT7-5 | Contains a T7 promoter upstream of the multiple cloning site; Ap | Laboratory stock |
| pT7-6 | Contains a T7 promoter upstream of the multiple cloning site; the multiple cloning site is in the opposite orientation from the pT7-5 vector; Ap | Laboratory stock |
CDC, Centers for Disease Control and Prevention; ATCC, American Type Culture Collection.
Enzymes, chemicals, and recombinant DNA techniques.
The antibiotics ampicillin, gentamicin (GEN), tetracycline (TET), kanamycin (KAN), chloramphenicol, spectinomycin (SPT), and streptomycin (STR) were used at concentrations of 100, 15, 15, 50, 20, 25, and 50 μg/ml, respectively, unless otherwise stated. Rifampin (RIF) was used at a concentration of 40 μg/ml for bacterial growth and 300 μg/ml during conjugation experiments. All of the antibiotics used were obtained from Sigma (St. Louis, Mo.). Restriction endonucleases and T4 DNA ligase were obtained from Promega (Madison, Wis.) and New England BioLabs (Beverly, Mass.). The Advantage cDNA PCR kit was purchased from Clontech (Palo Alto, Calif.). Chromosomal DNA from various A. hydrophila mutants was isolated with a QIAamp DNA Mini kit (Qiagen, Inc., Valencia, Calif.). The plasmid DNA and the DNA fragments from the agarose gel were prepared and purified with a QIAprep Miniprep Kit (Qiagen). All of the basic molecular biology techniques used in this study were previously described (7, 47).
Expression of the ast gene with a bacteriophage T7 promoter-polymerase system
A dual-plasmid T7 expression system developed by Tabor and Richardson (46) was used for the expression of the ast gene. The recipient E. coli strain HB101 contained a plasmid, pGP1-2, with a T7 RNA polymerase gene whose expression was regulated by a thermally inactivated repressor, cI857, and also a Km gene. The 4.8-kb SalI/BamHI DNA fragment containing the ast gene was cloned in Ap plasmid vectors pT7-5 and pT7-6, which had multiple cloning sites in opposite orientations and a promoter for the T7 RNA polymerase gene, located upstream of the multiple cloning sites. The recombinant pT7-5 and pT7-6 plasmids were transformed into E. coli HB101(pGP1-2) (39) and examined for the expression of the ast gene. Briefly, 2 ml of the recombinant E. coli clone harboring the ast gene was grown in Luria-Bertani (LB) medium (39, 42) at 30°C with 100 μg of ampicillin/ml and 40 μg of KAN/ml to an optical density at 600 nm (OD600) of 0.5. The cells were collected by centrifugation and washed three times with M9 medium (39, 42). The pellet was resuspended in 1 ml of M9 medium supplemented with 20 μg of thiamine/ml and 0.01% 18-amino-acid mixture (minus methionine and cysteine) and grown at 30°C for 60 min with shaking (180 rpm). To induce the gene for T7 RNA polymerase, the growth temperature of the culture was shifted to 42°C for 15 min and RIF (200 μg/ml) was added for an additional 10 min to inhibit E. coli endogenous RNA polymerase activity, followed by incubation at 30°C for 20 min. Newly synthesized proteins were labeled by adding 30 μCi of [35S]methionine-cysteine (ICN, Irvine, Calif.) during the last 5 min of incubation of the culture at 30°C. The labeled proteins were precipitated with 10% trichloroacetic acid on ice for 30 min. After centrifugation, the pellet was washed three times with cold 10% trichloroacetic acid and dissolved in 100 μl of the sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (42). Protein samples were subjected to electrophoresis and autoradiography (39).
For preparing cell lysate from an E. coli clone expressing the ast gene with the T7 expression system for biological activity measurement, 100 ml of LB medium was inoculated with the culture and the culture was grown at 30°C and shaken with appropriate antibiotics until an OD600 of 1.0 was reached. The culture was induced at 42°C for 25 min, and then 400 μg of RIF/ml was added. The temperature of the culture was reduced to 37°C for an additional 2 h, the cells were harvested and sonicated, and the cell lysate was examined for biological activity (15, 47).
DNA sequencing of the ast gene.
The entire 4.8-kb SalI/BamHI DNA fragment containing the ast gene was sequenced using a 373XL automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.) in the Protein Chemistry Core Laboratory, University of Texas Medical Branch. The new primers were designed based on the confirmed sequence of the ast gene, and both strands of the DNA were sequenced.
Development of single-knockout mutants of A. hydrophila SSU with truncation in either the alt or the ast gene.
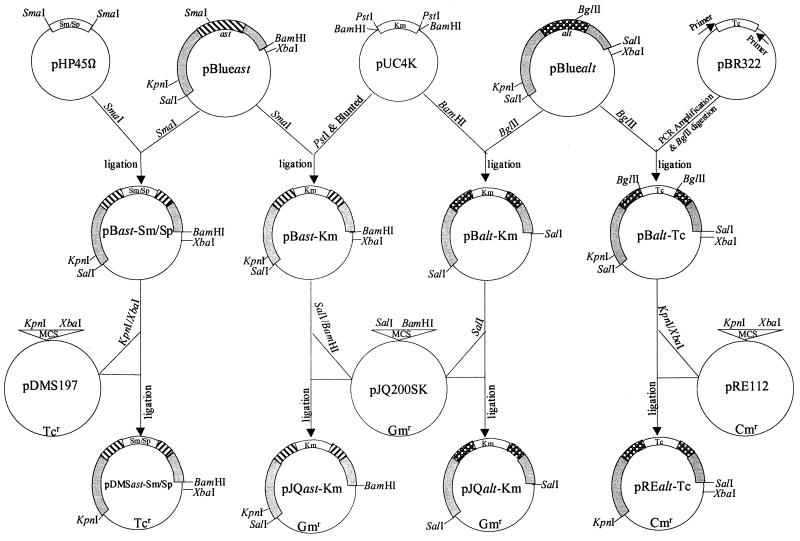
As shown in Fig. 1, the plasmid pBlue alt containing a 4.0-kb SalI DNA fragment with the alt gene from the chromosomal DNA of A. hydrophila was used to prepare the alt isogenic mutant (SSUΔalt). In the alt gene, there was a unique BglII restriction site; the plasmid pBlue alt was thus linearized with BglII enzyme (Fig. 1). A 1.2-kb Km gene cartridge was isolated from plasmid pUC4K (Amersham Pharmacia Biotech, Piscataway, N.J.) by using restriction enzyme BamHI, which bordered the Km gene cassette. This Km gene cassette was ligated to plasmid pBlue alt at the BamHI-compatible BglII restriction site to truncate the alt gene, which generated a new recombinant plasmid, pB alt-Km (Fig. 1). Subsequently, the SalI DNA fragment, which was now 5.2 kb in size due to the insertion of a 1.2-kb Km gene cassette, was removed from the plasmid pB alt-Km by SalI digestion and ligated to a suicide vector, pJQ200SK, at the SalI site, forming a new recombinant plasmid, pJQ alt-Km, in E. coli strain S17-1 (Fig. 1 and Table 1). This strategy to prepare an isogenic mutant provided 1.2 and 2.8 kb of the 5′ and 3′ DNA sequences flanking the truncated alt gene, respectively, to permit double-crossover homologous recombination.
FIG. 1.
Flow diagram showing the construction of various recombinant plasmids for the preparation of enterotoxin gene-deficient mutants of A. hydrophila SSU. The recombinant plasmid pBlue ast contained a 4.6-kb SalI/BamHI DNA fragment from the chromosome of A. hydrophila which harbored the ast gene. The ast gene was truncated at the SmaI restriction site by introducing either a Km gene cassette from plasmid pUC4K or a Sm-Sp gene cassette from plasmid pHP45Ω to generate recombinant plasmid pB ast-Km or pB ast-Sm/Sp, respectively. The truncated ast gene with its flanking sequences was cloned into suicide vector pJQ200SK or pDMS197, forming recombinant plasmid pJQ ast-Km or pDMS ast-Sm/Sp, respectively, for the generation of ast gene-deficient mutants of A. hydrophila. The recombinant plasmid pBlue alt contained a 4.0-kb DNA fragment from the chromosome of A. hydrophila which harbored the alt gene. The alt locus was truncated at the BglII restriction site by introducing either a Km gene cassette from plasmid pUC4K or a Tc gene cassette from plasmid pBR322 to generate recombinant plasmid pB alt-Km or pB alt-Tc, respectively. The truncated alt locus with its flanking sequences was cloned into suicide vector pJQ200SK or pRE112, forming recombinant plasmid pJQ alt-Km or pRE alt-Tc, respectively, for the generation of alt gene-deficient mutants of A. hydrophila. The striped bar represents the ast gene, while the dotted bar represents the alt gene. The gray bar represents sequences flanking the ast or alt gene. The open bar indicates the Km, Sm-Sp, or Tc gene cassette. These plasmids are not drawn to scale. MCS, multiple cloning sites. The primers used to amplify the Tc gene cassette had a BglII restriction site (Table 2).
To generate an SSUΔast mutant of A. hydrophila, the ast gene was cleaved at the unique SmaI restriction site within the 4.8-kb SalI/BamHI DNA fragment in plasmid pBlue ast (Fig. 1 and Table 1). Subsequently, the ast gene was truncated with the Km gene cassette, which was removed from the plasmid pUC4K by PstI digestion. The PstI restriction sites bordered the Km gene cassette. The ends of the Km gene cassette were made blunt with a PCR polishing kit (Stratagene), and the cassette was ligated at the blunted SmaI restriction site within the ast gene to create the pB ast-Km plasmid (Fig. 1). The truncated ast gene with its flanking sequences was removed by SalI/BamHI digestion of the plasmid pB ast-Km and ligated to the suicide vector pJQ200SK at the compatible restriction sites, forming a recombinant plasmid, pJQ ast-Km, in E. coli strain S17-1 (Fig. 1). This strategy provided 2.5 and 2.1 kb of the 5′ and 3′ DNA sequences flanking the truncated ast gene, respectively, to allow double-crossover homologous recombination.
The recombinant E. coli S17-1(pJQ alt-Km or pJQ ast-Km) strain (Fig. 1) was conjugated with Rifr A. hydrophila, as described previously for the development of an act isogenic mutant (SSUΔact) (47). The transconjugants were plated onto LB agar plates with RIF, KAN, and 5% sucrose to select double-crossover transconjugants (47). The cultures were identified as Aeromonas by a positive oxidase test to differentiate them from E. coli and by an automated identification system (Vitek, Hazelwood, Mo.) (47).
Construction of recombinant plasmids pDMS ast-Sm/Sp and pRE alt-Tc for developing double- and triple-knockout mutants of A. hydrophila SSU.
The pBlue ast plasmid, as described above, was linearized with SmaI restriction enzyme (Fig. 1). Subsequently, a 2.0-kb Sm-Sp gene cassette was isolated from plasmid pHP45Ω by digestion with SmaI enzyme, which bordered the Sm-Sp gene cassette, and the 2.0-kb Sm-Sp gene cassette then was inserted at the SmaI restriction site of the ast gene to create the pB ast-Sm/Sp plasmid (Fig. 1). Finally, a 5-kb KpnI/XbaI DNA fragment from this plasmid, containing the ast gene with a 2.0-kb Sm-Sp gene cassette, was cloned at the KpnI/XbaI restriction sites of a pDMS197 suicide vector containing a Tc gene to create a recombinant plasmid, pDMS ast-Sm/Sp (Fig. 1).
Likewise, the pBlue alt plasmid, as described previously, was linearized with the BglII restriction enzyme, which cleaved the alt gene, and truncated with a 1.3-kb Tc gene cassette obtained by PCR amplification from plasmid pBR322 by using specific primers (Fig. 1; Table 2). A 5.3-kb KpnI/XbaI DNA fragment containing the alt gene with the Tc gene cassette from plasmid pB alt-Tc was subcloned in the suicide vector pRE112 containing the Cm gene, to generate a recombinant plasmid, pRE alt-Tc (Fig. 1). The recombinant plasmids pDMS ast-Sm/Sp and pRE alt-Tc were transformed into E. coli SM10, as described previously (45, 47). Both E. coli strains, S17-1 and SM10, contained λpir, allowing replication of the suicide vectors only in these strains (24, 28).
TABLE 2.
Sequences of the primers used for amplification of various toxin genes and the antibiotic resistance cassettes
| Primer position | Primer sequencea | Location | Reference | Purpose |
|---|---|---|---|---|
| 5′ | 5′ CGTGGATCCATGCAAAAACTAAAAATAACTGGC 3′ with BamHI site | +1 to +24 of the act gene | 17 | Amplification of the act gene for probe preparation |
| 3′ | 5′ TTATTGATTGGCTGCTGGCTGCACGCT 3′ | +1456 to +1482 of the act gene | 17 | |
| 5′ | 5′ ATAGAGGAATTCCTCCATGATCGCCGGGCTGGTGGGCGCGGGCG 3′ | +72 to +103 of the alt gene | 20 | Amplification of the alt gene for probe preparation and identification of the alt isogenic mutant by PCR |
| 3′ | 5′ CATCCTAAGCTTTAAGCTTTCAACGCGCCATCGCCAACGCTCTC 3′ with EcoRI (5′ primer) and HindIII (3′ primer) sites | +1082 to +1114 of the alt gene | 20 | |
| 5′ | 5′ ATGCACGCACGTACCGCCATG 3′b | +1 to +21 of the ast gene | This study | Amplification of the ast gene for probe preparation and identification of the ast isogenic mutant by PCR |
| 3′ | 5′ GGACTTTTTCACCGCAGCGGGTT 3′b | +1886 to +1908 of the ast gene | This study | |
| 5′ | 5′ TAGAGATCTGAATTCTCATGTTTGACAGC 3′ with BglII site | −3 to +17 of plasmid pBR322 | 42 | Amplification of the Tc cassette for the truncation of the alt gene and probe preparation |
| 3′ | 5′ GCTAGATCTCCAAGGGTTGGTTTGCGCAT 3′ with BglII site | +1355 to +1374 of plasmid pBR322 | 42 | |
| 5′ | 5′ CTGAATTCACATGATCGTGGCCACCGACT 3′ with EcoRI site | −543 to −523 of the alt gene | Unpublished data | Amplification of the alt gene and its putative promoter region for complementation |
| 3′ | 5′ CTGAATTCTCAACGCGCCATCGCCAACGATCT 3′ with EcoRI site | +1084 to +1107 of the alt gene | 20 | |
| 5′ | 5′ AACTGCAGATGACCTCATAGTAGAC 3′ with PstI site | −532 to −514 of the ast gene | This study | Amplification of the ast gene and its putative promoter region for complementation |
| 3′ | 5′ AAGAATTCTCAGGACTTTTTCACCGCAGC 3′ with EcoRI site | +1891 to +1911 of the ast gene | This study | |
| 5′ | 5′ CGCTGAGGTCTGCCTCGTGAAGAAGGTGTT 3′ | +434 to +464 of the pUC4K plasmid | 18 | Amplification of the Km gene cassette for PCR identification of the generated mutants |
| 3′ | 5′ AAAGCCACGTTGTGTCTAAAATCTCTGATGT 3′ | +1613 to +1643 of the pUC4K plasmid | 18 | |
| 5′ | 5′ CTATGGCGCCATCAACAGCTCGCC 3′ | +423 to +446 of the ast gene | This study | Sequencing primers for further identification of the truncated ast gene in the mutants |
| 3′ | 5′ GCGATAGCTGAGCGGCTTGCCCTG 3′ | +553 to +576 of the ast gene | This study | |
| 5′ | 5′ CTCAACACCATCACCGACGTG 3′ | +307 to +327 of the alt gene | 20 | Sequencing primers for further identification of the truncated alt gene in the mutants |
| 3′ | 5′ GCTCAGGGCGAAGCCGCGCTC 3′ | +454 to +474 of the alt gene | 20 | |
| 5′ | 5′ GTCTGCAGCATGGCCGACGCCATCG 3′ with PstI site | −546 to −529 of the act gene | This study | Amplification of the act gene and its putative promoter region for complementation |
| 3′ | 5′ TGGAATTCTTATTGATTGGCTGCTGGCGT 3′ with EcoRI site | +2345 to +2365 of the act gene | This study |
Underlining indicates restriction enzyme sites in the primer.
Similar primers with NdeI (5′ primer) and XhoI (3′ primer) restriction enzyme sites were used for amplifying the coding region of the ast gene and cloning into the pET30a vector.
Construction of double-knockout mutants of A. hydrophila SSU.
To generate an alt- and ast-negative mutant of A. hydrophila (SSUΔalt,ast), E. coli SM10 λpir(pDMS ast-Sm/Sp) (Fig. 1) and the Rifr and Kmr SSUΔalt mutant, as developed above, were used for conjugation. Cultures that were resistant to KAN, STR-SPT, RIF, and 5% sucrose should have represented genuine double-crossover mutants. For developing an act- and ast-negative mutant of A. hydrophila (SSUΔact,ast), E. coli SM10 λpir(pDMS ast-Sm/Sp) (Fig. 1) and the Rifr and Kmr SSUΔact mutant, as previously developed (47), were used for conjugation. Cultures resistant to KAN, STR-SPT, RIF, and 5% sucrose were analyzed. For developing an act- and alt-negative mutant of A. hydrophila (SSUΔact,alt), E. coli SM10 λpir(pRE alt-Tc) (Fig. 1) and the Rifr and Kmr SSUΔact mutant, as previously developed (47), were used for conjugation. Cultures resistant to KAN, TET, RIF, and 5% sucrose should have represented double-crossover mutants.
Construction of a triple-knockout mutant of A. hydrophila SSU.
To generate an act-, alt-, and ast-negative mutant of A. hydrophila (SSUΔact,alt,ast), a double-knockout mutant (SSUΔact,ast), as developed above, and E. coli SM10 λpir(pRE alt-Tc) (Fig. 1) were used for conjugation. Cultures resistant to KAN, STR-SPT, TET, RIF, and 5% sucrose should have represented genuine triple-knockout mutants. The identity of various mutants developed was confirmed by PCR, Southern blot analysis, and DNA sequence analysis.
Southern blot analysis.
The chromosomal DNA from different enterotoxin gene-deficient mutants, as well as the wild-type A. hydrophila, was isolated, and an aliquot (10 μg) of the chromosomal DNA was digested with appropriate restriction enzymes and subjected to 0.8% agarose gel electrophoresis (42, 47). Next, the digested DNA was transferred to a nylon membrane (Gibco BRL, Gaithersburg, Md.) and baked at 80°C for 2 h. The blots were prehybridized and hybridized by using Quikhyb (Stratagene) at 68°C, as described by the manufacturer. The blots were hybridized using alt and ast gene probes. The full-length toxin genes for hybridization were obtained by PCR amplification from the chromosomal DNA of A. hydrophila SSU. The sequences of the primers used for PCR amplification of the toxin genes are shown in Table 2. The primers were synthesized commercially by Biosynthesis, Inc. (Lewisville, Tex.), and the program used for PCR was as follows: 94°C for 2 min (denaturation), followed by 30 cycles of 94°C for 1 min and 68°C for 3 min. The final extension was performed at 72°C for 7 min. The PCR-amplified products were isolated from the agarose gel and purified by using a QIAquick Gel Extraction kit (Qiagen). Similarly used probes for Southern blot analysis were the Km gene cassette (1.2 kb) obtained from the plasmid pUC4K by PstI restriction enzyme digestion, the Sm-Sp gene cassette (2.0 kb) recovered from plasmid pHP45Ω by SmaI restriction enzyme digestion, and the Tc gene cassette (1.3 kb) obtained by PCR amplification from the plasmid pBR322 using specific primers (Table 2). The suicide vector plasmids (pJQ200SK, pDMS197, and pRE112) were probed against linearized plasmids after digestion with restriction enzyme XbaI. The probes were labeled with [α-32P]dCTP (ICN) by using a random primer kit (Gibco BRL). The membranes were washed twice at 68°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0) (42, 47) plus 0.1% SDS for 20 min and then twice in 1× SSC plus 0.1% SDS for 20 min at 68°C. The blots were exposed to the X-ray film at −70°C for 2 to 12 h.
Northern blot analysis.
Wild-type A. hydrophila SSU and its various enterotoxin gene-deficient isogenic mutants were grown in LB medium at 37°C overnight. The next morning, 200 μl of the overnight cultures was added to 4 ml of the fresh LB medium in 50-ml sterilized disposable tubes and allowed to grow for another 3 h. The cells were collected by centrifugation, and the total RNA was isolated using the RNA isolation kit from Qiagen. The RNA samples (10 μg) were subjected to electrophoresis on a 1.2% formaldehyde-agarose gel with 1× MOPS buffer (0.2 M MOPS [morpholinepropanesulfonic acid] [pH 7.0], 0.005 M sodium acetate, 0.01 M EDTA, pH 8.0) (42, 47). The RNA was transferred to the nylon membrane, and after baking, the filters were prehybridized, hybridized, and washed as described for Southern blot analysis. The 32P-labeled various gene probes were used for hybridization. The amount of RNA in each lane was quantitated by scanning 23S or 16S rRNA bands after ethidium bromide staining of the gel, using the Gel Doc 2000 System (Bio-Rad Laboratories, Hercules, Calif.). All of the reagents used for Northern blot analysis were treated with diethyl pyrocarbonate (Sigma).
Purification of Ast and Western blot analysis.
The coding region of the ast gene (1,911 bp) generated by PCR amplification from the pBluescript ast plasmid with specific primers with 5′ NdeI and 3′ XhoI restriction sites (Table 2) was ligated to a bacteriophage T7 polymerase-promoter-based pET30a vector (Novagen, Madison, Wis.) at the NdeI/XhoI sites and transformed into E. coli HMS174(DE3). This vector had six histidine residues, which were fused in frame with the protein of interest at the C-terminal end for affinity purification on a nickel column. The recombinant E. coli clone (pT30 ast) was grown in LB medium (200 ml) with shaking (180 rpm) to an OD600 of 0.6 before induction with 1 mM isopropylthio-β-galactoside (IPTG) for 4 h at 37°C. The expression of the ast gene was monitored by SDS-12% PAGE and Coomassie blue staining, with uninduced culture as a control. For purification of Ast, the cells were sonicated after IPTG induction and the pellet was solubilized in 8 M urea in denaturing binding buffer (Invitrogen, Carlsbad, Calif.). The sample was loaded onto a nickel column (Invitrogen), and Ast was eluted with 150 to 200 mM imidazole. The identity of Ast was confirmed by microsequencing of Ast from the Immobilon-P membrane after SDS-PAGE (19) with a 494HT microsequencer (Applied Biosystems, Inc.) at the Protein Chemistry Core Laboratory, University of Texas Medical Branch.
The antibodies to Ast were generated in mice after intraperitoneal injection with polyacrylamide gel pieces containing Ast mixed with Freund's incomplete adjuvant. The animals were bled before immunization and every 2 weeks after immunization, with booster antigen given every week for 2 months. The antibody titer was determined by an enzyme-linked immunosorbent assay with Ast as the source of antigen (15). To examine expression of the ast gene in various enterotoxin gene-deficient mutants of A. hydrophila, the cultures were grown to an OD600 of 0.6. A 2-ml suspension of the culture was centrifuged, and the pellet was dissolved in 200 μl of the SDS sample buffer (42). An aliquot (5 μl) of each sample was subjected to SDS-12% PAGE and Western blot analysis (17). Specific polyclonal antibodies to Ast were used as primary antibodies for Western blot analysis, followed by appropriate secondary antibodies, which were labeled with horseradish peroxidase (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.). The blots were developed by using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.).
Complementation of SSUΔalt, SSUΔast, and SSUΔact mutants of A. hydrophila SSU.
By using specific primers (Table 2) with an EcoRI restriction site, a 1.5-kb DNA fragment containing the alt gene and its putative promoter region was amplified from the chromosomal DNA of A. hydrophila. It was then ligated to the vector pBR322 at the EcoRI restriction site to generate recombinant plasmid pBR alt (Table 1), which was first transformed into E. coli HB101, which carried a helper plasmid, pRK2013 (with the Km gene). Subsequently, via conjugation, the recombinant pBR alt plasmid with the helper plasmid pRK2013 was transformed into an alt gene-deficient mutant of A. hydrophila (SSUΔalt) (Table 1) that was generated during this study by double-crossover homologous recombination (Fig. 1). The transconjugants were screened on LB agar plates containing RIF, KAN, and TET. The presence of the pBR alt recombinant plasmid in the A. hydrophila SSUΔalt mutant was confirmed by plasmid isolation and restriction enzyme analysis.
By using a strategy similar to that described above and specific primers (Table 2), we amplified a 2.5-kb DNA fragment containing the ast gene and its putative promoter region from A. hydrophila chromosomal DNA. This DNA fragment was ligated to the vector pBR322 at the EcoRI/PstI restriction sites to generate the recombinant plasmid pBR ast (Table 1). Subsequently, the pBR ast plasmid was transformed into an ast gene -deficient mutant (SSUΔast), as prepared for the experiment described for Fig. 1.
For SSUΔact complementation, a pair of primers was designed (Table 2) to amplify a 2.0-kb DNA fragment containing the act gene and its putative promoter region from A. hydrophila chromosomal DNA. The amplified DNA fragment was subsequently inserted into vector pBR322 at EcoRI/PstI restriction sites to generate recombinant plasmid pBR act in E. coli HB101 with helper plasmid pRK2013 (Table 1). The pBR act plasmid was then transformed from E. coli into SSUΔact by conjugation as previously described. The wild-type A. hydrophila transformed with pBR322 vector alone served as a control.
CHO cell elongation assay.
The assay was performed as previously described (38). Briefly, the 96-well culture plates were seeded with 104 CHO cells/200 μl in F-12 medium, which contained penicillin (100 U/ml), STR (100 μg/ml), GEN (50 μg/ml), and fetal bovine serum (1%). The CHO cells were incubated at 37°C with 5% CO2 for 1 h. Subsequently, a twofold dilution of cell lysates containing Ast was added to the cells, and the plates were incubated for 24 h at 37°C. The cells were fixed with 70% methanol, stained with Giemsa stain, and examined for CHO cell elongation.
Hemolytic assay.
A twofold dilution of the culture filtrates from wild-type A. hydrophila and its various isogenic mutants was prepared in phosphate-buffered saline (PBS) and mixed with rabbit red blood cells (2%; Colorado Serum Co., Denver, Colo.) in a microtiter plate (47). The plate was incubated at 37°C for 1 h and subsequently at 4°C overnight. The hemolytic activity unit was defined as the reciprocal dilution of Act in the culture filtrate demonstrating 50% lysis of red blood cells. The culture filtrates were prepared by growing cultures in LB medium for 18 h at 37°C. After centrifugation, the culture filtrates were filter sterilized before measuring hemolytic activity (17).
Mouse and rat ligated ileal loop assay.
A diet-restricted, antibiotic-treated adult mouse model was used to evaluate the enterotoxic activity of various enterotoxin gene-deficient mutants of A. hydrophila SSU. Briefly, 20- to 25-g BALB/c mice (Taconic Farms, Inc., Germantown, N.Y.) were restricted in food intake by 20% for 3 weeks and then given STR (5 g/liter in drinking water) for 48 h prior to intraluminal challenge with the organism. The antibiotic was removed from water 6 h prior to surgery. The mice were anesthetized with halothane (Halocarbon Laboratories, River Edge, N.J.). An abdominal incision was made, and a single 5-cm segment of the small intestine was constructed with 00-size silk suture (18). Approximately 2 × 105 CFU (per 100 μl) of the test organism grown in LB medium was inoculated in the ligated small intestinal loop of each mouse (18). After 12 to 16 h of observation, the animals were euthanized by cervical dislocation, the intestinal loops were excised, and the fluid was collected in a microcentrifuge tube. The microcentrifuge tubes were centrifuged briefly, and the amount of luminal fluid was measured (18). When rats were used, the animals (70 to 75 g; Sprague-Dawley, Indianapolis, Ind.) were anesthetized with 35 mg of pentobarbital sodium (Nembutal)/kg of body weight intraperitoneally. After an abdominal incision, the rat intestine was ligated into 5-cm segments, as described above, and injected with 0.5 ml of the cell lysates from E. coli clones containing the ast gene. The animals were sacrificed with an overdose of pentobarbital sodium (100 mg/kg) after 12 h, and fluid accumulation was measured.
Colonization of the small intestine by A. hydrophila wild type and enterotoxin gene-deficient mutants.
Mice challenged with various bacteria in the lumen of the small intestinal loops were sacrificed at 2 and 12 to 16 h. The small intestinal segments of the animals were washed with PBS to remove unbound bacteria and homogenized in PBS. Various dilutions were plated on MacConkey agar plates with 10 μg of ampicillin (Remel, Lenexa, Kans.)/ml to selectively grow A. hydrophila. In some experiments, the tissues were homogenized in PBS and then mixed with the accumulated fluid after 12 to 16 h to determine the increase in the number of A. hydrophila organisms.
Statistical analysis.
Wherever appropriate, the data were analyzed with the multiple-group comparison Tukey test, and P values of ≤0.05 were considered significant.
Nucleotide sequence accession number.
The sequence of the ast gene has been submitted to GenBank with accession no. AF419157.
RESULTS
Expression of the ast gene from a 4.8-kb SalI/BamHI DNA fragment of the A. hydrophila SSU chromosome and DNA sequence analysis of the ast gene.
Both Act and Alt enterotoxins have previously been molecularly characterized in our laboratory (15, 16, 17, 20, 21, 22). However, although the gene encoding Ast has been cloned, we only recently sequenced the gene (accession no. AF419157). The sequences of these enterotoxin genes, as well as those of their flanking DNA, were needed to prepare isogenic mutants of A. hydrophila deficient in production of one or more of these enterotoxins. This, in turn, allowed us to delineate their precise role in causing gastroenteritis.
To obtain an estimate of the potential length of the ast gene ORF and the direction of ast gene transcription, we expressed a 4.8-kb SalI/BamHI DNA fragment containing the ast gene in E. coli with a bacteriophage T7 expression system using vectors pT7-5 and pT7-6 (Table 1). This system provided us with two advantages: (i) the expression of the genes on the cloned DNA fragment could be exclusively monitored by [35S]methionine labeling of the proteins, as E. coli protein synthesis was shut down by RIF, which did not affect T7 RNA polymerase activity, and (ii) pT7-5 and pT7-6 vectors had multiple cloning sites in opposite orientations, thereby allowing us to determine the direction of the ast gene transcription.
Our data indicated two radiolabeled polypeptides of 32 and 71 kDa when a 4.8-kb SalI/BamHI DNA fragment was expressed from vector pT7-6 in the direction of SalI to BamHI. No radioactive band was detected when we used a pT7-5 vector to insert this fragment in the opposite orientation (data not shown). Our DNA sequence analysis of the 4.8-kb SalI/BamHI DNA fragment revealed this fragment to be 4,623 bp with three ORFs in the direction of SalI to BamHI at nucleotide positions 10 to 894 (encoding a 295-amino-acid-long polypeptide), 1007 to 1861 (encoding a 284-amino-acid-long polypeptide), and 1964 to 3874 (encoding a 636-amino-acid-long polypeptide) with approximate molecular sizes of 32, 31, and 71 kDa, respectively.
The cell lysate from this E. coli clone promoted CHO cell elongation (titer, 1:256) and caused fluid secretion (2.8 ± 0.2 ml/5 cm) in the rat ligated small intestinal loops. The cell lysate from an E. coli clone with vector alone or with the 4.6-kb SalI/BamHI DNA fragment inserted in the opposite orientation in vector pT7-5 did not cause any CHO cell elongation or fluid secretion and served as a negative control. Cholera toxin (1 μg) and PBS were used as positive and negative controls, respectively, for the CHO cell and loop assay. The fluid secretion caused by cholera toxin was 3.2 ± 0.3 ml/5 cm of the loop. A total of 10 rats were used, each with an appropriate negative and positive control and the test samples. These data suggested Ast to be either 31 to 32 or 71 kDa in size.
We subsequently cloned a 3.6-kb BamHI fragment from the chromosomal DNA of another clinical isolate of A. sobria, which, when expressed with the T7 expression system (using the pT7-6 vector) in the correct orientation, exhibited enterotoxic activity similar to that seen with the 4.6-kb SalI/BamHI fragment from A. hydrophila SSU on CHO cells, as well as in rat ligated ileal loops. No biological activity was detected in those clones in which the 3.6-kb DNA fragment was inserted in the opposite orientation. The DNA sequence analysis of this 3.6-kb fragment revealed only one ORF at nucleotide positions 943 to 2853, which encoded a 636-amino-acid-long polypeptide similar to Ast from A. hydrophila SSU. A 91% homology was noted between Ast from these two isolates of Aeromonas at the amino acid level. These data indicated Ast to be 71 kDa in size. The DNA sequence (943 bp) upstream of the 5′ end of the ast gene showed only 5% homology between 4.6-kb SalI/BamHI and 3.6-kb BamHI DNA fragments, while the homology was 87% in the region spanning 749 bp downstream of the 3′ end of the ast gene. These data indicated a significant divergence in sequences flanking the ast gene in different species of Aeromonas. The DNA sequence of the ast gene, along with its deduced amino acid sequence, is available in GenBank (accession no. AF419157).
The highly purified Ast exhibited a size of 71 kDa after SDS-PAGE, and the NH2-terminal sequence (5 amino acid residues sequenced) of the purified Ast matched the DNA-derived amino acid sequence. The availability of the sequence of the entire 4.6-kb SalI/BamHI fragment allowed us to develop an ast isogenic mutant of A. hydrophila to define its role in causing gastroenteritis.
Characterization of SSUΔalt and SSUΔast mutants of A. hydrophila SSU.
The strategies used to develop alt and ast isogenic mutants are depicted in Fig. 1. Conjugation of wild-type A. hydrophila SSU with E. coli S17-1 harboring either pJQ alt-Km or pJQ ast-Km with a truncated alt or ast gene, respectively, should have resulted in transconjugants that were resistant to RIF, KAN, and 5% sucrose but sensitive to GEN. Such mutants should have undergone genuine double-crossover homologous recombination, resulting in the replacement of native alt and ast genes with truncated alt and ast genes and concomitant loss of the suicide vector with Gm and sacB genes.
To confirm the identify of these isogenic mutants, the chromosomal DNA was isolated and subjected to PCR and Southern blot analysis. For PCR analysis, specific primers generated to either the alt and ast genes or the Km gene cassette were used (Table 2). These primers detected 1.1- and 1.9-kb DNA fragments from the chromosome of wild-type A. hydrophila, which represented the correct sizes of the alt and ast genes, respectively (20). However, the size of the alt and ast genes was larger by 1.2 kb in the isogenic mutants, due to the insertion of the Km gene cassette. Similarly, the primers to the Km gene cassette detected a 1.2-kb DNA fragment from the isogenic mutants but not from the chromosomal DNA of wild-type A. hydrophila (data not shown). The PCR data then were confirmed by performing Southern blot analysis.
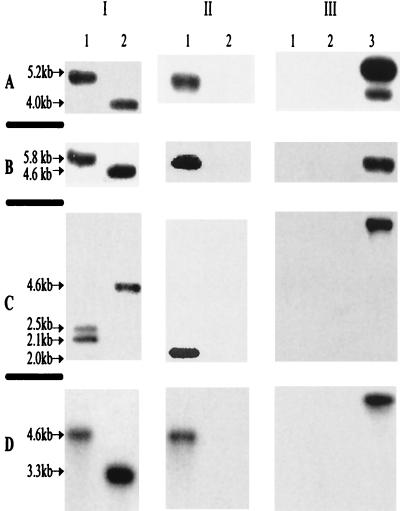
As depicted in Fig. 2A, the size of the chromosomal DNA fragment from wild-type A. hydrophila digested with SalI restriction enzyme was 4.0 kb when an alt-specific gene probe was used (Fig. 2A-I, lane 2). However, the size of the SalI DNA fragment from the mutant SSUΔalt was 5.2 kb due to the insertion of a Km gene cassette (Fig. 2A-I, lane 1). A similarly sized DNA fragment was detected in the digested chromosomal DNA of the mutant strain when the Km gene cassette was used as a probe (Fig. 2A-II, lane 1). This probe did not react with the digested DNA from the wild-type A. hydrophila (Fig. 2A-II, lane 2). No band was detected in the digested chromosomal DNA of both the mutant (Fig. 2A-III, lane 1) and wild-type A. hydrophila (Fig. 2A-III, lane 2), when the suicide vector pJQ200SK was used as a probe. The probe reacted with the pJQ200SK plasmid digested with XbaI restriction enzyme and served as a positive control (Fig. 2A-III, lane 3). These data indicated that the mutant strain SSUΔalt had completely lost the suicide vector sequence as a result of double-crossover homologous recombination.
FIG. 2.
Confirmation of the identity of the enterotoxin gene-deficient mutants of A. hydrophila SSU based on Southern blot analysis. (A) Chromosomal DNA from SSUΔalt mutant (lanes 1) and wild-type A. hydrophila (lanes 2) was digested with SalI restriction enzyme. Suicide vector pJQ200SK digested with restriction enzyme XbaI was used in lane 3. (B) Chromosomal DNA from the SSUΔast mutant (lanes 1) and wild-type A. hydrophila (lanes 2) was digested with SalI/BamHI restriction enzyme. Suicide vector pJQ200SK digested with restriction enzyme XbaI was used in lane 3. (C) Chromosomal DNA from the SSUΔalt,ast or SSUΔact,ast mutant (lanes 1) and wild-type A. hydrophila (lanes 2) was digested with SalI/BamHI restriction enzyme. Suicide vector pDMS197 digested with restriction enzyme XbaI was used in lane 3. (D) Chromosomal DNA from the SSUΔact,alt or SSUΔact,alt,ast mutant (lanes 1) and wild-type A. hydrophila (lanes 2) was digested with KpnI/XbaI restriction enzyme. Suicide vector pRE112 digested with restriction enzyme XbaI was used in lane 3. Different enterotoxin genes (alt [A-I and D-I] and ast [B-I and C-I]), different antibiotic gene cassettes (Km cassette [A-II and B-II], Sm-Sp cassette [C-II], and Tc cassette [D-II]), and different suicide vectors (pJQ200SK [A-III and B-III], pDMS197 [C-III], and pRE112 [D-III]) were used as probes. In panel A, lane 3, the two bands were due to incomplete digestion of the vector pJQ200SK.
Similarly, we digested chromosomal DNA from mutant SSUΔast and wild-type A. hydrophila with restriction enzymes SalI/BamHI and subjected them to Southern blot analysis (Fig. 2B). As shown in Fig. 2B-I, lane 2, the digested chromosomal DNA from wild-type A. hydrophila exhibited a band of 4.6 kb when an ast gene-specific probe was used. The size of this fragment from the mutant strain SSUΔast was 5.8 kb due to the insertion of a Km gene cassette (Fig. 2B-I, lane 1). Although the 5.8-kb DNA fragment from the SSUΔast mutant reacted with the Km cassette gene probe (Fig. 2B-II, lane 1), it failed to react with chromosomal DNA from the wild-type A. hydrophila as expected (Fig. 2B-II, lane 2). Neither digested chromosomal DNA from the wild-type strain nor that from the mutant strain of A. hydrophila reacted with the suicide vector probe, as shown in Fig. 2B-III (lanes 1 and 2). Lane 3 in Fig. 2B-III represented a positive control showing the suicide vector probe hybridizing to itself.
Characterization of SSUΔalt,ast, SSUΔact,ast, and SSUΔact,alt double-knockout mutants and an SSUΔact,alt,ast triple-knockout mutant of A. hydrophila
Since we used the same plasmid, pDMS ast-Sm/Sp, for the generation of mutants SSUΔalt,ast and SSUΔact,ast (Fig. 1), the Southern blot patterns for these two mutants were similar, and therefore, a representative blot for only one of the mutants is shown here. The chromosomal DNA from these isogenic mutants and wild-type A. hydrophila was digested with restriction enzymes SalI/BamHI and hybridized with various probes. In Fig. 2C-I, the ast-specific gene probe was used. Two bands of 2.1 and 2.5 kb were detected in the mutant strain (lane 1), whereas one 4.6-kb DNA fragment was detected in the wild-type A. hydrophila (Fig. 2C-I, lane 2). The detection of two bands in the mutant strain was expected, as the Sm-Sp gene cassette was flanked by a BamHI site. Therefore, digestion of the chromosomal DNA from the mutant strain with SalI/BamHI enzymes removed the Sm-Sp gene cassette and divided the ast gene into two DNA fragments of 2.1 and 2.5 kb. In Fig. 2C-II, lane 1, a 2.0-kb band representing the antibiotic gene cassette was detected in the mutant but not in the wild-type A. hydrophila (lane 2) when a Sm-Sp gene cassette was used as a probe. In Fig. 2C-III, lanes 1 and 2, the chromosomal DNA from both the mutant and the wild-type A. hydrophila did not react with the pDMS197 vector probe, indicating loss of the suicide vector from the chromosome of the mutant.
Because the same plasmid, pRE alt-Tc, was used for generation of mutants SSUΔact,alt and SSUΔact,alt,ast (Fig. 1), the Southern blot patterns for these two mutants were similar, and therefore, a representative blot for only one of the mutants is shown here. The chromosomal DNA from these two mutant strains as well as that from the wild-type A. hydrophila was digested with restriction enzymes KpnI/XbaI. In Fig. 2D-I, the alt gene was used as a probe, which detected a 4.6-kb DNA fragment in the mutant strain (lane 1). This fragment was larger by 1.3 kb than the one seen in wild-type A. hydrophila (Fig. 2D-I, lanes 1 and 2) due to the insertion of the Tc gene cassette. The size of the DNA fragment detected in the wild-type A. hydrophila was 3.3 kb (Fig. 2D-I, lane 2). In Fig. 2D-II, lane 1, a similarly sized DNA fragment (4.6 kb) was detected in the mutant strain when a Tc gene cassette was used as a probe, whereas this probe failed to react with the wild-type A. hydrophila chromosomal DNA (Fig. 2D-II, lane 2). As expected, neither the chromosomal DNA from the mutant nor that from the wild-type A. hydrophila reacted with the suicide vector probe (Fig. 2D-III, lanes 1 and 2). The Southern blot data obtained with the double- and triple-knockout mutants were confirmed by PCR analysis using toxin gene-specific primers and by DNA sequence analysis of the PCR product using primers to the toxin genes (Table 2). The DNA sequence analysis revealed insertion of antibiotic resistance genes at the correct location in these mutants (data not shown).
Northern blot analysis to demonstrate loss of transcription of the corresponding enterotoxin genes in the isogenic mutants of A. hydrophila SSU.
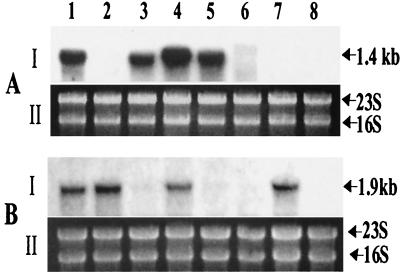
Total RNA from various mutants and wild-type A. hydrophila was isolated and subjected to Northern blot analysis by using act-, alt-, and ast-specific gene probes. As shown in Fig. 3A, an act gene transcript of 1.4 kb was detected in the wild-type A. hydrophila (lane 1) and in mutants SSUΔalt (lane 3), SSUΔast (lane 4), and SSUΔalt,ast (lane 5), in which the act gene was intact. A similar level of hemolytic activity was noted in the culture filtrates of these mutants (data not shown). However, in mutants SSUΔact (lane 2), SSUΔact,alt (lane 6), SSUΔact,ast (lane 7), and SSUΔact,alt,ast (lane 8), no act gene transcript was detected, indicating the successful truncation of the act gene in these mutants. Such mutants also did not exhibit any hemolytic activity.
FIG. 3.
The transcripts for the enterotoxin genes act and alt were eliminated in the indicated mutants, based on Northern blot analysis. Total RNA from wild-type A. hydrophila and its enterotoxin gene-deficient mutants was isolated and hybridized with the act gene probe (A-I) and the alt gene probe (B-I), as described in Materials and Methods. Lane 1, wild-type A. hydrophila; lane 2, mutant SSUΔact; lane 3, mutant SSUΔalt; lane 4, mutant SSUΔast; lane 5, mutant SSUΔalt,ast; lane 6, mutant SSUΔact,alt; lane 7, mutant SSUΔact,ast; lane 8, mutant SSUΔact,alt,ast. The RNA loaded in each lane was quantitated by scanning 16S and 23S rRNA bands after ethidium bromide staining of the gel (A-II and B-II).
In Fig. 3B, we used an alt-specific gene probe, and a 1.9-kb transcript was detected in wild-type A. hydrophila (lane 1) and in mutants SSUΔact (lane 2), SSUΔast (lane 4), and SSUΔact,ast (lane 7). No alt-specific transcript was detected in mutants SSUΔalt (lane 3), SSUΔalt,ast (lane 5), SSUΔact,alt (lane 6), and SSUΔact,alt,ast (lane 8). Surprisingly, no ast gene transcription was detected in wild-type A. hydrophila and its various enterotoxin gene-deficient mutants (data not shown). These data possibly suggested (i) a very low level of the transcript for the ast gene, (ii) a short half-life of the ast mRNA, and (iii) rapid degradation of the ast gene transcript.
Western blot analysis to demonstrate the presence of Ast in wild-type A. hydrophila SSU and its isogenic mutants.
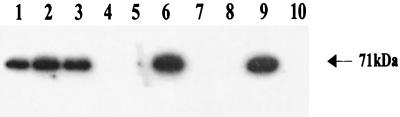
Data presented in Fig. 4 clearly show the presence of Ast antigen in wild-type A. hydrophila (lane 1) and all of the isogenic mutants that had an intact ast gene (lanes 2, 3, and 6) in Western blots probed with Ast-specific antibodies. The Ast-specific band was missing from the isogenic mutants SSUΔast (lane 4), SSUΔalt,ast (lane 5), SSUΔact,ast (lane 7), and SSUΔact,alt,ast (lane 8). Purified Ast was used as a positive control (lane 9), and cell lysates from E. coli served as a negative control (lane 10) to demonstrate the specificity of Ast antibodies.
FIG. 4.
Western blot analysis showing that the ast gene expression was eliminated in its corresponding gene-deficient mutant of A. hydrophila. The cell lysates from different enterotoxin-deficient mutants of A. hydrophila were subjected to SDS-12% PAGE, and subsequently the proteins were transferred to a nitrocellulose membrane for Western blot analysis as described in Materials and Methods. Lane 1, wild-type A. hydrophila; lane 2, SSUΔact; lane 3, SSUΔalt; lane 4, SSUΔast; lane 5, SSUΔalt,ast; lane 6, SSUΔact,alt; lane 7, SSUΔact,ast; lane 8, SSUΔact,alt,ast; lane 9, purified Ast (0.1 μg) as a positive control; lane 10, cell lysate from E. coli as a negative control.
Evaluation of various isogenic mutants of A. hydrophila in a mouse model.
A diet-restricted, antibiotic-treated adult mouse model was used to evaluate different isogenic mutants of A. hydrophila for enterotoxic activity. This model provided optimal fluid secretion after A. hydrophila challenge. In animals fed a normal diet and challenged with wild-type A. hydrophila (5 × 105 CFU), a mean fluid secretion of 175 ± 55 μl/5 cm of the loop was recorded over a 12- to 16-h observation period. The fluid accumulation was 912 ± 28 μl/5 cm in animals which were food restricted but not treated with STR. In food-restricted and STR-treated mice, the fluid accumulation increased to 1,378 ± 38 μl/5 cm. A total of 10 mice in each group were used in the above-mentioned study. We noted that, with the SSUΔalt,ast mutant, the fluid secretion occurred rapidly, with maximum fluid levels noted in 4 h. This mutant induced fluid secretion as a result of Act production, as the alt and ast genes were deleted from this strain. However, the fluid secretion evoked by Alt and Ast in an act isogenic mutant of A. hydrophila was minimal within 4 to 6 h, becoming maximal only after 12 h. Therefore, we opted to observe fluid secretion by various enterotoxin gene-deficient mutants of A. hydrophila after 12 to 16 h of observation.
As shown in Table 3, single-knockout mutants, such as SSUΔact, SSUΔalt, and SSUΔast, reduced fluid secretion by 64, 48, and 43%, respectively, compared to fluid secretion evoked by the wild-type A. hydrophila. The enterotoxic activity of mutants SSUΔalt, SSUΔast, and SSUΔact was restored after complementation. The wild-type A. hydrophila with pBR322 vector alone was used as a control in the complementation experiments. The differences in fluid accumulation evoked by various mutants not only were significant compared to the wild-type bacterium but also exhibited significance when these mutants were compared to one another by using Tukey multiple-group comparison analysis (Table 3). The only mutants which demonstrated no statistically significant difference in fluid accumulation were SSUΔalt and SSUΔast, which culminated in 445 ± 28 and 485 ± 22 μl of fluid/5 cm of the loop, respectively. It was noted that, compared to fluid secretion of 1,378 ± 38 μl/5 cm of the loop observed in a different experiment (as stated above) using diet-restricted, antibiotic-treated animals, the amount of fluid accumulated was less in the experiments presented in Tables 3 and 4, indicating variation in animals purchased at different times.
TABLE 3.
Ability of wild-type A. hydrophila SSU, single-knockout mutants, and their complemented strains to cause fluid secretion in a mouse model
| Organism injected (2 × 105/100 μl)a | No. of BALB/c mice tested | Fluid accumulation (μl/5-cm loop ± SD) | % Reduction in fluid secretion |
|---|---|---|---|
| WT A. hydrophila | 25 | 850 ± 50c | 0 |
| SSUΔact | 25 | 300 ± 29b,c | 64 |
| SSUΔalt | 25 | 445 ± 28b,c,d | 48 |
| SSUΔast | 25 | 485 ± 22b,c,d | 43 |
| WT A. hydrophila(pBR322) | 25 | 812 ± 44 | 0 |
| SSUΔalt(pBalt) | 25 | 820 ± 24 | 0 |
| SSUΔast(pBast) | 25 | 805 ± 20 | 0 |
| SSUΔact(pBact) | 25 | 868 ± 25 | 0 |
The number of organisms in each loop after 12 to 16 h was increased by 2 logs. WT, wild type.
All groups were significantly different from wild-type A. hydrophila at P < 0.001 by multiple-group comparison Tukey test.
All groups were significantly different from each other at P < 0.001 by multiple-group comparison Tukey test.
These groups were not statistically different from each other (P = 0.056).
TABLE 4.
Ability of wild-type A. hydrophila SSU, double-knockout mutants (SSUΔalt,ast, SSUΔact,ast, and SSUΔact,alt), and triple-knockout mutant (SSUΔact,alt,ast) to cause fluid secretion in a mouse model
| Organism injected (2 × 105/100 μl)a | No. of BALB/c mice tested | Fluid accumulation (μl/5-cm loop ± SD) | % Reduction in fluid secretion |
|---|---|---|---|
| WT A. hydrophila | 25 | 858 ± 28b | 0 |
| SSUΔalt,ast | 25 | 552 ± 26b | 36 |
| SSUΔact,ast | 25 | 330 ± 28b | 62 |
| SSUΔact,alt | 25 | 230 ± 18b | 73 |
| SSUΔact,alt,ast | 25 | NDc | 100 |
The number of organisms in each loop after 12 to 16 h was increased by 2 logs. WT, wild type.
All groups were significantly different from each other at P < 0.001 by multiple-group comparison Tukey test.
ND, not detected. The limit of detection was less than 50 μl.
In the SSUΔalt,ast mutant, the combined reduction in fluid secretion due to Alt and Ast was 36%, and Act accounted for 64% of the total fluid accumulated (Table 4). For the SSUΔact,ast mutant, which would cause fluid secretion only through Alt production, 38% of the enterotoxic activity was noted, compared to that observed for the wild-type A. hydrophila. In a double-knockout mutant in which act and alt genes were deleted, fluid secretion totaling 230 ± 18 μl/5 cm of the loop was measured, indicating the contribution of Ast to be 27% (Table 4). No detectable fluid secretion was noted in the mutant strain in which all three enterotoxin genes were deleted. Once again, the differences in fluid accumulation in these various mutants were statistically significant compared either to the wild-type bacterium or among one another with the Tukey test. All of these mutants colonized the small intestine of mice to the same extent [(1.0 ± 0.6) × 105 to (1.2 ± 0.6) × 105 CFU/ml] as that noted for the wild-type A. hydrophila (1.1 × 105 ± 0.8 × 105 CFU/ml) after 2 h, and their number increased by approximately 2 logs after 12 to 16 h of incubation.
DISCUSSION
We have performed, for the first time, an extensive case-control human study at the International Center for Diarrheal Diseases Research, Dhaka, Bangladesh, which included children younger than 5 years of age, to demonstrate the extent of Aeromonas infection in these children. We indeed established an association of Aeromonas with diarrhea and showed that enterotoxins were involved in Aeromonas-associated gastroenteritis (3). Our recent studies also indicated that bloody diarrhea was most commonly associated with the production of the cytotoxic enterotoxin Act, while nonbloody diarrhea was correlated with the elaboration of the cytotonic enterotoxins Alt and Ast (3; unpublished data). Diarrheal isolates harboring one, two, or all three of the enterotoxin genes were recovered from patients, although most of the Aeromonas isolates contained two of the three enterotoxin genes in various combinations. These studies were, therefore, designed to delineate precisely the roles of individual enterotoxins in evoking diarrhea by developing mutants with various combinations of deletions of enterotoxin genes.
Although the genes encoding Act and Alt were previously sequenced in our laboratory (17, 20), the DNA sequence of the ast gene was not available and was needed to generate an ast isogenic mutant. Based on the expression of a 4.6-kb SalI/BamHI DNA fragment in E. coli with the T7 expression system, biological activity measurement and subsequent DNA sequence analysis revealed that Ast was 71 kDa in size. The size of Ast was further confirmed by its molecular characterization from another clinical isolate of A. sobria and by the development of an ast isogenic mutant. The SSUΔast mutant truncated within the ast gene indeed exhibited a reduced enterotoxic activity, and biological activity associated with Ast was restored after complementation (Table 3).
Our initial strategy was to use purified enterotoxins individually or in various combinations to demonstrate their role in evoking fluid secretion and to determine any synergism among these enterotoxins. However, we noted that, when the ast gene was hyperexpressed with the T7 polymerase-promoter-based pT30a vector system, most of the Ast was membrane bound, requiring harsh treatment for solubilization (e.g., 8 M urea), thereby resulting in the loss of biological activity. We encountered a similar solubility problem when act and alt genes were hyperexpressed in E. coli (20, 25). However, we circumvented the Act and Alt solubility problem by expressing the corresponding genes in A. salmonicida and A. hydrophila strains, lacking act and alt genes, respectively, by using a broad-host-range pMMB66 vector with an IPTG-inducible tac promoter (22; unpublished data). This vector system allowed Act and Alt proteins to be secreted out into the medium in a biologically active form (22; unpublished data). The act and alt gene-negative strains of Aeromonas were needed to prevent homologous recombination between the toxin gene on the recombinant pMMB66 plasmid and on the chromosome.
We were unable to express the ast gene by using this vector, because until recently we did not have an Aeromonas mutant with a complete deletion of the ast gene to prevent homologous recombination between the toxin gene on the recombinant pMMB66 plasmid and that on the chromosome. These potent enterotoxins are produced in small amounts from the bacterium, and hyperexpression of these toxin genes is essential for generating meaningful data on their relative contributions in evoking fluid secretion. We therefore opted to prepare isogenic mutants deficient in various enterotoxin genes to define their role in causing diarrhea in a murine model.
Inactivation of the coding region of genes by insertion of antibiotic resistance markers is a general strategy for the construction of defined mutants (9, 24, 36, 44). The use of suicide vectors is fundamental to this technique, allowing the creation of deletions or insertions in specific genes on chromosomes (24). However, once the suicide vector is integrated into the chromosome, it is necessary to remove the vector DNA, resulting in replacement of the wild-type allele with a mutant allele. A suicide vector carrying a conditional lethal gene, such as a sacB gene, that discriminates between the integration of the vector and double-recombination events has been widely used (45). Therefore, all of the suicide vectors used in this study (pJQ200SK, pDMS197, and pRE112) (Fig. 1 and Table 1) harbored the sacB gene. The sacB gene, which encodes levansucrase, probably polymerizes levan, a product of catabolism of sucrose in the periplasm of gram-negative bacteria, which is toxic to bacteria when they are grown in the presence of sucrose (23). Vectors pDMS197 and pRE112 had a conditional R6K ori and required π protein for replication, while pJQ200SK had a P15A ori from plasmid pACYC184 (24, 28).
In this study, six isogenic mutants of A. hydrophila, in which three enterotoxin genes were deleted in various combinations, were prepared. The biological activity of the single-knockout strains (SSUΔact, SSUΔalt, and SSUΔast) of A. hydrophila could be restored by complementation, suggesting no polar effects of marker-exchange mutagenesis on genes downstream of those encoding the enterotoxins. Since double- and triple-enterotoxin-gene-knockout strains of A. hydrophila were prepared using one of the single-enterotoxin-gene-knockout strains, we did not expect polar effects. However, complementation of double- and triple-enterotoxin-gene-knockout strains of A. hydrophila was difficult to perform to completely rule out the possibility of any polar effects in such mutants. We determined the exact insertional sites of the antibiotic resistance cassettes within the enterotoxin genes on the chromosomes of various isogenic mutants and confirmed the absence of frameshift mutations within the toxin genes. All of these mutants (single, double, and triple knockout) exhibited similar growth rates and the ability to colonize and multiply in the small intestine, which further suggested the absence of polar effects in these mutants. However, we should not overlook the possibility of polar effects in the double- and triple-knockout strains of A. hydrophila, which could have some impact on the fluid accumulation results. Such polar effects might constitute a limitation of insertional mutagenesis.
We noted previously and during present studies that the frequency of double crossover was very low (0.01%) when the mutants SSUΔact, SSUΔalt, and SSUΔast were prepared using suicide vector pJQ200SK (47). We therefore employed recently developed suicide vectors pDMS197 and pRE112 (24) for generating double- and triple-knockout mutants of A. hydrophila, which increased the frequency of obtaining double-crossover mutants by 10-fold. Further, it was noted that, when the pJQ200SK suicide vector was used, some false-positive mutants were observed, especially during the preparation of double- and triple-knockout strains of A. hydrophila that were truncated in two and three enterotoxin genes, respectively. The majority of the false-positive mutants still had a portion of the suicide vector plasmid integrated in the chromosomal DNA of A. hydrophila, based on Southern blot data (data not shown). These mutants, however, exhibited the correct phenotype (i.e., resistance to sucrose and sensitivity to GEN). Our subsequent analysis of these mutants indicated that Gmr and sacB genes were indeed lost from these cultures (data not shown). In some instances, we could also detect free recombinant pJQ200SK plasmid, suggesting its replication to some extent in A. hydrophila, which could also interfere with obtaining genuine double-crossover mutants. The frequency of obtaining false-positive mutants was significantly reduced with pMDS197 and pRE112 vectors.
The correct identity of all of the isogenic mutants was confirmed by Southern, Northern, and Western blot analysis (Fig. 2 to 4). In Northern blot analysis, all of the mutants exhibited the correct pattern of inhibition of the corresponding enterotoxin gene transcripts when act and alt gene probes were used (Fig. 3). However, it was noted that the alt mRNA level was significantly lower than that of act mRNA (Fig. 3A and B). These data suggested (i) low levels of the alt gene expression, under studied conditions, and (ii) instability or shorter half-life of the alt mRNA. It will be intriguing to determine in the future whether alt gene expression could be increased during in vivo conditions. Interestingly, we were unable to detect a transcript for the ast gene, even in the wild-type A. hydrophila, although the Ast protein could be seen by Western blot analysis in wild-type A. hydrophila and its isogenic mutants with an intact ast gene (Fig. 4). Our data might be suggestive of a short half-life for the ast mRNA or its rapid degradation and need confirmation. This possibility was based on our observation that we could detect the ast gene transcript in significant amounts from E. coli when the toxin gene was hyperexpressed using a pET30a vector system (data not shown). However, we cannot rule out the possibility that the ast gene expression requires interaction of A. hydrophila with the host cell, and this is under investigation.
We first developed a mouse model to evaluate the fluid secretory potential of wild-type A. hydrophila and its various enterotoxin gene-deficient mutants, based on studies showing that malnourished animals were more susceptible to infection than were adequately nourished animals (37). Further, STR disrupted the aerobic and anaerobic characteristics of the cecal flora and was particularly effective at decreasing colonization resistance (43). Deletion of the enterotoxin genes individually indicated that Act, Alt, and Ast contributed to the fluid secretory response by 64, 48, and 43%, respectively (Table 3). Data obtained with SSUΔact,ast and SSUΔact,alt mutants, which caused fluid secretion via the production of Alt and of Ast, respectively, conclusively proved their contribution to be 38 and 27%, respectively (Table 4). These data coincided with our observation that mice immunized with Alt and Ast reduced fluid secretion caused by A. hydrophila challenge by 39 and 30%, respectively 20; unpublished data).
Based on the data presented in Tables 3 and 4 for SSUΔact and SSUΔalt,ast mutants, it was evident that Act was the major enterotoxin contributing to the fluid secretory response (64%), followed by Alt and Ast. Our data also suggested some interaction among these various enterotoxins (Tables 3 and 4). The exact molecular basis of this interaction among the various enterotoxins is not known yet and needs further study. The inability of the triple-knockout mutant to exhibit any detectable enterotoxic response confirmed the role of all three of the enterotoxins (Act, Alt, and Ast) in A. hydrophila-induced diarrhea. The presence of various combinations of enterotoxins in different Aeromonas isolates could increase or decrease the expression of specific enterotoxin genes and thus could dictate the severity of diarrhea. The availability of purified enterotoxins would help in further delineating interactions among these enterotoxins.
In conclusion, we have demonstrated for the first time the contribution of each of the three enterotoxins of A. hydrophila in causing gastroenteritis in a murine model by developing isogenic mutants. Our data tend to suggest some interaction among these enterotoxins in vivo that could lead to altered fluid secretion. Such attenuated strains of A. hydrophila with deletions of the act, alt, and ast genes could be attractive candidates for vaccine development.
Acknowledgments
Jian Sha and E. V. Koslova contributed equally to this work.
This work was supported by a grant from the National Institutes of Health (AI41611). Jian Sha, a postdoctoral fellow, was supported by the McLaughlin postdoctoral fellowship.
The editorial assistance of Mardelle Susman is greatly appreciated. We thank B. Chatuev of our department for providing plasmids pDMS197 and pRE112 and E. coli strain SM10.
Editor: J. T. Barbieri
REFERENCES
- 1.Abbott, S. L., S. Seli, M. Catino, Jr., M. A. Hartley, and J. M. Janda. 1998. Misidentification of unusual Aeromonas species as members of the genus Vibrio: a continuing problem. J. Clin. Microbiol. 36:1103-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alavandi, S. V., M. S. Subashini, and S. Ananthan. 1999. Occurrence of haemolytic and cytotoxic Aeromonas species in domestic water supplies in Chennai. Indian J. Med. Res. 110:50-55. [PubMed] [Google Scholar]
- 3.Albert, M. J., M. Ansaruzzaman, K. Talukdar, A. K. Chopra, I. Kuhn, M. Rahman, A. S. G. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altwegg, M., L. G. Martinetti, J. Luthy-Hottenstein, and M. Rohrbach. 1991. Aeromonas-associated gastroenteritis after consumption of contaminated shrimp. Eur. J. Clin. Microbiol. Infect. Dis. 10:44-45. [DOI] [PubMed] [Google Scholar]
- 5.Annapurna, E., and S. C. Sanyal. 1977. Enterotoxicity of Aeromonas hydrophila. J. Med. Microbiol. 10:317-323. [DOI] [PubMed] [Google Scholar]
- 6.Asao, T., Y. Kinoshita, S. Kozaki, T. Uemura, and G. Sakaguchi. 1984. Purification and some properties of Aeromonas hydrophila hemolysin. Infect. Immun. 46:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 8.Barnett, T. C., S. M. Kirov, M. S. Strom, and K. Sanderson. 1997. Aeromonas ssp. possess at least two distinct type IV pilus families. Microb. Pathog. 23:241-247. [DOI] [PubMed] [Google Scholar]
- 9.Becker, A., M. Schmidt, W. Jager, and A. Puhler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 10.Brandi, G. M., F. Sisti, F. Giardini, G. F. Schiavano, and A. Albano. 1999. Survival ability of cytotoxic strains of motile Aeromonas spp. in different types of water. Lett. Appl. Microbiol. 29:211-215. [DOI] [PubMed] [Google Scholar]
- 11.Buchanan, R. L., and S. A. Palumbo. 1985. Aeromonas hydrophila and Aeromonas sobria as potential food poisoning species: a review. J. Food Safety 7:15-29. [Google Scholar]
- 12.Chakraborty, T., B. Huhle, H. Bergbauer, and W. Goebel. 1986. Cloning, expression, and mapping of the Aeromonas hydrophila aerolysin gene determinant in Escherichia coli K-12. J. Bacteriol. 167:368-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty, T., M. A. Montenegro, S. C. Sanyal, R. Helmuth, E. Bulling, and K. N. Timmis. 1984. Cloning of enterotoxin gene from Aeromonas hydrophila provides conclusive evidence of production of a cytotonic enterotoxin. Infect. Immun. 46:435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty, T., V. Husslein, B. Huhle, H. Bergbauer, T. Jarchau, H. Hor, and W. Goebel. 1988. Genetics of hemolysins in Aeromonas hydrophila. Zentbl. Bakteriol. Mikrobiol. Hyg. Abt. 1 Suppl. 17:215-222. [Google Scholar]
- 15.Chopra, A. K., and C. W. Houston. 1989. Purification and partial characterization of a cytotonic enterotoxin produced by Aeromonas hydrophila. Can. J. Microbiol. 35:719-727. [DOI] [PubMed] [Google Scholar]
- 16.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129-1137. [DOI] [PubMed] [Google Scholar]
- 17.Chopra, A. K., C. W. Houston, J. W. Peterson, and G.-F. Jin. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513-523. [DOI] [PubMed] [Google Scholar]
- 18.Chopra, A. K., J. H. Hung, X.-J. Xu, K. Burden, D. W. Niesel, M. Rosenbaum, and J. W. Peterson. 1999. Role of Salmonella enterotoxin in overall virulence of the organism. Microb. Pathog. 27:155-171. [DOI] [PubMed] [Google Scholar]
- 19.Chopra, A. K., J. W. Peterson, P. Chary, and R. Prasad. 1994. Molecular characterization of an enterotoxin from Salmonella typhimurium. Microb. Pathog. 16:85-98. [DOI] [PubMed] [Google Scholar]
- 20.Chopra, A. K., J. W. Peterson, X.-J. Xu, D. H. Coppenhaver, and C. W. Houston. 1996. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog. 21:357-377. [DOI] [PubMed] [Google Scholar]
- 21.Chopra, A. K., R. Pham, and C. W. Houston. 1994. Cloning and expression of putative cytotonic enterotoxin-encoding genes from Aeromonas hydrophila. Gene 139:87-91. [DOI] [PubMed] [Google Scholar]
- 22.Chopra, A. K., X.-J. Xu, D. Ribardo, M. Gonzalez, K. Kuhl, J. W. Peterson, and C. W. Houston. 2000. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 68:2808-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards, R. A., H. K. Linda, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 25.Ferguson, M. R., X.-J. Xu, C. W. Houston, J. W. Peterson, D. H. Coppenhaver, V. L. Popov, and A. K. Chopra. 1997. Hyperproduction, purification, and mechanism of action of the cytotoxic enterotoxin produced by Aeromonas hydrophila. Infect. Immun. 65:4299-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanninen, M. L., P. Oivanen, and V. Hirvela-Koski. 1997. Aeromonas species in fish, fish-eggs, shrimp and freshwater. Int. J. Food Microbiol. 34:17-26. [DOI] [PubMed] [Google Scholar]
- 27.Hanninen, M. L., S. Salmi, L. Mattila, R. Taipalinen, and A. Siitonen. 1995. Association of Aeromonas spp. with traveller's diarrhoea in Finland. J. Med. Microbiol. 41:26-31. [DOI] [PubMed] [Google Scholar]
- 28.Harayama, S., M. Tsuda, and T. Lino. 1980. High frequency mobilization of the chromosome of Escherichia coli by a mutant of plasmid RP4 temperature sensitive for maintenance. Mol. Gen. Genet. 180:47-56. [DOI] [PubMed] [Google Scholar]
- 29.Howard, S. P., and J. T. Buckley. 1986. Molecular cloning and expression in Escherichia coli of the structural gene for the hemolytic toxin aerolysin from Aeromonas hydrophila. Mol. Gen. Genet. 204:289-295. [DOI] [PubMed] [Google Scholar]
- 30.Kirov, S. M., E. K. Ardestani, and L. J. Hayward. 1993. The growth and expression of virulence factors at refrigeration temperature by Aeromonas strains isolated from food. Int. J. Food Microbiol. 20:159-168. [DOI] [PubMed] [Google Scholar]
- 31.Kirov, S. M., L. A. O'Donovan, and K. Sanderson. 1999. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect. Immun. 67:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirov, S. M., T. C. Barnett, C. M. Pepe, M. S. Strom, and M. J. Albert. 2000. Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 68:4040-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ljungh, A., and T. Wadstrom. 1985. Review. Aeromonas and Plesiomonas as possible causes of diarrhea. Infection 13:169-173. [DOI] [PubMed] [Google Scholar]
- 34.Majeed, K. N., A. F. Egan, and I. C. MacRae. 1990. Production of exotoxins by Aeromonas ssp. at 5°C. J. Appl. Bacteriol. 69:332-337. [DOI] [PubMed] [Google Scholar]
- 35.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishibuchi, M., K. Kumagai, and J. B. Kaper. 1991. Contribution of the tdh1 gene of Kanagawa phenomenon-positive Vibrio parahaemolyticus to production of extracellular thermostable direct hemolysin. Microb. Pathog. 11:453-460. [DOI] [PubMed] [Google Scholar]
- 37.Nzegwu, H. C., and R. J. Levin. 1994. Role of the enteric nervous system in the maintained hypersecretion induced by enterotoxin STa in the nutritionally deprived intestine. Gut 35:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad, R., A. K. Chopra, J. W. Peterson, R. Pericas, and C. W. Houston. 1990. Biological and immunological characterization of a cloned cholera toxin-like enterotoxin from Salmonella typhimurium. Microb. Pathog. 9:315-329. [DOI] [PubMed] [Google Scholar]
- 39.Prasad, R., A. K. Chopra, P. Chary, and J. W. Peterson. 1992. Expression and characterization of the cloned Salmonella typhimurium enterotoxin. Microb. Pathog. 13:109-121. [DOI] [PubMed] [Google Scholar]
- 40.Prentki, P., and H. M. Krish. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 41.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Sanderson, K., F. M. Ghazali, and S. M. Kirov. 1996. Colonization of streptomycin-treated mice by Aeromonas species. J. Diarrhoeal Dis. Res. 14:27-32. [PubMed] [Google Scholar]
- 44.Segada, L. M., G. M. Carlone, L. L. Gheesling, and A. J. Lesse. 2000. Characterization of P1-deficient isogenic mutant of Haemophilus influenzae biogroup aegyptius associated with Brazilian purpuric fever. Microb. Pathog. 28:145-155. [DOI] [PubMed] [Google Scholar]
- 45.Steinmetz, M., D. Le Cog, S. Aymerich, G. Gonzy-Treboul, and P. Gay. 1985. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol. Gen. Genet. 200:220-228. [DOI] [PubMed] [Google Scholar]
- 46.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, X.-J., M. R. Ferguson, V. L. Popov, C. W. Houston, J. W. Peterson, and A. K. Chopra. 1998. Role of a cytotoxic enterotoxin in Aeromonas-mediated infections: development of transposon and isogenic mutants. Infect. Immun. 66:3501-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada, S., S. Matsushita, S. Dejsirilet, and Y. Kudoh. 1997. Incidence and clinical symptoms of Aeromonas-associated traveller's diarrhoea in Tokyo. Epidemiol. Infect. 119:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]