Abstract
In this study we determined the complete nucleotide sequence of Shiga toxin 2e-encoding bacteriophage φP27, isolated from the Shiga toxin-producing Escherichia coli patient isolate 2771/97. φP27 is integrated as a prophage in the chromosomal yecE gene. This integration generates identity segments of attL and attR sites with lengths of 11 nucleotides. The integrated prophage genome has a size of 42,575 bp. We identified 58 open reading frames (ORFs), each with a length of >150 nucleotides. The deduced proteins of 44 ORFs showed significant homologies to other proteins present in sequence databases, whereas 14 putative proteins did not. For 29 proteins, we could deduce a putative function. Most of these are related to the basic phage propagation cycle. The φP27 genome represents a mosaic composed of genetic elements which are obviously derived from related and unrelated phages. We identified five short linker sequences of 22 to 151 bp in the φP27 sequence which have also been detected in a couple of other lambdoid phages. These linkers are located between functional modules in the phage genome and are thought to play a role in genetic recombination. Although the overall DNA sequence of φP27 is not highly related to other known phages, the data obtained demonstrate a typical lambdoid genome structure.
Shiga toxin (Stx)-producing Escherichia coli (STEC) frequently causes diarrhea, hemorrhagic colitis, and the hemolytic-uremic syndrome (HUS) in humans (47). STEC that causes such human diseases is referred to as enterohemorrhagic E. coli. The expression of Stx is believed to be the major pathogenicity factor of this organism. STEC contains genes encoding one or more members of the major Stx groups, Stx1 and Stx2. Stx2 comprises a family of related proteins with high sequence similarity, which includes Stx2 and its variants, designated Stx2c, Stx2d, Stx2e, and Stx2f (23, 40, 51, 57, 58). Pathogenic STEC usually contains genes encoding Stx1, Stx2, or both, whereas E. coli isolates containing genes encoding Stx2 variants are more frequently isolated from asymptomatic carriers (Stx2d) or from animals such as pigs (Stx2e) or pigeons (Stx2f) (57). Stx2c is also produced by human isolates but mostly in strains that also contain genes encoding Stx1, Stx2, or both. Stx2e-producing E. coli is frequently isolated from weaning pigs suffering from edema disease, which involves systemic vascular damage as a result of intestinal infection with STEC that is adapted to swine. The characteristic clinical outcome of edema disease is neurological impairment and death. Like HUS, edema disease often has a prodromal phase of diarrhea (15). Stx2e-producing E. coli has also sporadically been isolated from patients with diarrhea and HUS. These isolates belonged to serogroups O101 and O9 that have not been reported in STEC strains associated with edema disease (19). Apparently, the stx genes of STEC are generally phage borne (41, 44, 55, 62, 70). In a recent publication, the phage origin of a stx2e gene sequenced from human E. coli isolate 2771/97 was described and the stx-flanking region of φP27 and the phage morphology were characterized. Furthermore, it showed that only 1 out of 11 Stx2e-producing STEC isolates harbors an infectious Stx2e-encoding phage. From the other isolates no Stx phages could be induced (42).
The aim of the present study was to determine the nucleotide sequence of Stx2e-encoding phage φP27, to describe its genome organization, and to compare it with that of other Stx phages. Furthermore, we were interested in the general genetic localization of stx2e genes in E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Shiga toxin 2e-producing E. coli ONT:H− strain 2771/97 was isolated from a patient with diarrhea and was described previously (42). T9 and T21 are E. coli DH5α transductants lysogenized with φP27. E. coli DH5α (GibcoBRL, Eggenstein, Germany) was used as the host for cloning experiments. Stx2e-producing strains 55/89 (O2:H5), 3054/97 (O60:H2), EH186 (O60:H−), EH60 (O101:H9), 24059/97 (ONT:H−), 3229/98 (O60:H−), and 3357/98 (O60:H−) were isolated from patients with diarrhea; strains 24066/97 (ONT:H−) and 26275/97 (ONT:H−) were from asymptomatic carriers; and strains 2392/98 (O60:H−) and 626/98 (O26:H−) originated from porcine meat. The two strains isolated from the feces of pigs, E57 (O138:K81) and ED-53 (O101:H−), were described earlier (42). Plasmid pK18 was used as a cloning vector (53). Plasmid pIM10 contains functional recA of E. coli (21) and was kindly provided by Gabriele Blum-Oehler and Jörg Hacker, Würzburg, Germany.
Preparation of φP27 phage DNA.
A freshly grown single colony of the lysogen T21(pIM10) was suspended in 200 ml of Luria-Bertani broth and incubated with vigorous shaking until an optical density at 600 nm of 0.5 was reached. After adjusting the culture with mitomycin C (Sigma-Aldrich, Deisenhofen, Germany) to a final concentration of 0.5 μg/ml, the flask with the bacterial suspension was wrapped in aluminum foil and incubated overnight. The phage particles were separated from the cell debris by centrifugation (23,000 × g, 30 min, 4°C) followed by filtration through a funnel filter (Filtrak, Niederschlag, Germany). To remove bacterial nucleic acids, DNase I and RNase A (Sigma-Aldrich) were added to a final concentration of 0.5 μg/ml (each). After an incubation at 37°C for 45 min, sodium chloride was added to a final concentration of 5.8% (wt/vol) and dissolved and the solution was incubated on ice for 1 h. After a centrifugation step (23,000 × g, 10 min, 4°C), the phage particles were precipitated by adjusting the supernatant to 10% polyethylene glycol 8000 (wt/vol). After dissolving the polyethylene glycol 8000 at room temperature, the mixture was incubated on ice for 1 h. Phage particles were harvested by centrifugation (25,000 × g, 30 min, 4°C). The resulting phage pellet was dried at room temperature and dissolved in 1 ml of SM buffer (5.8 g of NaCl, 2 g of MgSO4·7H2O, 50 ml of 1 M Tris-HCl [pH 7.5], and 0.1 g of gelatin in a final volume of 1 liter of double-distilled H2O). To release phage DNA, 1 ml of twofold-concentrated proteinase K buffer (20 ml of 1 M Tris-HCl [pH 8.0], 20 ml of 0.5 M EDTA [pH 8.0], and 100 ml of 10% sodium dodecyl sulfate [wt/vol] in a final volume of 1 liter of double-distilled H2O) and proteinase K (Merck GmbH, Darmstadt, Germany) at a final concentration of 20 mg/ml were added and the suspension was incubated for 1 h at 56°C. The lysate was deproteinized twice by phenol-chloroform (1:1 [vol/vol]) extraction. Then the phage DNA was collected by ethanol precipitation. The resulting pellet was washed with ethanol (70% [vol/vol]) and dried at room temperature. Finally, the DNA was dissolved in 200 μl of double-distilled H2O.
DNA techniques.
DNA sequencing was performed with an automated DNA sequencer (ABI Prism 377; Perkin-Elmer/Applied Biosystems, Weiterstadt, Germany) by using an ABI Prism BigDye Terminator Cycle Sequencing kit (Perkin-Elmer/Applied Biosystems). Random EcoRI, BamHI, SalI, SphI, KpnI, and PstI fragments of φP27 were cloned in pK18, and initial sequence information of these clones was obtained with universal and reverse primers for pUC/M13 vectors. These sequences were used to create customized oligonucleotides for primer walking. Genomic DNA of phage φP27 was used as the template for primer walking.
PCR was conducted with the GeneAmp 9600 PCR System (Perkin-Elmer/Applied Biosystems). Briefly, one freshly grown colony of the respective strain was suspended in 50 μl of 0.85% sodium chloride solution. The amplification reaction was performed in a total volume of 50 μl containing 5 μl of bacterial suspension, 30 pmol of each primer, 5 μl of 10-fold Taq DNA polymerase buffer, 3 μl of MgCl2 stock solution, and 2 U of Amplitaq DNA polymerase (Perkin-Elmer/Applied Biosystems).
Whole bacterial DNA was isolated as described by Heuvelink et al. (31). DNA-DNA hybridization was performed according to the manufacturer's recommendation (Roche Diagnostics, Mannheim, Germany). An int probe was generated by PCR with primers 2771-12 (5′-GAAAACCATCTGGGCAC-3′) and 2771-21 (5′-GCACCACATCAAGGTTC-3′). The generation of PCR-derived hybridization probes was described earlier (56). PCR and sequence analysis of aroE were performed with primers aro+1 (5′-CGGATGAGCTTACTGAAC-3′) and aro-2 (5′-CAATAAATGCCTGGATGAATGAG-3′). Oligonucleotide primers were purchased from Sigma-ARK GmbH, Darmstadt, Germany.
Sequence assembly and analysis.
Sequence data were assembled into contigs by using PREGAP 4 and GAP 4 of the Staden package (65). Searches for open reading frames (ORFs) and predictions of translation start positions were performed online with the WebGeneMark.hmm software (5). Searches for homologous DNA and protein sequences were conducted with the BLAST software (1) against the nonredundant GenBank database (http://www.ncbi.nlm.nih.gov/blast/blast/). Predicted φP27 protein sequences were also compared with Prosite (10) and the Protein Families (Pfam) database for conserved motifs (4). φP27 protein sequences were further compared with proteins found in BLAST searches by using the program GeneDoc, version 2.6.01 (www.psc.edu/biomed/genedoc). Transmembrane regions were predicted with TMpred (32). tRNA genes and tRNA structure were determined with tRNAscan-SE (36). A compendium of online tools at the German Cancer Research Center (DKFZ), Heidelberg, Germany (http://genome.dkfz-heidelberg.de/) was used to perform further DNA analysis.
Nucleotide sequence accession number.
The complete sequence of bacteriophage φP27 has been deposited in the EMBL and GenBank database libraries and given the accession no. AJ298298.
RESULTS
Cloning and sequencing of the φP27 genome.
In order to determine the complete nucleotide sequence of φP27, we isolated phage DNA from the E. coli transductant T21(pIM10) (42). DNA fragments of φP27 restricted with either EcoRI, BamHI, SalI, SphI, KpnI, or PstI were randomly cloned in pK18, and initial sequences were determined by using universal and reverse primers for pUC/M13 vectors. Using this sequence information, walking primers were designed to determine the whole sequence. For this purpose, isolated genomic DNA of φP27 was used as a template.
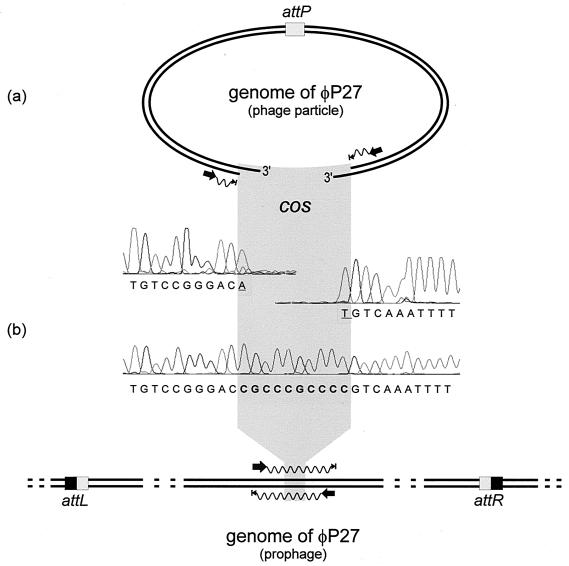
The assembly of all sequence information obtained revealed a linear phage genome and did not result in a circular permutation of the phage sequence. The absence of direct terminal repeats upon sequence assembly indicated that φP27 DNA is restricted by a site-specific mechanism for DNA packaging and not by using a head full mechanism as postulated for E. coli O157:H7 Stx phage 933W (52). In phage λ, site-specific packaging results in the formation of cohesive ends (cos sites) (18). In the prophage state, cos sites are covalently joined together. We used two primers (Fig. 1b) to amplify the region containing the putative cos sites of the φP27 prophage. We compared the DNA sequence of this PCR product with those of the corresponding regions of the DNA isolated from free phage particles (Fig. 1a). From the latter, we revealed runoff sequences where the sequencing reaction stopped abruptly, indicating the end of the template. Alignment of the sequences of the PCR product and phage ends revealed a gap of 10 nucleotides in the latter sequence (Fig. 1). This demonstrates that φP27 possesses cos sites and that these ends contain 3′ single-strand extensions (5′-CGCCCGCCCC-3′).
FIG. 1.
Scheme of the sequencing procedure used for the identification of cos sites in the φP27 genome. (a) Sequencing primer binding sites were selected close to the ends of the linear φP27 genome to obtain runoff sequences. Electropherograms of the runoff sequences are presented. (b) In the upper part, the DNA sequence of the corresponding region sequenced from the prophage is shown. The sequencing procedure is explained in the lower part. Black arrows indicate binding sites and the directions of primers. Waves depict cycle sequence reactions that have been performed directly from the phage DNA template (a) and from a PCR product (b). Arrowheads with bars indicate the stops of sequencing reactions. The phage attachment site (attP) and the attL and attR sites generated by insertion of the phage are highlighted by boxes. Underlined nucleotides were added by the Taq polymerase independent of the template.
Determination of the chromosomal phage integration site.
In order to identify the integration site of φP27 in the bacterial chromosome, we looked for a DNA fragment at the prophage-host chromosome border containing both phage DNA and bacterial DNA. Integration of lambdoid phages into the bacterial chromosome generally occurs by site-specific recombination between the phage attP and the bacterial attB sites (12). As a result of integration, the prophage is flanked by short direct repeats (identity segments of attL and attR), separating both phage and host chromosome. One of the phage-host border sequences is generally neighbored by a phage integrase (int) gene (11).
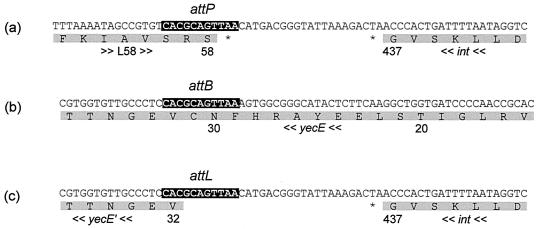
We restricted bacterial DNA of E. coli 2771/97 and phage DNA of φP27 with EcoRV and hybridized the restriction fragments with a probe specific for the φP27 integrase gene (int). This revealed a fragment of 2.2 kb in the genomic DNA of E. coli 2771/97 and a 3.0-kb fragment in the DNA isolated from phage particles. Due to the integration event, changes of restriction fragment lengths have been expected. We performed an inverse PCR approach for further analysis. Chromosomal DNA of E. coli 2771/97 was digested with EcoRV, and restriction fragments were treated with T4 DNA ligase and used as templates for PCR. Two primers were designed, which are orientated in opposite directions outwards the integrase gene. This revealed a PCR product with the expected length of 2.2 kb which was subsequently sequenced. By sequence analysis of the PCR product, we identified the phage integrase gene close to a gene identical to chromosomal E. coli yecE (Fig. 2). The prophage is integrated in the 5′ region of the yecE gene. According to WebGeneMark.hmm, the phage provides yecE with a new translational start. This putatively leads to the production of a truncated YecE, missing its 31 N-terminal amino acids.
FIG. 2.
Determination of the phage integration site. (a) The attP site of φP27 is depicted, including parts of ORF L58 and int. (b) The corresponding attB site in the chromosomal yecE gene is shown. (c) Depiction of the DNA sequence of one of the borders of the integrated prophage. According to WebGeneMark.hmm, integration of the prophage into yecE leads to a truncated protein. The upper lines are the nucleotide sequences, and the lower lines are the amino acid sequences. The numbers below the amino acid sequence mark the amino acid positions of the respective protein. yecE′ labels the truncated amino acid sequence, asterisks indicate stop codons, and arrows indicate the transcription orientation. Black boxes in the nucleotide sequences indicate the identity segment of attachment sites.
For characterization of the second host-phage border, we designed a PCR with one primer specific for chromosomal yecD and the other one for phage ORF L57 (see below). We revealed a PCR product with a size of 550 bp containing a part of ORF L57 and complete L58, the remaining part of yecE, and the 3′ region of yecD. This confirmed that the chromosomal integration site of φP27 is the yecE gene. We were able to determine the identity segments of the attL and attR sites with the sequence 5′-CACGCAGTTAA-3′ (Fig. 2).
The yecE gene has not been reported as an integration site for phages before. yecE is located in a gene cluster flanking the aspartate tRNA ligase (aspS) gene in E. coli K-12 and E. coli O157 (6, 49). The gene order is aspS, yecD, yecE, yecN, yecO, yecP, and bisZ, with the latter encoding a biotin sulfoxide reductase 2 (6). For the yec genes, no functions have yet been described. However, orthologous genes have been annotated in Salmonella enterica serovar Typhimurium and Yersinia pestis, and also the deduced amino acid sequence is significantly homologous to a putative cytoplasmic protein found in S. enterica serovar Typhi, Y. pestis, and Vibrio cholerae El Tor (27, 45, 46). YecE contains sequence motifs which categorize this protein as a member of the Pfam family DUF72 (4), which includes proteins of Bacillus subtilis and some members of the Archaea.
DNA characteristics and search for ORFs.
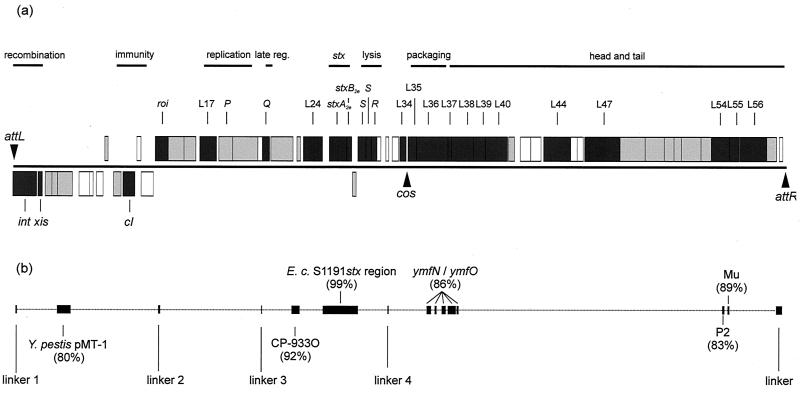
The DNA sequence of the φP27 prophage consists of 42,575 bp starting with attL and ending with attR (Fig. 3; Table 1). The overall G+C content of the φP27 genome is 49.3 mol%.
FIG. 3.
Schematic illustration of the φP27 genome. (a) Boxes depict ORFs determined by WebGenemark.hmm. ORFs directed in rightward orientation are drawn above the black line; ORFs directed in the leftward direction are drawn below the line. Boxes in dark gray represent ORFs encoding proteins described in Results. Boxes in light gray mark ORFs, the deduced proteins of which show homology to known proteins which are described only in Table 1. Empty boxes label ORFs encoding proteins without known homologies. (b) Black boxes and vertical bars represent regions with nucleotide sequence homologies to other sequences, including linkers. Homology values are depicted by boxes. late reg., late regulation.
TABLE 1.
Characteristics of φP27 ORFs and deduced proteins
| ORF label | Gene name | Directionb | Left end position no. | Right end position no. | No. of amino acids | Function of deduced protein | e value | % Identitya | Accession no. and sequence similarityc |
|---|---|---|---|---|---|---|---|---|---|
| attL | 1 | 11 | |||||||
| L01 | int | − | 30 | 1343 | 437 | Integrase | 0.0 | 70 (All) | BAB19626; putative integrase, prophage VT1-Sa (E. coli) |
| 0.0 | 70 (All) | AAG57258; putative integrase, prophage CP-933V (E. coli) | |||||||
| L02 | xis | − | 1399 | 1635 | 78 | Excisionase | 3e−16 | 53 (66) | NP_049462; putative excisionase, phage 933W (E. coli) |
| 3e−16 | 53 (66) | NP_050501; putative excisionase, prophage VT2-Sa (E. coli) | |||||||
| L03 | − | 1794 | 2159 | 121 | 1e−22 | 63 (80) | BAB36434; hypothetical protein (E. coli RIMD 0509952) | ||
| 2e−13 | 60 (60) | AAG57256; hypothetical protein, prophage CP-933V (E. coli) | |||||||
| L04 | − | 2068 | 2511 | 147 | 7e−04 | 34 (100) | NP_050134; hypothetical protein, prophage φadh (Lactobacillus gasseri) | ||
| L05 | − | 2480 | 3307 | 275 | Protease | e−124 | 79 (All) | NP_047914; putative serine protease (Y. pestis) | |
| L06 | − | 3669 | 4307 | 212 | |||||
| L07 | − | 4247 | 4450 | 67 | |||||
| L08 | − | 4632 | 4997 | 121 | |||||
| L09 | + | 5106 | 5285 | 59 | 5e−06 | 78 (28) | AAG55461; hypothetical protein, prophage CP-933M (E. coli) | ||
| L10 | − | 5575 | 5991 | 138 | Repressor | 1e−06 | 37 (69) | AAC74643; DicA repressor (E. coli) | |
| L11 | cI | − | 6095 | 6751 | 218 | CI repressor | 1e−45 | 42 (All) | NP_059606; repressor C2, phage P22 (S. enterica) |
| L12 | + | 6863 | 7090 | 75 | |||||
| L13 | − | 7104 | 7775 | 223 | |||||
| L14 | roi | + | 7876 | 8583 | 235 | DNA binding | 1e−32 | 55 (136) | NP_112063; DNA-binding protein Roi, phage HK620 (E. coli) |
| 5e−32 | 52 (140) | AAD04652; Roi, phage H-19B (E. coli) | |||||||
| L15 | + | 8636 | 9460 | 274 | Nucleic acid binding | 5e−20 | 31 (194) | B82549; hypothetical protein XF2506 (X. fastidiosa) | |
| 5e−14 | 33 (127) | AAG55918; putative antirepressor, prophage CP-933N (E. coli) | |||||||
| 8e−14 | 38 (118) | NP_046925; antirepressor AntB, phage N15 (E. coli) | |||||||
| L16 | + | 9457 | 10116 | 219 | 9e−31 | 48 (153) | S34345; hypothetical protein 179 (Shigella flexneri) | ||
| 1e−12 | 43 (112) | AAG54596; putative regulator, prophage CP-933I (E. coli) | |||||||
| L17 | + | 10334 | 11260 | 308 | DNA binding | 9e−31 | 42 (196) | AAK16983; hypothetical protein, prophage CP-933P (E. coli) | |
| 2e−28 | 33 (227) | AAG55470; hypothetical protein, prophage CP-933M (E. coli) | |||||||
| L18 | P | + | 11391 | 12149 | 252 | DNA replication | 2e−24 | 31 (207) | BAB12748; DNA replication protein DnaC (Buchnera aphidicola) |
| 1e−21 | 27 (210) | AAG55471; DNA replication factor, prophage CP-933M (E. coli) | |||||||
| L19 | + | 12146 | 13537 | 463 | Helicase | 6e−87 | 39 (447) | NP_059611; helicase, phage P22 (S. enterica) | |
| 2e−86 | 39 (451) | NP_037740; gp55, prophage HK97 (E. coli) | |||||||
| L20 | + | 13549 | 13791 | 80 | |||||
| L21 | Q | + | 13791 | 14171 | 126 | Late antiterminator | 6e−25 | 50 (119) | AAG55890; Q antiterminator, prophage CP-933N (E. coli) |
| 7e−24 | 47 (120) | O48429; antitermination protein Q, prophage H-19B (E. coli) | |||||||
| 3e−23 | 44 (126) | CAB39299; antitermination protein Q, phage 933W (E. coli) | |||||||
| L22 | + | 14266 | 15495 | 409 | 6e−04 | 32 (127) | CAA22431; putative chromatin assembly factor (Schizosaccharomyces pombe) | ||
| L23 | + | 15711 | 15908 | 65 | 5e−29 | 98 (63) | AAG56133; hypothetical protein, prophage CP-933O (E. coli) | ||
| 4e−28 | 96 (63) | BAB35615; hypothetical protein (E. coli RIMD 0509952) | |||||||
| 1e−19 | 97 (49) | AAG55892; hypothetical protein, prophage CP-933N (E. coli) | |||||||
| L24 | dam | + | 16059 | 17117 | 352 | DNA methylase | 0.0 | 86 (All) | AAG56134; adenine methyltransferase, prophage CP-933O (E. coli) |
| 0.0 | 86 (All) | BAB35203; DNA methylase (E. coli RIMD 0509952) | |||||||
| 2e−81 | 55 (282) | NP_046948; adenine-specific methylase, phage N15 (E. coli) | |||||||
| ileZ | + | 17158 | 17233 | tRNA | |||||
| arqO | + | 17335 | 17411 | tRNA | |||||
| L25 | stxA2e | + | 17502 | 18461 | 319 | rRNA N-glycosidase | 0.0 | 99 (All) | CAA57173; Stx2e, A subunit (E. coli) |
| L26 | stxB2e | + | 18474 | 18737 | 87 | Receptor binding | 3e−44 | 100 (All) | CAA57176; Stx2e, B subunit (E. coli) |
| L27 | − | 18788 | 18985 | 65 | 2e−28 | 96 (61) | CAC05562; hypothetical protein (E. coli T4/97) | ||
| 5e−25 | 96 (55) | CAC05572; hypothetical protein (E. coli H.I.8) | |||||||
| L28 | S | + | 19114 | 19548 | 144 | Holin | 8e−06 | 36 (86) | AAF80841; ORF 89, prophage D3 (Pseudomonas aeruginosa) |
| 0.002 | 28 (123) | BAA36235; holin ORF 9, phage φCTX (P. aeruginosa) | |||||||
| L29 | S | + | 19538 | 19813 | 91 | Holin | 0.005 | 34 (85) | H83531; hypothetical protein PA0909 (P. aeruginosa) |
| 0.049 | 32 (86) | BAA36236; holin ORF 10, phage φCTX (P. aeruginosa) | |||||||
| L30 | R | + | 19816 | 20193 | 125 | Endolysin | 3e−25 | 48 (All) | CAA09701; endolysin gp 19, phage PS3 (S. enterica serovar Typhimurium) |
| 3e−12 | 38 (119) | AAC38580; peptidoglycan lytic enzyme (Listeria monocytogenes) | |||||||
| L31 | + | 20136 | 20351 | 71 | |||||
| L32 | + | 20630 | 20785 | 51 | |||||
| L33 | + | 20985 | 21332 | 115 | |||||
| L34 | + | 21394 | 21744 | 116 | 1e−37 | 62 (All) | CAB58450; hypothetical protein ORF 7 (Xenorhabdus nematophilus) | ||
| 2e−11 | 39 (111) | AAB59284; putative holin, prophage φ105 (B. subtilis) | |||||||
| cos | 21806 | 21815 | |||||||
| L35 | + | 21860 | 22330 | 156 | Terminase small subunit | 2e−11 | 31 (138) | AAG50266; hypothetical protein, phage GMSE-1 (Sodalis) | |
| 2e−07 | 31 (96) | AAG55950; hypothetical protein, prophage CP-933C (E. coli) | |||||||
| 2e−07 | 31 (96) | BAB35020; terminase small subunit (E. coli RIMD 0509952) | |||||||
| L36 | + | 22345 | 24057 | 570 | Terminase large subunit | e−178 | 77 (293) | P75978; hypothetical protein YmfN (E. coli) | |
| e−141 | 45 (All) | NP_061498; terminase, phage D3 (P. aeruginosa) | |||||||
| L37 | + | 24069 | 24251 | 60 | 2e−27 | 95 (All) | P75979; hypothetical protein YmfR (E. coli) | ||
| L38 | + | 24251 | 25492 | 413 | Portal protein | 5e−63 | 84 (135) | P75980; hypothetical protein YmfO (E. coli) | |
| 4e−52 | 33 (372) | NP_108600; head portal protein (M. loti) | |||||||
| 1e−44 | 32 (375) | NP_037699; portal protein, phage HK97 (E. coli) | |||||||
| L39 | + | 25470 | 26120 | 216 | Prohead protease | 2e−24 | 40 (172) | AAF13181; putative prohead protease (Rhodobacter capsulatus) | |
| 2e−18 | 35 (196) | NP_037665; head maturation protease, prophage HK022 (E. coli) | |||||||
| L40 | + | 26134 | 27357 | 407 | Major capsid protein | 2e−73 | 36 (All) | NP_108602; phage major capsid protein gp36 (M. loti) | |
| 2e−58 | 34 (All) | AAF27364; phage φC31 gp36-like protein (H. influenzae) | |||||||
| L41 | + | 27404 | 27727 | 107 | 6e−04 | 29 (104) | AAG55948; hypothetical protein, prophage CP-933C (E. coli) | ||
| L42 | + | 28067 | 28630 | 187 | |||||
| L43 | + | 28627 | 29190 | 187 | |||||
| L44 | + | 29358 | 30854 | 498 | Tail sheath protein | 5e−78 | 35 (All) | P44233; putative tail sheath protein, prophage FLUMU (H. influenzae) | |
| 7e−75 | 35 (All) | NP_050643; sheath protein gpL, phage Mu (E. coli) | |||||||
| L45 | + | 30854 | 31210 | 118 | |||||
| L46 | + | 31210 | 31539 | 109 | |||||
| L47 | + | 31624 | 33573 | 649 | Tail length determinator | 9e−41 | 26 (493) | F82769; tail protein XF0730 (X. fastidiosa) | |
| 3e−13 | 22 (473) | NP_046782; putative tail length determinator gpT, phage P2 (E. coli) | |||||||
| L48 | + | 33589 | 34980 | 463 | DNA binding | 1e−23 | 26 (450) | P71389; DNA circulation protein, prophage FLUMU (H. influenzae) | |
| 3e−8 | 23 (479) | NP_050647; DNA circulation protein N, phage Mu (E. coli) | |||||||
| L49 | + | 34977 | 36032 | 351 | 4e−40 | 30 (All) | NP_050648; tail protein P, phage Mu (E. coli) | ||
| L50 | + | 36032 | 36565 | 177 | Baseplate assembly | 3e−12 | 31 (170) | AAF41502; putative baseplate assembly protein V (Neisseria meningitidis) | |
| 3e−11 | 26 (174) | NP_050649; baseplate assembly protein gp45, phage Mu (E. coli) | |||||||
| L51 | + | 36571 | 36984 | 137 | 4e−14 | 40 (130) | BAB38409; hypothetical protein (E. coli RIMD 0509952) | ||
| 8e−11 | 41 (112) | NP_050650; gp46, phage Mu (E. coli) | |||||||
| L52 | + | 36977 | 38059 | 360 | 3e−78 | 55 (260) | P75981; hypothetical protein YmfP, prophage E14 (E. coli) | ||
| 1e−29 | 31 (330) | NP_050651; gp47, phage Mu (E. coli) | |||||||
| L53 | + | 38059 | 38649 | 196 | 2e−59 | 55 (194) | P75982; hypothetical protein YmfQ, prophage E14 (E. coli) | ||
| 1e−05 | 26 (182) | NP_050652; gp48, phage Mu (E. coli) | |||||||
| L54 | + | 38636 | 39634 | 332 | Tail fiber protein | 2e−33 | 39 (247) | A42463; hypothetical protein Bcv (Shigella boydii) | |
| 7e−20 | 31 (305) | NP_046775; putative tail fiber protein gpH, phage P2 (E. coli) | |||||||
| L55 | + | 39637 | 40185 | 182 | Tail fiber assembly | 3e−37 | 40 (All) | C42463; hypothetical protein B177 (S. boydii) | |
| 2e−29 | 36 (All) | NP_050654; tail fiber assembly protein U, phage Mu (E. coli) | |||||||
| L56 | + | 40209 | 41681 | 490 | Tail fiber protein | 2e−20 | 32 (235) | BAB35654; putative tail fiber protein (E. coli RIMD 0509952) | |
| 3e−20 | 32 (235) | AAK16943; putative tail fiber protein, prophage CP-933P (E. coli) | |||||||
| L57 | + | 41678 | 42244 | 188 | 5e−29 | 35 (All) | AAF63233; ORF 191A, prophage P-EibA (E. coli) | ||
| L58 | + | 42399 | 42575 | 58 | |||||
| attR | 42565 | 42575 |
Numbers in parentheses represent the whole number of amino acids from which the sequence identity is calculated. All, whole length identity. Empty fields in the table indicate that no homologous sequences were available.
−, lower strand; +, upper strand.
The GenBank database was used for homolog searches.
A search for ORFs with WebGeneMark.hmm revealed 58 ORFs larger than 150 nucleotides. We labeled these ORFs consecutively from L01 to L58. The characteristics of these ORFs and their corresponding predicted proteins are described in Table 1. BlastP searches in the EMBL and GenBank databases with the deduced proteins as query sequences revealed significant similarities (expect [e] values of <10−3) for 44 ORFs. For the remaining 14 ORFs, low or no homologies were detected. In a total of 29 cases, BlastP results suggested a possible function for the putative φP27 proteins (Table 1).
Description of selected ORFs.
The deduced protein of L01 is closely related to members of the integrase-recombinase family (3). The highest similarities are found to putative integrases of Stx-converting phages VT1-Sakai (74), CP-933V (49), 933W (52), and VT2-Sakai (37) (Table 1). The deduced amino acid sequence of L02 demonstrates significant sequence similarity to the putative excisionases of phages 933W and VT2-Sakai (Table 1). The protein deduced from ORF L11 is homologous to various repressor proteins of the λ CI type (54). The most related protein is the corresponding regulator of phage P22, there termed C2 (71). The λ CI repressor inhibits almost all phage gene expression by binding to DNA operator sequences. For this purpose, CI proteins of many bacteriophages contain strong helix-turn-helix (HTH) motifs in their amino-terminal domains (24). Analysis of the deduced amino acid sequence of L11 with the program of Dodd and Egan (17) indicated a strong HTH motif starting at position 17. Similar to the situation in λ, the putative cI gene of φP27 is flanked by putative operator sequences OL and OR on both sides. Each of the operators consists of three adjacent suboperators with imperfect dyad symmetry (OL1, bp 5997-6010; OL2, bp 6026-6039; OL3, bp 6061-6074; OR3, bp 6776-6789; OR2, bp 6800-6813; OR1 bp 6832-6845).
The L14 protein is highly homologous to the Roi proteins of bacteriophages HK620, H-19B, VT2-Sa, HK022, HK97, and 933W (16, 33, 37, 43, 52). In HK022, Roi causes phage propagation dependent on the integration host factor (IHF). Roi binds to a sequence within its own coding gene (14), which is closely linked to an IHF binding site. The search with an IHF binding consensus sequence (5′-AATCAANNNNTTA-3′) revealed a putative binding site within the L14 coding sequence (base pair positions 8028 to 8040).
DNA replication in λ involves a replication complex including phage proteins gp O and gp P and a variety of host proteins, including primase (DnaG) and helicase (DnaB) (69). Phage λ contains four 20-bp direct repeat sequences in the coding region of the O gene. These repeats are recognized by the O protein to initiate replication (22). In the O gene coding region of phage VT2-Sakai, two 30-bp direct repeats were identified which are thought to be involved in the initiation of replication in this phage (37). The search of direct repeats in the φP27 region associated with phage replication revealed two 17-bp direct repeats (5′-AAAACATGATCCGCAAG-3′) which are located in ORF L17 at positions 10816 to 10832 and 10840 to 10856, respectively. The N-terminal part of the L17 protein contains a putative HTH motif (similar to the SMART family HTH_GNTR), and this region is homologous to a putative replication protein (BAB35699) from E. coli O157:H7 (26). These findings together with the occurrence of a direct repeat let us suggest that the gene product of L17 might be involved in φP27 DNA replication.
L21 encodes a protein homologous to Q-like proteins of phages CP-933N, APSE-1, H-19B, and 933W (Table 1) (43, 49, 52, 72). In λ, the Q gene encodes a product of the delayed early region. Q functions as a transcription antiterminator that regulates expression of late-phase genes by modifying transcription complexes by binding to a DNA sequence (qut) near a late promoter pR′ (20).
The region from L22 to L31 of the φP27 genome was already the subject of a recent publication and was described previously in detail (42). It includes the genes encoding an adenine-specific DNA methylase (L24), the Stx2e subunits (L25 and L26), and a lysis cassette including two holins (L28 and L29) and an endolysin (L30).
The L34 protein demonstrates weak homology to a putative holin of Bacillus phage φ105 (Table 1).
We have shown that φP27 creates cohesive ends during DNA packaging and that these cos sites extend from base pair positions 21806 to 21815 in the prophage. The two ORFs following downstream can be the φP27 terminase components for the small (L35) and the large (L36) subunits according to their location and homology search results. In lambdoid phages, the terminase complex interacts with the portal of the prohead, consisting of an oligomer of the portal protein, for example, 12 subunits in phage HK97 (29). According to Blast results, L38 may be the portal protein of phage φP27. Once packaging is complete, another protein interacts with the DNA-filled head in order to complete the head. In λ, one of these is gpW, a small (68 amino acids) highly positive charged protein with an isoelectric point (pI) of 10.8, forming either a plug at the connector to prevent ejection of the DNA, or binding directly to the DNA (50). In the φP27 genome, we detected a corresponding ORF (L37), putatively encoding a small protein (60 amino acids) with a calculated pI value of 9.6 and thereby comparable to the pI of gpW. Despite lacking overall sequence similarity to gpW-like proteins, L37 could be the head-tail joining protein of φP27.
The deduced protein of ORF L40 shows homology to the major capsid proteins of phages found in gram-negative and -positive bacterial species such as Mesorhizobium loti, Caulobacter crescentus (AAK24747), Haemophilus influenzae, E. coli (AAG55944) and Streptomyces spp. (NP_047927). The deduced protein of L39 is homologous to the head maturation proteases of HK022 and HK97 (33), which are necessary to cleave the N termini from assembled head proteins.
L44 encodes a protein which is homologous to the tail sheath protein of phages FLUMU and Mu (Table 1) (68). Phages with contractile tails contain such sheath proteins. The existence of a tail sheath protein in φP27 is indicative of the presence of a contractile tail. This is in contrast to most other lambdoid phages, where flexible noncontractile tails are common. None of the other Shiga toxin-converting phages investigated so far contains such a sheath protein.
The protein determining the length of the tail in phage λ is the tape measure protein. By deletion or formation of small duplications of genes encoding tape measure proteins of phage λ (28, 34, 35) and lactococcal phage TP901-1 (48), it was shown that the length of the phage tail was proportional to the size of the respective tape measure protein. The L47 protein demonstrates homology to the tape measure protein of a phage detected in Xylella fastidiosa (Table 1). In phage TP901-1, a deletion mutant with a reduction in the length of the tape measure protein shows also a comparative reduction of the tail length (48). With a length of 649 amino acids, the deduced tape measure protein of φP27 has about the same length as the deletion product in TP901-1 and the tail of φP27 has the corresponding length of about 95 nm.
According to BlastP results, the φP27 ORF L54 encodes long side tail fibers and L55, a protein putatively needed for the assembly of the fibers (Table 1). Interestingly, the deduced protein of ORF L56 also demonstrates sequence similarity to phage tail fiber proteins. The sequence similarity is located in the N-terminal part of the L56 protein, and the remaining part is not homologous to known proteins. This raises the question of whether φP27 has two sets of tail fibers to broaden its host range as discussed for the phage K1-5 (59).
Genome structure.
The overall genetic organization of phage φP27 is similar to that of other lambdoid phages. The left-most part of φP27 is devoted to site-specific integration and excision (Fig. 3; Table 1). Downstream there is a region putatively responsible for immunity. Following a cluster of genes with proposed functions in phage replication, the late-phase region starts with a gene encoding a Q-like antiterminator. A lysis cassette and genes for DNA packaging, as well as head and tail genes, are located downstream of the antiterminator gene (Fig. 3). The genes encoding Stx2e are located between the genes encoding the late antiterminator Q and the lysis cassette. Most of the deduced proteins of ORFs L44 to L55 demonstrate homology to proteins of phage Mu and show the same gene order (data not shown). With the exception of the area encoding Stx2e, the φP27 genome has only low-level sequence homology to those of other phages. However, five short sequences attracted attention because they could be detected with high sequence identities in other lambdoid phages. These regions range from 22 to 151 bp, and we refer to them as linkers 1 to 5 (Table 2; Fig. 3).
TABLE 2.
Length and location of linker sequences of φP27 and comparison with those of other bacteriophages
| Phage | No. of identical base pairs/total base pairs in linker (position)a:
|
||||
|---|---|---|---|---|---|
| 1 (145-266) | 2 (8070-8169) | 3 (3787-13834) | 4 (20872-20897) | 5 (42246-42575) | |
| CP-933V | 89/109 (81) | 22/22 (100) | 85/95 (89) | ||
| Sa-VT1 | 89/109 (81) | 22/22 (100) | 85/95 (89) | ||
| Sa-VT2 | 41/47 (87) | 38/41 (92) | 41/47 (87) | 22/22 (100) | |
| 31/35 (88) | |||||
| 933V | 41/47 (87) | 37/41 (90) | 41/47 (87) | 22/22 (100) | |
| CP-933O | 61/74 (82) | 24/25 (96) | 84/92 (91) | ||
| HK620 | 70/83 (84) | ||||
| H19-B | 70/83 (84) | 41/47 (87) | 24/24 (100) | ||
| APSE-1 | 81/99 (81) | ||||
| HK022 | 33/35 (94) | ||||
| HK97 | 34/38 (89) | 81/87 (93) | |||
| P22 | 25/26 (96) | ||||
| CP-933C | 27/29 (93) | ||||
| CP-933N | 39/42 (92) | 25/25 (100) | |||
| CP-933U | 25/25 (100) | ||||
| CP-933M | 25/25 (100) | ||||
| phage 7888 | 25/25 (100) | ||||
| CP-933R | 24/24 (100) | 72/81 (88) | |||
| CP-933P | 130/151 (86) | ||||
| 45/50 (90) | |||||
| λ | 64/72 (88) | ||||
| phage P-EibC | 75/89 (84) | ||||
Linker positions are base pair positions in the φP27 sequence. Values in parentheses are percentages of overall sequence identity.
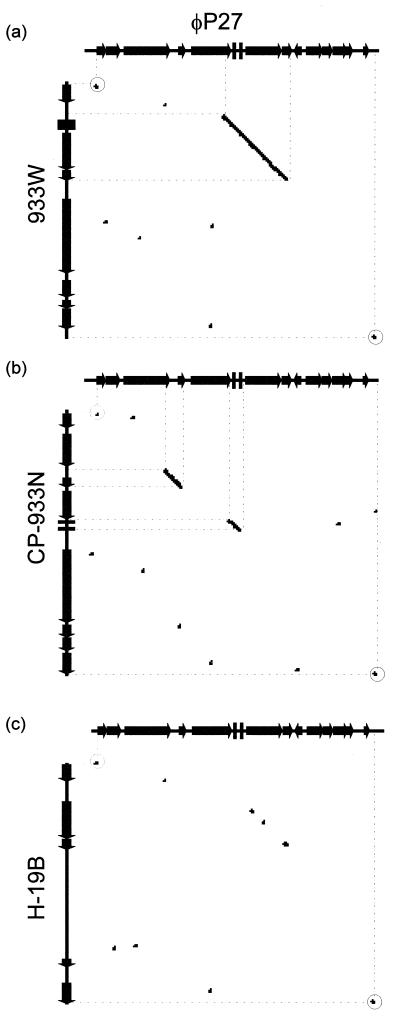
We performed a detailed inspection of the region located between linkers 3 and 4 by investigating the homology to the corresponding regions of Stx phages 933W (stx2) and H19-B (stx1) and the Stx-negative phage CP-933N. This region starts at the beginning of gene Q and ends directly behind the phage lysis cassette. We performed a DNA homology dot plot analysis with the Staden software package (Fig. 4). A comparison of φP27 and 933W (Fig. 4a) demonstrates regions of high homology exclusively in the linker sequences (Fig. 4) and in the Shiga toxin region, including the preceding tRNA genes. Comparison of φP27 and H-19B (Fig. 4c) demonstrates only low homologies in the stx region. Whereas stx2 (933W) and stx1 (H19-B) share approximately 59% overall nucleotide sequence identity, stx2 (933W) and stx2e (φP27) share 90% overall identity and stx1 and stx2e share 60% overall identity. Interestingly, Stx-negative phage CP-933N (Fig. 4b) and φP27 contain two regions of higher homology. The first one comprises the tRNA genes, and the second one comprises L23 (Fig. 4).
FIG. 4.
Dot blot matrix of the stx2e-flanking region of φP27 (genes Q to L32) and corresponding regions of phages 933W (a), CP-933N (b), and H-19B (c). At every point in the matrix where the two sequences are identical a dot is placed. A diagonal stretch of dots indicates regions where the two sequences are similar. Dots with circles represent linkers 3 (left side) and 4 (right side).
Localization of stx2e genes in Stx2e-producing E. coli strains.
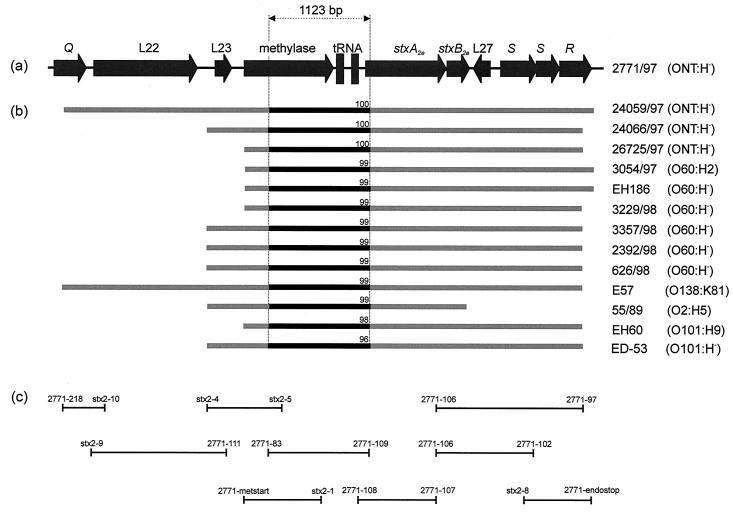
In a previous work, Muniesa et al. isolated φP27 from STEC 2771/97, described its stx-flanking region, and demonstrated that only 2771/97, out of 11 Stx2e-producing E. coli strains, harbored an inducible Stx2e phage (42). Here we used the sequence information obtained from φP27 and performed PCR and sequencing approaches to characterize the stx2e-flanking regions of the 10 remaining strains mentioned above and three additional stx2e-positive E. coli strains.
Seventeen PCR primers (Table 3) were used in multiple PCR approaches to amplify and analyze the region between Q and R of all strains included in this study. This strategy is depicted in Fig. 5. To confirm the identity of the PCR products, initial sequencing was performed with the same primers as employed for PCR. In 12 of 13 strains, we found an association between stx2e and R. Interestingly, in only 2 of 13 strains, stx2e was linked to φP27 Q-like sequences. In all strains, φP27-like sequences extend from stx2e to the methylase gene (L24).
TABLE 3.
Oligonucleotide primers used for characterization of stx2e-flanking regions
| Primer designation | Primer sequence (5′-3′) |
|---|---|
| 2771-218 | AGTGATGATGACGGACTG |
| stx2-10 | CACTTTTCACGCAACCATAAC |
| stx2-9 | AGGTATTTCTATGGGCGG |
| 2771-111 | GCCAGACCCGCCATAAC |
| 2771-83 | GAACGCTTTAACGTGCTG |
| 2771-109 | CGAAGAAAAACCCAGTAACAG |
| stx2-4 | CAAAAAGCGGCACAGGAC |
| stx2-5 | CCCTTTGCCGCATAGCC |
| 2771-metstart | ATGGCTAACACTGTAAAAATA |
| stx2-1 | CCCCATGAAAAAATCTGCAAC |
| 2771-108 | CCTTCTAAGCAATCGGTC |
| 2771-107 | GATGGCAATTCAGTATAACGG |
| 2771-106 | GAGTCAACCAGAATGTCAG |
| 2771-97 | CCTGCTTAAATGCCTGTG |
| 2771-102 | CTTAACATTGCCGCCTCC |
| stx2-8 | CAGGGTTTGACAGTGGG |
| 2771-endostop | TCACCACTTCAACTGAAAGTG |
FIG. 5.
Schematic illustration of the PCR and sequencing approach used for characterization of stx2e-flanking regions of different STEC isolates. (a) Map of the stx2e-flanking region of φP27 spanning from Q to R. (b) Bars in light gray indicate the regions analyzed by PCR and initial sequencing with the same primers as employed for PCR. Black bars between dotted vertical lines depict regions which have been completely sequenced and used for the phylogenetic analysis depicted in Fig. 6. Numbers on black bars show percent DNA identity of the fragments compared to the corresponding fragment of φP27. (c) Size and location of PCR products and applied primers.
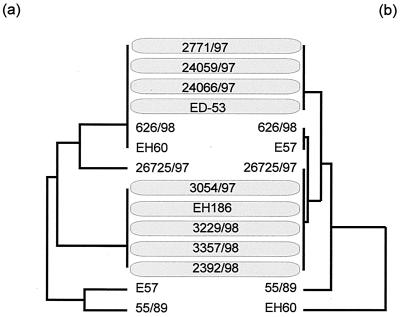
We determined the sequence of a 1,123-bp fragment of the PCR products described above (Fig. 5). A comparison of these sequences with the corresponding φP27 segment demonstrated a high degree of homology which is depicted in Fig. 5. To investigate clonal relationships among the strains, we performed a phylogenetic tree analysis. The same was performed for a host housekeeping gene. A 415-bp fragment of aroE which encodes the shikimate 5-dehydrogenase was amplified (2), sequenced, and analyzed from all investigated strains. The phylogenetic trees constructed from phage sequences and aroE sequences are similar (Fig. 6) and distribute most of the investigated strains in two large clusters. These clusters are serotype dependent. Whereas cluster 1 contains human isolates with serotype ONT:H− and one isolate with serotype O101:H−, cluster 2 mostly contains isolates of serotype O60:H−. This observation let us suggest that phages and housekeeping genes have undergone a coevolution and indicates an uptake of the Stx2e phages early in the evolution of this particular STEC group.
FIG. 6.
Phylogenetic analyses and comparison of sequences obtained from the 1,123-bp 5′ stx2e-flanking region and the housekeeping gene aroE from particular STEC isolates. We used GrowTree (HUSAR software package) to create a phylogenetic tree from a distance matrix created by the program by using the unweighted pair group method with arithmetic averages. The left side contains aroE sequences (a), and the right side contains phage sequences (b).
DISCUSSION
STEC comprises a heterogeneous group of pathogenic bacteria capable of producing potent cytotoxins which cause necrosis and apoptosis of eukaryotic target cells (67). STEC isolated from humans is found in more than 60 E. coli serogroups, and such isolates contain, besides stx, a diverse array of accessory virulence genes (9, 56). Most of the determinants responsible for pathogenicity and virulence of STEC are located on mobile genetic elements such as plasmids, pathogenicity islands, and bacteriophages (8, 9, 25, 60). Whereas the stx genes of some early STEC isolates have been shown to be carried in the genome of some lambdoid bacteriophages (60, 63), recent studies indicate that stx is generally phage borne (41, 44, 55, 62, 70). It could also be shown that the stx-flanking regions of Stx phages from different STEC isolates are heterogeneous in sequence (70).
Here we present the complete genome sequence of Stx2e-encoding phage φP27 and compare it with that of other lambdoid phages. Albeit the homology to other phages is low on DNA sequence level, the functional structure of lambdoid phages is conserved in φP27.
Bacteriophage φP27 contains a small genome. It consists of only 42,575 bp of DNA compared with 48,502 bp of phage lambda (58 versus 66 ORFs, respectively). There are some major differences in the gene composition of both phages. The longest ORF in φP27 comprises 1,950 bp, whereas the longest λ ORF consists of 3,399 bp. However, the average ORF length of 662 to 664 bp is similar in both phage genomes. It appears that φP27 lacks some genes that are present in λ. Whereas 30 genes are located between int and Q in phage λ, φP27 harbors only 18 ORFs in this region. This region is approximately 3 kb shorter than the corresponding λ region. We have not been able to find nin-like or ren-like genes as well as an N-like gene in φP27. Also, the region between the endolysin genes and int, which contains the machinery for head and tail structures, is shorter in φP27 (20,760 bp) than in λ (approximately 27,000 bp). However, the region including Q and the endolysin gene is 6,402 bp in φP27 and only 2,541 bp in λ. Although the overall genetic structure is conserved in both phages, they differ in the presence or absence of particular genes or gene groups.
Lambdoid bacteriophages are composed of functional genetic modules which carry the information for recombination, replication, host lysis, head and tail proteins, and assembly of phage particles (11). These genetic modules can be exchanged between lambdoid bacteriophages by homologous or illegitimate recombination. Hendrix et al. (30) have hypothesized that all lambdoid bacteriophages existing in nature in different habitats may serve as a gene pool for the development of new bacteriophages. Lambdoid phages appear to be genetic mosaics with different members sharing different sets of genes (7, 11, 66). The genome of lambdoid phages has been divided into 11 major segments of functionally clustered genes (13). Thus, they have a modular type of genomic organization. The order of these modules is identical in different lambdoid phages, but the corresponding module-encoded proteins can be heterogeneous.
We detected five short sequences in the range from 22 to 151 bp in the φP27 genome which were also found in other phages. Casjens et al. (13) suggested that such short regions of homology (e.g., conserved sites such as promoters or terminators) are recombination hot spots. According to Shen and Huang (61), for homologous regions smaller than 23 to 27 bp, recombination becomes inefficient. Thus, the five DNA segments of φP27 regions, referred to as linkers 1 to 5, may be large enough to be involved in homologous recombination between different phages. A similar phenomenon has been suggested for lactococcal bacteriophages (7). Four of the linkers are located between functional modules, at sites where recombination between two phages would make sense in order to move whole functional modules. A new phage arising after exchange of complete functional modules is likely to form a viable phage: interacting proteins are transmitted together and genes that encode DNA binding proteins remain linked with the DNA elements they are adapted to. For example, a recombination event at linker 3 would transfer the late regulator Q together with its site of action (qut) and a functional lysis cassette to a new descendant. A recombination at linker 4 would transfer a cos site together with the DNA packaging machine and phage head and tail proteins. On the other hand, an exchange at linker 5 would combine a phage receptor specificity carried in the phage tail region with a new integration site, attP, for prophage integration.
We suggest that exchanges at these linkers have occurred in the evolution of phage φP27. Evidence for such events may be found in the G+C distribution of the φP27 genome. From attL to linker 2, the G+C content is 44.29 mol%; from linker 2 to linker 3, the G+C content increases to 50.57 mol%. In the region between linker 3 and linker 4, the G+C content decreases to 45.93 mol%, and the last area following linker 4 comprises a G+C content of 52.05 mol% (data not shown).
Due to the fact that experiments failed to obtain Stx2e-transducing phages from Stx2e-producing E. coli strains, this variant was thought to be chromosomally encoded rather than phage borne (38, 73). With the isolation of phage φP27 (42), this opinion had to be modified. E. coli strain 2771/97 was the only Stx2e producer out of 11 test strains capable of producing an Stx2e-converting phage (42). We were interested in the genetic background of stx genes from the remaining 10 strains. Therefore, we investigated stx2e-flanking regions of these 10 and another 3 stx2e-containing strains. Using sequence information obtained from phage φP27, we analyzed the linkage of stx2e genes and φP27-like genes by PCR. In all but one strain (55/89), the stx2e genes are located at the same distance, upstream of a φP27-like endolysin, representing a part of the phage lysis cassette. In the other direction, two strains show a linkage between a φP27-like late antiterminator Q and the toxin genes (24059/97 and E57). In all other stains, φP27-like sequences can at least be traced to the start of the DNA methylase. Thus, it becomes clear that stx2e genes seem to be generally located on the genome of φP27-related phages at the same positions shown for all other toxin variants investigated so far.
The reason why we could not isolate infectious phage particles from most of the strains is at present not known. It is conceivable that parts of the respective prophages are deleted, as shown in Shigella dysenteriae (39), that insertion elements prevent phage propagation as suggested for VT-2 Sakai (37), or that defective phage particles are produced that are not able to successfully infect indicator strains. We sequenced a DNA fragment of 1,123 bp from all stx2e-positive strains which contains a part of the methylase gene and the tRNA coding area. This was performed in order to investigate the clonal relationship between the phage sequences found upstream of the investigated stx2e genes. For most of the strains, the sequences are very similar. Interestingly, the resulting phylogenetic tree correlates well with that of aroE sequences. The similar phylogenetic profile lets us suggest that the prophages were acquired by their bacterial host early in the evolution of this specific STEC group, and the related trees indicate specific adaptation of phages to their hosts. STEC can be isolated from a large number of sources. They seem to have adapted to specific ecological niches. Since stx genes seem to occur ubiquitously in E. coli, selection advantages of stx carriers may be postulated. Some studies demonstrated that mobile genetic elements can impose fitness costs on the bacteria carrying them. Therefore, it may not be plausible that genes on mobile genetic elements, once they have been acquired, will be sequestered as chromosomal genes. Following a hypothesis of Smith (64), mobile genetic elements of obligate pathogenic bacteria prevent a population of pathogens from losing their pathogenicity genes and help to regain the virulence of so-called cheater strains (64). Future work on Stx phages is needed to fully understand the role of these mobile genetic elements in STEC evolution.
Acknowledgments
This work was supported by grant Schm 1360/1-2 from the Deutsche Forschungsgemeinschaft.
Editor: A. D. O'Brien
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anton, I. A., and J. R. Coggins. 1988. Sequencing and overexpression of the Escherichia coli aroE gene encoding shikimate dehydrogenase. Biochem. J. 249:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos, P., A. Landy, K. Abremski, J. B. Egan, E. Haggard-Ljungquist, R. H. Hoess, M. L. Kahn, B. Kalionis, S. V. Narayana, L. S. Pierson III, et al. 1986. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 5:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, V. J. Collado, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Brondstedt, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 8.Brunder, W., and H. Karch. 2000. Genome plasticity in Enterobacteriaceae. Int. J. Med. Microbiol. 290:153-165. [DOI] [PubMed] [Google Scholar]
- 9.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145:1005-1014. [DOI] [PubMed] [Google Scholar]
- 10.Bucher, P., and A. Bairoch. 1994. A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:53-61. [PubMed] [Google Scholar]
- 11.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 12.Campbell, A. M. 1992. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174:7495-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens, S., G. Hatfull, and R. Hendrix. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin. Virol. 3:383-397. [Google Scholar]
- 14.Clerget, M., and F. Boccard. 1996. Phage HK022 Roi protein inhibits phage lytic growth in Escherichia coli integration host factor mutants. J. Bacteriol. 178:4077-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornick, N. A., I. Matise, J. E. Samuel, B. T. Bosworth, and H. W. Moon. 2000. Shiga toxin-producing Escherichia coli infection: temporal and quantitative relationships among colonization, toxin production, and systemic disease. J. Infect. Dis. 181:242-251. [DOI] [PubMed] [Google Scholar]
- 16.Dhillon, T. S., A. P. Poon, D. Chan, and A. J. Clark. 1998. General transducing phages like Salmonella phage P22 isolated using a smooth strain of Escherichia coli as host. FEMS Microbiol. Lett. 161:129-133. [DOI] [PubMed] [Google Scholar]
- 17.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feiss, M., W. Widner, G. Miller, G. Johnson, and S. Christiansen. 1983. Structure of the bacteriophage lambda cohesive end site: location of the sites of terminase binding (cosB) and nicking (cosN). Gene 24:207-218. [DOI] [PubMed] [Google Scholar]
- 19.Franke, S., D. Harmsen, A. Caprioli, D. Pierard, L. H. Wieler, and H. Karch. 1995. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J. Clin. Microbiol. 33:3174-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman, D. I., and D. L. Court. 1995. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 18:191-200. [DOI] [PubMed] [Google Scholar]
- 21.Fuchs, S., I. Mühldorfer, A. Donohue-Rolfe, M. Kerényi, L. Emödy, A. Rossen, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27:13-23. [DOI] [PubMed] [Google Scholar]
- 22.Furth, E. F., and S. H. Wickner. 1983. Lambda DNA replication, p. 145-173. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Gannon, V. P., C. Teerling, S. A. Masri, and C. L. Gyles. 1990. Molecular cloning and nucleotide sequence of another variant of the Escherichia coli Shiga-like toxin II family. J. Gen. Microbiol. 136:1125-1135. [DOI] [PubMed] [Google Scholar]
- 24.Gussin, G. N., A. D. Johnson, C. O. Pabo, and R. T. Sauer. 1983. Repressor and Cro protein: structure, function, and role of lysogenization, p. 92-121. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genome comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 27.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrix, R. W. 1988. Tail length determination in double-stranded DNA bacteriophages. Curr. Top. Microbiol. Immunol. 136:21-29. [DOI] [PubMed] [Google Scholar]
- 29.Hendrix, R. W., and R. L. Duda. 1998. Bacteriophage HK97 head assembly: a protein ballet. Adv. Virus Res. 50:235-288. [DOI] [PubMed] [Google Scholar]
- 30.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heuvelink, A. E., K. N. van-de, J. F. Meis, L. A. Monnens, and W. J. Melchers. 1995. Characterization of verocytotoxin-producing Escherichia coli O157 isolates from patients with haemolytic uraemic syndrome in Western Europe. Epidemiol. Infect. 115:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann, K., and W. Stoffel. 1993. TMbase-A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166. [Google Scholar]
- 33.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 34.Katsura, I. 1987. Determination of bacteriophage lambda tail length by a protein ruler. Nature 327:73-75. [DOI] [PubMed] [Google Scholar]
- 35.Katsura, I., and R. W. Hendrix. 1984. Length determination in bacteriophage lambda tails. Cell 39:691-698. [DOI] [PubMed] [Google Scholar]
- 36.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makino, K., K. Yokoyama, Y. Kubota, C. H. Yutsudo, S. Kimura, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, T. Iida, K. Yamamoto, M. Onishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 38.Marques, L. R., J. S. M. Pieris, S. J. Cryz, and A. D. O'Brien. 1987. Escherichia coli strains isolated from pigs with edema disease produce a variant of Shiga-like toxin II. FEMS Microbiol. Lett. 44:33-38. [Google Scholar]
- 39.McDonough, M. A., and J. R. Butterton. 2000. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058-1069. [DOI] [PubMed] [Google Scholar]
- 40.Melton-Celsa, A. R., S. C. Darnell, and A. D. O'Brien. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizutani, S., N. Nakazono, and Y. Sugino. 1999. The so-called chromosomal verotoxin genes are actually carried by defective prophages. DNA Res. 6:141-143. [DOI] [PubMed] [Google Scholar]
- 42.Muniesa, M., J. Recktenwald, M. Bielaszewska, H. Karch, and H. Schmidt. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neely, M. N., and D. I. Friedman. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255-1267. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 45.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaiha, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 46.Parkhill, J., B. W. Wren, G. Dougan, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaiha, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, A. V. Karlyshev, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plaque. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 47.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen, M., S. Ostergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 49.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. Blattner. 2001. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 50.Perucchetti, R., W. Parris, A. Becker, and M. Gold. 1988. Late stages in bacteriophage lambda head morphogenesis: in vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology 165:103-114. [DOI] [PubMed] [Google Scholar]
- 51.Pierard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plunkett, G., D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 54.Ptashne, M. 1992. A genetic switch: phage λ and higher organisms. Blackwell Scientific Publications & Cell Press, Cambridge, Mass.
- 55.Rietra, P. J., G. A. Willshaw, H. R. Smith, A. M. Field, S. M. Scotland, and B. Rowe. 1989. Comparison of Vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J. Gen. Microbiol. 135:2307-2318. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt, H., C. Geitz, P. I. Tarr, M. Frosch, and H. Karch. 1999. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J. Infect. Dis. 179:115-123. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scholl, D., S. Rogers, S. Adhya, and C. R. Merril. 2001. Bacteriophage K1-5 encodes two different tail fiber proteins, allowing it to infect and replicate on both K1 and K5 strains of Escherichia coli. J. Virol. 75:2509-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scotland, S. M., H. R. Smith, G. A. Willshaw, and B. Rowe. 1983. Vero cytotoxin production in strain of Escherichia coli is determined by genes carried on bacteriophage. Lancet ii:216. [DOI] [PubMed] [Google Scholar]
- 61.Shen, P., and H. V. Huang. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, H. R., N. P. Day, S. M. Scotland, R. J. Gross, and B. Rowe. 1984. Phage-determined production of vero cytotoxin in strains of Escherichia coli serogroup 0157. Lancet i:1242-1243. [DOI] [PubMed] [Google Scholar]
- 63.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 64.Smith, J. 2001. The social evolution of bacterial pathogens. Proc. R. Soc. Lond. Ser. B 268:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 66.Susskind, M. M., and D. Botstein. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taguchi, T., H. Uchida, N. Kiyokawa, T. Mori, N. Sato, H. Horie, T. Takeda, and J. Fujimoto. 1998. Verotoxins induce apoptosis in human renal tubular epithelium derived cells. Kidney Int. 53:1681-1688. [DOI] [PubMed] [Google Scholar]
- 68.Takeda, S., T. Sasaki, A. Ritani, M. M. Howe, and F. Arisaka. 1998. Discovery of the tail tube gene of bacteriophage Mu and sequence analysis of the sheath and tube genes. Biochim. Biophys. Acta 1399:88-92. [DOI] [PubMed] [Google Scholar]
- 69.Taylor, K., and G. Wegrzyn. 1995. Replication of coliphage lambda DNA. FEMS Microbiol. Rev. 17:109-119. [DOI] [PubMed] [Google Scholar]
- 70.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van-der-Wilk, F., A. M. Dullemans, M. Verbeek, and J. F. van-den-Heuvel. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262:104-113. [DOI] [PubMed] [Google Scholar]
- 73.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yokoyama, K., K. Makino, Y. Kubota, S. Kimura, C. H. Yutsudo, K. Kurokawa, K. Ishii, S. Hattori, I. Tatsuno, H. Abe, M. Yoh, T. Iida, M. Ohnishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 2000. Complete nucleotide sequence of the prophage VT1-Sakai carrying the Shiga toxin 1 genes of the enterohemorrhagic Escherichia coli O157:H7 strain derived from the Sakai outbreak. Gene 258:127-139. [DOI] [PubMed] [Google Scholar]