Abstract
The transcriptional activity and allele variation of the 28-kDa outer membrane protein gene (p28) of Ehrlichia chaffeensis were analyzed to determine the mechanism of the antigenic variation of the 28-kDa outer membrane proteins. Reverse transcriptase PCR amplification of mRNA indicated that 16 of the 22 members of the p28 multigene family were transcribed. Amino acid sequence analysis indicated that the p28-19 protein was produced in vitro in the Arkansas strain. The p28-19 gene and its promoter region were sequenced and compared in 12 clinical isolates of E. chaffeensis to determine allele variation. The variation of the p28-19 gene among the isolates is limited to three types represented by strains Arkansas, 91HE17, and Sapulpa, respectively. These results indicate that the majority of the p28 genes are active genes and that antigenic variation of the E. chaffeensis 28-kDa proteins may result from differential expression of the p28 gene family members rather than gene conversion.
Ehrlichiae are small, obligately intracellular, gram-negative bacteria that reside in the endosomes of host cells. Tick-borne ehrlichiae are maintained in nature by causing prolonged or persistent infection in their natural animal hosts (1, 2, 5, 6, 11, 19). Prolonged ehrlichial infection in human beings has been reported for Ehrlichia chaffeensis (7, 17) and Anaplasma phagocytophila 9; H. W. Horowitz, J. Raffalli, R. B. Nadelman, J. Wu, and G. P. Wormser, Letter, Lancet 351:650, 1998). E. canis also causes chronic human infection (15). Persistent infection by E. chaffeensis and E. canis may be caused by immune evasion through antigenic variation of the 28-kDa outer membrane proteins. There are at least 22 copies (alleles) of the 28-kDa-protein gene (p28) in the chromosome of E. chaffeensis, the pathogen of human monocytotropic ehrlichiosis (13). The alleles have 20 to 80% homology and are organized in a single locus in the genome (22). The p28 alleles are predicted to encode proteins ranging from 25 to 32 kDa (22). Although all of the p28 alleles are complete genes, it is not clear whether they are active genes or pseudogenes. Characterization of the transcriptional activities of the p28 alleles will reveal the mechanism of the 28-kDa protein's antigenic variation.
The objectives of this study were to investigate the transcriptional activity of each of the p28 alleles in a single isolate of E. chaffeensis, to investigate the variation of the p28-19 allele in different E. chaffeensis isolates, and to illustrate the model of antigenic variation of the 28-kDa proteins.
MATERIALS AND METHODS
Ehrlichia isolates.
All of the E. chaffeensis isolates studied were from monocytotropic ehrlichiosis patients and have been reported previously (3, 4, 8, 10, 14, 18). The isolates were cultivated in DH82 cells with 5% bovine calf serum-supplemented minimal essential medium at 37°C. Ehrlichiae were harvested when 100% of the cells were infected (approximately 5 days postinfection). The cells were centrifuged for 20 min at 12,100 × g to pellet the ehrlichiae. The pellet was suspended in SPK buffer (0.2 M sucrose and 0.05 M potassium phosphate buffer, pH 7.4) (20) and sonicated twice for 10 s each time on ice at 40 W with an Ultrasonic Processor (Sonic & Materials Inc., Newtown, Conn.). The suspension was centrifuged at 200 × g for 10 min to remove cell debris. The supernatant was centrifuged through a 30% Percoll column at 63,000 × g for 30 min. One-third of the solution from the bottom of a centrifuge tube was collected by aspiration with a pipette. One volume of SPK was added to the collected solution, and the mixture was centrifuged at 12,100 × g for 20 min to collect the ehrlichiae.
Selection of an E. chaffeensis clone by limiting dilution.
E. chaffeensis strain Arkansas-infected DH82 cells from a 150-cm2 flask were sonicated at 40 W for 10 s. The lysate was centrifuged at 200 × g for 5 min, and the supernatant was filtered through a 0.45-μm-pore-size syringe-driven filter to obtain a single-cell suspension of E. chaffeensis. The filtered lysate was diluted in cell culture medium in 10-fold increments from 10−1 to 10−6, and each dilution was added to four wells of a 24-well plate with 0.1 ml in each well. The cells were incubated at 37°C with 5% CO2, and the medium was changed every 3 days. On days 7, 14, 21, and 28 postinoculation, cells were examined by PCR and Diff-Quik staining for E. chaffeensis infection. For PCR, 500 μl of supernatant of the cell culture from each well was centrifuged at 15,800 × g for 10 min in a microcentrifuge. The pellet was resuspended in 50 μl of distilled water. Ten microliters of the suspension was used to amplify the p28-19 gene with primers 1819f (GTG GCA AAA GAA TGT AGC AAT AAG) and 1336r (GCT GTT GTG TAA CTG TAG ACT GGT). PCR amplification was performed for 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min with a final extension of 7 min. For Diff-Quik staining, 200 μl of supernatant from each well was centrifuged onto a slide with a Cytospin centrifuge. The slides were stained and examined for E. chaffeensis morulae.
DNA and RNA preparation.
Genomic DNA was prepared from Percoll gradient-purified ehrlichiae by using an IsoQuick nucleic acid extraction kit (ORCA Research, Inc., Bothell, Wash.) in accordance with the instructions of the manufacturer. A QIAquick PCR purification kit (Qiagen, Inc., Santa Clarita, Calif.) was used to purify PCR products for DNA sequencing. A High Pure Plasmid Isolation Kit (Roche, Indianapolis, Ind.) was used to purify plasmid DNA. Total RNA was extracted from E. chaffeensis-infected DH82 cells by using an Rneasy Mini kit (Qiagen, Valencia, Calif.).
Analysis of E. chaffeensis strain Arkansas p28 transcription by RT-PCR.
The transcription of various p28 genes was assessed in the cultured Arkansas isolate of E. chaffeensis by reverse transcriptase (RT) PCR. Primers were designed to amplify 12 of the p28 genes (p28-1′, p28-1, p28-2, p28-3, p28-4, p28-5, p28-6, p28-7, p28-8, p28-9, p28-16, and p28-19) and 15 of the intergenic regions (sp1′, sp1, sp2, sp3, sp4, sp5, sp6, sp7, sp8, sp9, sp10, sp11, sp12, sp13, and sp18) (Table 1). The intergenic regions were spanned by primers designed from the sequences in neighboring p28 genes such that amplification would occur only if the two genes were transcribed as a single mRNA. Primers for the amplification of p28-10, p28-11, p28-12, p28-13, p28-14, p28-15, p28-17, p28-18, p28-20, and p28-21 were reported previously (22). Primer function was assessed by PCR amplification by 25 cycles of 94° for 30 s, 50° for 1 min, and 72° for 30 s with a final extension of 5 min. Two reactions were then set up for each primer pair, one RT-PCR and one PCR using the RNA template as a negative control. The RT-PCRs were carried out with the Roche Titan One-Step RT-PCR system (Roche Biochemicals, Indianapolis, Ind.). The control PCRs were carried out with the Roche PCR Master Kit (Roche Biochemicals). The reaction mixtures were incubated at 50° for 30 min. Amplification was then carried out for 10 cycles of 94° for 30 s, 50° for 1 min, and 72° for 30 s, followed immediately by 25 cycles of 94° for 30 s, 50° for 1 min, and 72° for 30 s extended by an extra 5 s for each cycle, followed by a 5-min final extension.
TABLE 1.
Primers for amplification of 28-kDa protein-encoding genes and their intergenic sequences
| Gene | Primer | Sequence |
|---|---|---|
| p28-1′ | p1′f | ATC GGA TAT TCA GGA GGA GGT CT |
| p1′r | CAA GGG GGT ACT GAA CTG ATA GG | |
| p28-1 | p1f | CAA AGG AAA TAA GCG GTG GTA A |
| p1r | GGC TTG CTG TTT GGT GGT GTT | |
| p28-2 | p2f | ACC TGA AGC GTT GGA AGA GC |
| p2r | GGG AAC CCG GAA GAT GAG TAA | |
| p28-3 | p3f | ATT AGG GAT AGG TTT GTC TGT |
| p3r | CTT TGA AGT AAT GGG ATA GC | |
| p28-4 | p4f | CAA ATC AAA CGC TTA TAA CCT ACA G |
| p4r | GCC CTA CAA GAT CTT CAC CAA T | |
| p28-5 | p5f | AAC CCT CTG ACA GTA ATC CTA AAA A |
| p5r | TCC CAA CTT CAC CAC CAA AAT | |
| p28-6 | p6f | ACA GGG CAG TAT AGA CCA GGA GTA |
| p6r | TTC ACC ACC AAA ATA AGC AAC AT | |
| p28-7 | p7f | ATT CAA CAT TCC TTA TAA CAC AAA A |
| p7r | AGA ATA ATG GAT AAC TGA TAC CTA CCT T | |
| p28-8 | p8f | TTC TAG CCC TAC CTC AAA CAA C |
| p8r | TGC GTA TAC AGG TCC AAA ATC | |
| p28-9 | p9f | CAC ATC TGA GGC CTC TTC TAC A |
| p9r | ATT CCT GCG CAT ACA TAC G | |
| p28-16 | p16f | CGC TTC GCA TTT TGG AGT TTT |
| p16r | GTG CCG AGA AGA TGG GAC AGA | |
| p28-19 | p19f | GAA GCG CAA TAT CCA ACT CCT C |
| p19r | TGA CCA ATA AAC ACA GAA G | |
| Intergenic region 1-2 | sp1f | TAT CCA TCA CCA ACA CCA CCA AAC AG |
| sp1r | GCA AAG ATA ATG AAG GCA CAA | |
| Intergenic region 2-3 | sp2f | TAT GTC TTG GGG TTG GTG GTA A |
| sp2r | TGT ATG CTC CTT CAA ATT CTA TCC TAA | |
| Intergenic region 3-4 | sp3f | ATA TGT ATA GGC GTT GGT GGA GAT TTT |
| sp3r | CTG TAG GTT ATA AGC GTT TGA TTT GAG TA | |
| Intergenic region 4-5 | sp4f | CTC AAA TCA AAC GCT TAT AAC CTA CAG T |
| sp4r | TAC AAC ATC AAA ATC CCC ATA AGA GC | |
| Intergenic region 5-6 | sp5f | ACG TAT GTA TAG GAG TTG GAG GAG |
| sp5r | AAG TGT TAG GAT CGA TAG AAG ACG | |
| Intergenic region 6-7 | sp6f | TAT GTG CAG GTA TTG GTG AAG ATT |
| sp6r | AAC GTG GTA TAG AAG CCG CAT TTA | |
| Intergenic region 7-8 | sp7f | AAG GGT TGA GGT AGA ATG GTC |
| sp7r | CCT AAC TTG CCT TGG TAT GCT A | |
| Intergenic region 8-9 | sp8f | TTC TAG CCC TAC CTC AAA CAA C |
| sp8r | TGC GTA TAC AGG TCC AAA ATC | |
| Intergenic region 9-10 | sp9f | TCA AGG CAA GGT AGG TAT TAG TTA TTC T |
| sp9r | CCT CCA GGG TTT TTA GCA TCA A | |
| Intergenic region 10-11 | sp10f | ATA GGG GTG GAT GCA ATA GAA T |
| sp10r | TAG CGT AGA AGA ATC CAA GTG C | |
| Intergenic region 11-12 | sp11f | TGC CCT AAA CAC AAA GAA ACA T |
| sp11r | GTC CAC TAA CAT ATA ACC CAA ACT G | |
| Intergenic region 12-13 | sp12f | TTA ATG ACG CAC CCA AAG TT |
| sp12r | TTC GCC ACC AAA GTA TGC | |
| Intergenic region 13-14 | sp13f | ATG GCT GCT ATG ATT TTT CTT C |
| sp13r | CTG TAT CTT CCG CTT TAG GTT C | |
| Intergenic region 1′ | sp1′f | TTA ATG CAG ACA ACT ATG GTG GTG |
| sp1′r | GAA GTT TTG ATA GGT GTT GTC TTG | |
| V7, V9 isolate p28-19 | V2f | GCA GTG CAA TAC CTC ACA CTC A |
| V2r | ATG TCC GCC GAC AAA GAC AGA AG | |
| V5, Jax isolate p28-19 | V5f | GCA GTG CAA TAT CTC ACA CCA C |
| p19r | TGA CCA ATA AAC ACA GAA G | |
| V1, V6 isolate p28-19 | p19f | GAA GCG CAA TAT CCA ACT CCT C |
| p19r | TGA CCA ATA AAC ACA GAA G |
Analysis of p28-19 transcription in E. chaffeensis isolates by RT-PCR.
Primers were designed to amplify the p28-19 gene from several different isolates of E. chaffeensis (Table 1). Isolates V7 and V9 were amplified with primers V2f and V2r. Isolates V5 and Jax were amplified with primers V5f and p19r. Isolates V1 and V6 were amplified with p19f and p19r (22). Control RT-PCRs and PCRs were set up as detailed above and amplified by the same protocols.
N-terminal sequencing of p28 of E. chaffeensis.
The proteins of purified E. chaffeensis strain Arkansas were separated on a 12% polyacrylamide gel (15 by 20 cm). The proteins were transferred onto a polyvinylidene difluoride membrane and stained with Coomassie brilliant blue R. Each of the five protein bands from 25 to 32 kDa was excised from the membrane. The N-terminal sequences of the proteins were determined with the Applied Biosystems 494/HT PROCISE Sequencing System in the Protein Chemistry Laboratory at the University of Texas Medical Branch.
Sequencing of the p28-19 allele in E. chaffeensis isolates.
Two primers were designed from the DNA sequences (GenBank accession no. AF062761) of E. chaffeensis strain Arkansas to amplify the p28-19 alleles in the clinical isolates of E. chaffeensis. Forward primer 1819f is derived from the intergenic sequence 70 bp downstream of the stop codon of p28-18. Reverse primer 1336r is complementary to the intergenic sequence 293 bp downstream of the stop codon of p28-19. The primer pair is predicted to amplify a 1.4-kb DNA fragment including the p28-19 gene and DNA sequences flanking the gene on both the 5′ and 3′ ends. The primers are specific to p28-19 and do not anneal to the DNA sequences of other p28 multigenes. PCR amplification was performed as described above.
PCR products were cloned into vector pCR4 TOPO and sequenced by using the primers complementary to the vector DNA sequence, except for V5, which was sequenced directly by using PCR primers. Sequencing was performed with an ABI Prism 377 DNA sequencer (Perkin-Elmer Applied Biosystems) in the Protein Chemistry Laboratory at the University of Texas Medical Branch.
Cloning of the intergenic sequences of p28 genes into pKK232-8 to evaluate promoter function.
The pKK232-8 vector (Amersham Pharmacia Biotech, Piscataway, N.J.) contains the chloramphenicol acetyltransferase (cat) gene without a promoter. If a promoter sequence is cloned into the pKK232-8 vector, the promoter will drive the transcription of the cat gene. Thus, Escherichia coli transformed with a plasmid containing a promoter preceding the cat gene would have a chloramphenicol resistance phenotype. The entire intergenic sequences upstream of the p28-14 and p28-19 genes were amplified by PCR using primer pairs p14sf (ATT GCT CAA CCA TAA AAT AAT GGG A) and p14sr (GTT AAT AAA CCT TTT ATA AAA G) and 1819f and 1819r (ATA ACC TAA TAG TGA CAA ATA AAT), respectively. The PCR products were first cloned into vector pBlue-TOPO (Invitrogen, Carlsbad, Calif.) to obtain the BamHI and HindIII restriction sites for subsequent cloning of the insert into the pKK232-8 vector. The recombinant plasmid was digested with BamHI and HindIII to release the insert. After gel electrophoresis, the insert was excised from the agarose gel and purified by using a Gel Extraction kit (Qiagen, Inc.). The DNA fragment was cloned into BamHI/HindIII-digested, alkaline phosphatase-treated vector pKK232-8. The plasmid containing the intergenic sequence upstream of the p28-14 gene was designated pKK1314, and the plasmid containing the intergenic sequence upstream of the p28-19 gene was designated pKK1819. To characterize the precise location of the p28-19 gene promoter, further deletion of part of the insert in the pKK1819 plasmid was done by restriction enzyme digestion or PCR. Plasmid pKK1819 was digested with SpeI and ligated again with T4 ligase to delete 36 bp on the 5′ end of the insert, and the derived recombinant plasmid was designated pKK1819S. The intergenic sequence was also amplified by using primer 1819f2 (GAC TTG CTT TTA TAT GAC ACT TC) and primer 1819r, which amplified a product of 60 bp. The PCR product was cloned into vector pCR pBlue-TOPO (Invitrogen). The insert was digested with BamHI and HindIII and cloned directionally into pKK232-8. The resulting plasmid was designated pKK70.
cat expression in E. coli was measured by determining the growth curve of E. coli. A single colony of E. coli was inoculated into 2 ml of ampicillin-containing Luria-Bertani (LB) broth and shaken at 37°C overnight. One milliliter of the E. coli culture was mixed with 20 ml of LB medium containing ampicillin (final concentration of 100 μg/ml), and then the culture medium was divided equally into two parts. Chloramphenicol was added to one part to a final concentration of 34 μg/ml. The bacterial suspensions were distributed into a 96-well plate with 100 μl in each well. Each sample was prepared in triplicate. The plates were shaken at 250 rpm at 37°C. The optical density of the E. coli samples at a wavelength of 600 Å was read every 0.5 h for 3 h.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been assigned GenBank accession numbers AF077732, AF077733, AF077734, AF393388, AF393389, AF393390, AF393391, AF393392, AF393393, AF393394, and AF393395.
RESULTS
Limiting-dilution selection of a clone of E. chaffeensis.
At day 14 postinoculation, E. chaffeensis morulae were observed by Diff-Quik staining and E. chaffeensis DNA was detected by PCR amplification in all of the wells of cells inoculated with the 10−1 and 10−2 dilutions of E. chaffeensis, in two of the four wells of cells inoculated with the 10−3 dilution, and in one of the four wells inoculated with the 10−4 dilution. No cells inoculated with the 10−5 and 10−6 dilutions were positive for ehrlichial morulae or PCR amplification of ehrlichial DNA in a 4-week course of assays for E. chaffeensis. Clone 31 from the 10−3 dilution and clone 41 from the 10−4 dilution of E. chaffeensis were cultivated in cell culture for transcriptional analysis of the p28 multigene family.
Transcription of E. chaffeensis strain Arkansas p28 genes.
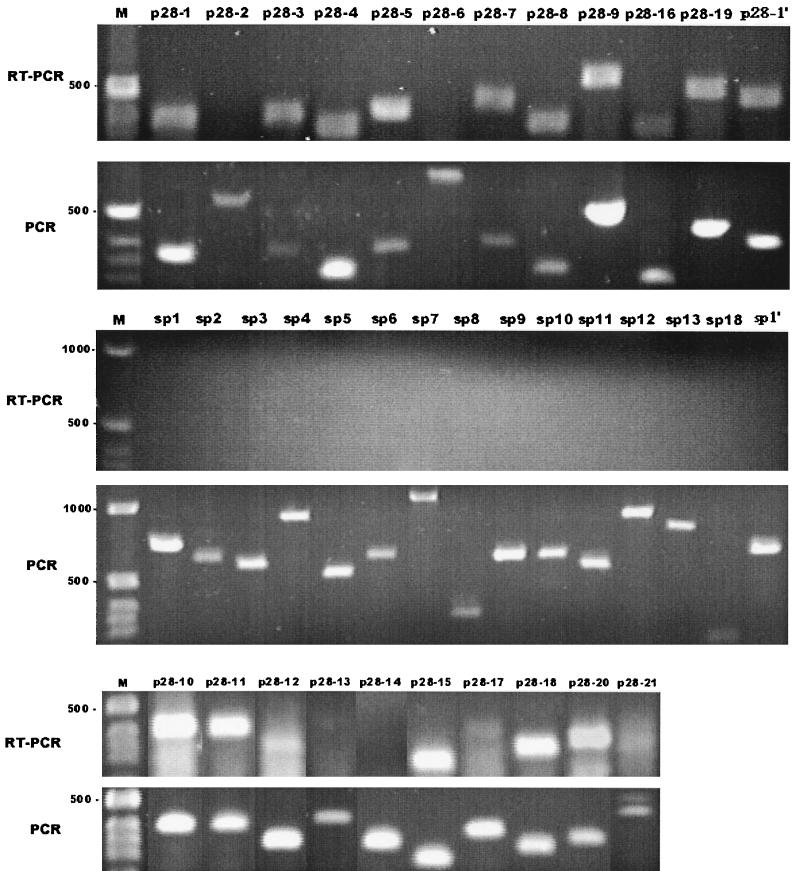
Our examination of the RNA of E. chaffeensis clone 41 by RT-PCR revealed the transcription of 16 of 22 p28 genes (Fig. 1). The p28-2, p28-6, p28-13, p28-14, p28-17, and p28-21 genes could not be detected by RT-PCR amplification. This experiment was repeated, and PCRs performed simultaneously on E. chaffeensis isolate DNA indicated that the amplification conditions were appropriate for amplification of the genes studied. All of the intergenic or spacer regions examined could not be detected by RT-PCR (Fig. 1). Amplification of template RNA with Taq polymerase was negative for all primer pairs (data not shown). Subsequently, we examined the transcription of p28 genes in clone 31 and in the uncloned mixed population of E. chaffeensis by RT-PCR. The transcription patterns of clone 31 and the mixed population of E. chaffeensis were identical to the pattern of clone 41 (data not shown).
FIG. 1.
Transcriptional analysis of the p28 genes and their intergenic sequences (sp) by RT-PCR. PCR amplification of the DNA template with Taq polymerase is shown as a positive control. On the left of each panel is a DNA standard(s) in base pairs. M, 1-kb DNA ladder markers.
Amino acid sequencing.
To determine which of the p28 proteins were produced by E. chaffeensis, all five proteins ranging from 25 to 32 kDa were subjected to N-terminal amino acid sequencing. The N-terminal amino acids of two proteins were successfully sequenced. The sequence was DRVNIGTVGXLGKQGRRLV for band 1, which was identical to histone H3 and might have been derived from the host cells. The N-terminal sequence of the protein from band 3 was DPAGSGINGNFYISGKYMP. This sequence is identical to that of the protein encoded by p28-19 after removal of the signal peptide. The results indicate that this protein is the product of the p28-19 gene.
The promoter of the p28 genes.
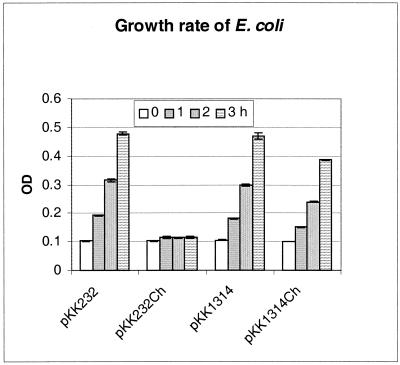
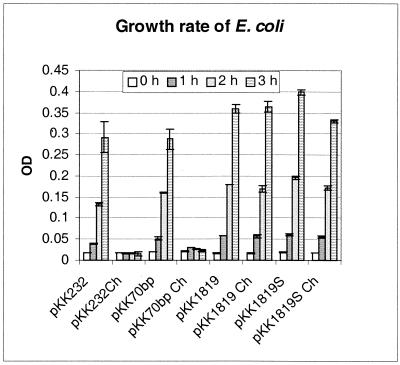
The promoter function of the intergenic sequences upstream of the p28-14 and p28-19 genes was characterized. These genes were chosen because they were representatives of a nontranscribed gene (p28-14) and a transcribed gene (p28-19). Chloramphenicol minimally inhibited the growth of E. coli transformed by the plasmid containing the intergenic sequence upstream of the p28-14 gene (Fig. 2). Chloramphenicol had no effect on the growth of E. coli transformed by plasmid pKK1819 containing the intergenic sequence upstream of the p28-19 gene (Fig. 3). The precise location of the promoter of the p28-19 gene was further characterized by nested deletion of the insert in plasmid pKK1819. pKK1819S was derived from pKK1819 by SpeI digestion to delete 36 bp from the 5′ end of the intergenic sequence. pKK70 contains 60 bp from the 3′end of the intergenic sequence. The DNA fragments in pKK1819, pKK1819S, and pKK70 start from the same point at the 3′ end but end at different points at the 5′ end. Chloramphenicol had no effect on the growth of E. coli containing plasmid pKK1819 and only slightly inhibited the growth of E. coli containing pKK1819S. The growth of E. coli containing pKK70 was completely inhibited by chloramphenicol (Fig. 3). The experiments were repeated three times, and the results were similar. These results indicate that the intergenic sequences upstream of the p28-14 and p28-19 genes both have promoter functions. The promoter of the p28-19 gene is within the 237-bp DNA sequence upstream of the ATG initiation codon of p28-19. Either part or all of the promoter sequences are located 70 bp upstream of the ATG codon since the 70-bp DNA sequence has no promoter function.
FIG. 2.
Growth rate of E. coli. E. coli harboring plasmid pKK232-8 or pKK1314 was cultivated in LB medium containing ampicillin or LB medium containing both ampicillin and chloramphenicol (Ch). The standard deviation of the value at each time point was obtained from triplicates of one experiment. OD, optical density.
FIG. 3.
Growth curves of E. coli. E. coli harboring plasmid pKK1819, pKK1819S, pKK70, or pKK232-8 was cultivated in LB medium containing ampicillin or LB medium containing both ampicillin and chloramphenicol (Ch). The standard deviation of the value at each time point was obtained from triplicates of one experiment. OD, optical density.
Allele variations of the p28-19 gene of 12 E. chaffeensis isolates.
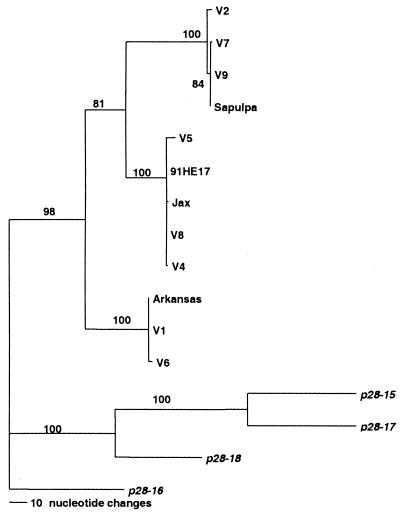
DNA sequencing indicated that both the promoter region and the coding sequences of p28-19 varied among the clinical isolates of E. chaffeensis studied (Fig. 4A and B). The coding sequences had more variations than the promoter, and the variations were present throughout the coding sequences. The mutations included base pair changes, insertions, and deletions. Although varied, both the promoters and the coding regions were readily classified into three genetic groups. Group I included isolates Sapulpa, V2, V7, and V9; group II included isolates 91HE17, V4, V5, V8, and Jax; and group III included isolates Arkansas, V1, and V6 (Fig. 5). The variation within each group was less than 0.5%, and the variation among the groups was 10.6 to 14.5%. The genetic groups did not correlate with the geographic distribution of the organisms. The organisms from one geographic area were not necessarily similar to each other, such as isolates V1 to V9, which were from central Tennessee and adjacent southern Kentucky. In some cases, isolates from different geographic areas were found to belong to the same genetic group. For example, organisms in the 91HE17 genetic group were from Tennessee (V4, V5, and V8), Florida (Jax), and Arkansas (91HE17). In fact, all three groups contained organisms from different geographic areas. The variations in the promoter regions of these isolates were also of three types, which corresponded very well to each genetic group. The organisms in the same group had the same promoter sequences, and no organisms in different groups had the same promoter sequences. Compared with all 22 members of the p28 multigene family of strain Arkansas, the analyzed DNA sequences from all of the isolates studied had the highest level of homology to p28-19 and less homology to other alleles of p28.
FIG. 4.
(A) Clustal alignment of the promoter region of the p28-19 gene of the E. chaffeensis isolates studied. Low-passage isolates V1, V4, and V7 represent the three genetically defined groups Arkansas, 91HE17, and Sapulpa, respectively. (B) Clustal alignment of the coding region of the p28-19 gene of E. chaffeensis isolates. V1, V4, and V7 represent the three groups Arkansas, 91HE17, and Sapulpa, respectively.
FIG. 5.
Phylogenetic analysis of the alleles of p28-19 of the p28 multigene family of E. chaffeensis isolates. p28-15 to p28-18, which are the closest genes to p28-19, were used as the outgroup. Bootstrap values are shown at the nodes of the branches.
p28-19 transcription in selected isolates of E. chaffeensis.
RNA was purified from E. chaffeensis isolates V1, V5, V6, V7, V9, and Jax, and RT-PCR was performed to evaluate specific p28-19 transcription with genetic group-specific primers. Transcription of p28-19 was detected via RT-PCR amplification in all of the isolates tested (data not shown).
DISCUSSION
In the Arkansas isolate examined by RT-PCR, mRNA was detected in 16 alleles of the p28 gene of E. chaffeensis, indicating that multiple genes were transcribed. These mRNAs were transcribed from clone 41, which was derived from an endpoint dilution of E. chaffeensis. At the endpoint dilution, on average, there was 0.25 organism per well. In the titer prior to the endpoint, there was 0.75 organism per well. In dilutions beyond the endpoint, there were no organisms present in the wells. To produce a single-cell organism, E. chaffeensis-infected DH82 cell lysate was filtered with a bacterial filter. The pore size of the filter (0.45 μm) is smaller than the diameter of E. chaffeensis (0.5 μm) and therefore would only allow single-organism squeezing into the filter. Thus, the chance of clone 41 being derived from multiple organisms is very unlikely.
The lack of detectable transcripts of p28-2, p28-6, p28-13, p28-14, p28-17, and p28-21 was unlikely to have resulted from low quality or quantity of the templates because the same batch and the same amount of RNA were positive by RT-PCR using other p28 gene primers. These results indicate that these p28 genes of the Arkansas strain were not transcribed in DH82 cell culture. These genes were not transcribed under the conditions investigated but may be transcribed under other conditions since all p28 genes are complete genes (22). The fact that p28-14 had a promoter but was not transcribed in our study further indicated that the silent genes could be transcribed under other conditions. No detectable transcript levels were detected for the intergenic sequences, indicating that the p28 genes were transcribed monocistronically. Because transcripts were not detected by RT-PCR for the p28-13, p28-14, p28-17, and p28-21 genes, these genes were transcribed neither monocistronically nor polycistronically with their neighboring genes. These results are consistent with a previous study (22). A recent report indicated that E. canis p28 genes are polycistronically transcribed (13). The differences between our results and the previous results suggest that the p28 genes of E. chaffeensis and E. canis are transcribed differently.
We attempted to sequence all five proteins ranging from 25 to 32 kDa because the predicted molecular sizes of all p28 proteins are in this range. The predominant p28 protein produced in strain Arkansas was from the p28-19 gene. Our results are consistent with those of a previous study (12) in which the amino acid sequence of the p28-19 gene product was obtained. RT-PCR analysis indicated that the p28-19 gene was transcribed in all of the isolates examined. The inconsistency between the number of p28 proteins expressed as determined by amino acid sequencing and the number of genes transcribed as determined by RT-PCR indicates that some p28 proteins are expressed but are difficult to sequence and/or that the p28 genes may be transcribed but are not necessary expressed. It is possible that posttranscriptional regulation is involved in the expression of p28 genes. It will be interesting to investigate the regulation of p28 protein expression.
We further sequenced the p28-19 gene in 12 clinical isolates of E. chaffeensis for comparison of allele variation to investigate whether gene conversion is involved in the antigenic variation of the p28 protein. Allelic variation of p28-19 of E. chaffeensis has been observed previously among the few clinical isolates available (16, 21). The question that remains to be answered is whether the variations are stable or variation is a continuous event that results from gene conversion and is involved in the short-term antigenic variation of E. chaffeensis to evade the host immune system. The p28 genes have been hypothesized to participate in the short-term antigenic variation of E. chaffeensis through gene recombination (16). However, our results did not support this hypothesis. Although the DNA sequences of p28-19 in 12 clinical isolates of E. chaffeensis varied, the variation was limited to three types. Thus, each variant was apparently stable. Otherwise, we would not expect to find identical stable variants in the expression site among the isolates from different geographic areas. The homology of the p28-19 gene that we sequenced from all of the isolates we studied was much higher for the p28-19 gene than for other alleles of the p28 multigene family of strain Arkansas. These results indicate that the sequences that we obtained from the 12 isolates were a counterpart of the p28-19 gene.
Our results indicate that most of the p28 alleles are actively transcribed and that there is very limited allelic variation of a gene expressed among the E. chaffeensis isolates we studied. From these results, we conclude that differential expression, rather than conversion, of the p28 gene may be involved in the antigenic variation of the p28 protein.
Acknowledgments
This study was supported by a grant from the National Institute of Allergy and Infectious Diseases (AI 45871).
Editor: J. T. Barbieri
REFERENCES
- 1.Andrew, H. R., and R. A. Norval. 1989. The carrier status of sheep, cattle and African buffalo recovered from heartwater. Vet. Parasitol. 34:261-266. [DOI] [PubMed] [Google Scholar]
- 2.Breitschwerdt, E. B., B. C. Hegarty, and S. I. Hancock. 1998. Sequential evaluation of dogs naturally infected with Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia equi, Ehrlichia ewingii, or Bartonella vinsonii. J. Clin. Microbiol. 36:2645-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, S. M., X. J. Yu, V. L. Popov, E. L. Westerman, F. G. Hamilton, and D. H. Walker. 1997. Genetic and antigenic diversity of Ehrlichia chaffeensis: comparative analysis of a novel human strain from Oklahoma and previously isolated strains. J. Infect. Dis. 175:856-863. [DOI] [PubMed] [Google Scholar]
- 4.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson, J. E., J. E. Childs, K. L. Biggie, C. Moore, D. Stallknecht, J. Shaddock, J. Bouseman, E. Hofmeister, and J. G. Olson. 1994. White-tailed deer as a potential reservoir of Ehrlichia spp. J. Wildl. Dis. 30:162-168. [DOI] [PubMed] [Google Scholar]
- 6.Dawson, J. E., and S. A. Ewing. 1992. Susceptibility of dogs to infection with Ehrlichia chaffeensis, causative agent of human ehrlichiosis. Am. J. Vet. Res. 53:1322-1327. [PubMed] [Google Scholar]
- 7.Dumler, J. S., W. L. Sutker, and D. H. Walker. 1993. Persistent infection with Ehrlichia chaffeensis. Clin. Infect. Dis. 17:903-905. [DOI] [PubMed] [Google Scholar]
- 8.Dumler, J. S., S. M. Chen, K. Asanovich, E. Trigiani, V. L. Popov, and D. H. Walker. 1995. Isolation and characterization of a new strain of Ehrlichia chaffeensis from a patient with nearly fatal monocytic ehrlichiosis. J. Clin. Microbiol. 33:1704-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumler, J. S., and J. S. Bakken. 1996. Human granulocytic ehrlichiosis in Wisconsin and Minnesota: a frequent infection with the potential for persistence. J. Infect. Dis. 173:1027-1030. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, B. E., T. P. Monson, J. S. Dumler, C. C. Morris, A. B. Westbrook, J. L. Duncan, J. E. Dawson, K. G. Sims, and B. E. Anderson. 1992. Identification of Ehrlichia chaffeensis morulae in cerebrospinal fluid mononuclear cells. J. Clin. Microbiol. 30:2207-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrus, S., T. Waner, I. Aizenberg, J. E. Foley, A. M. Poland, and H. Bark. 1998. Amplification of ehrlichial DNA from dogs 34 months after infection with Ehrlichia canis. J. Clin. Microbiol. 36:73-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohashi, N., N. Zhi, Y. Zhang, and Y. Rikihisa. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohashi, N., Y. Rikihisa, and A. Unver. 2001. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paddock, C. D., J. W. Sumner, G. M. Shore, D. C. Bartley, R. C. Elie, J. G. McQuade, C. R. Martin, C. S. Goldsmith, and J. E. Childs. 1997. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J. Clin. Microbiol. 35:2496-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez, M., Y. Rikihisa, and B. Wen. 1996. Ehrlichia canis-like agent isolated from a man in Venezuela: antigenic and genetic characterization. J. Clin. Microbiol. 34:2133-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy, G. R., and C. P. Streck. 1999. Variability in the 28-kDa surface antigen protein multigene locus of isolates of the emerging disease agent Ehrlichia chaffeensis suggests that it plays a role in immune evasion. Mol. Cell. Biol. Res. Commun. 1:167-175. [DOI] [PubMed] [Google Scholar]
- 17.Roland, W. E., G. McDonald, C. W. Caldwell, and E. D. Everett. 1995. Ehrlichiosis--a cause of prolonged fever. Clin. Infect. Dis. 20:821-825. [DOI] [PubMed] [Google Scholar]
- 18.Standaert, S. M., T. Yu, M. A. Scott, J. E. Childs, C. D. Paddock, W. L. Nicholson, J. Singleton, Jr., and M. J. Blaser. 2000. Primary isolation of Ehrlichia chaffeensis from patients with febrile illnesses: clinical and molecular characteristics. J. Infect. Dis. 181:1082-1088. [DOI] [PubMed] [Google Scholar]
- 19.Telford, S. R., J. E. Dawson, P. Katavolos, C. K. Warner, C. P. Kolbert, and D. H. Persing. 1996. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc. Natl. Acad. Sci. USA 93:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss, E., J. C. Williams, G. A. Dasch, and Y. H. Kang. 1989. Energy metabolism of monocytic Ehrlichia. Proc. Natl. Acad. Sci. USA 86:1674-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu, X. J., J. W. McBride, and D. H. Walker. 1999. Genetic diversity of the 28-kilodalton outer membrane protein gene in human isolates of Ehrlichia chaffeensis. J. Clin. Microbiol. 37:1137-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, X. J., J. W. McBride, X. Zhang, and D. H. Walker. 2000. Characterization of the complete transcriptionally active Ehrlichia chaffeensis 28 kDa outer membrane protein multigene family. Gene 248:59-68. [DOI] [PubMed] [Google Scholar]