Abstract
The high-pathogenicity island (HPI) of yersiniae encodes an iron uptake system represented by its siderophore yersiniabactin (Ybt). The HPI is present in yersiniae with high levels of pathogenicity—i.e., Yersinia pestis, Y. pseudotuberculosis, and Y. enterocolitica biogroup (BG) 1B—but absent in Y. enterocolitica strains with low (BG 2 to 5) and no (BG 1A) levels of pathogenicity and has been shown to be an important virulence factor. Comparison of the HPI in Y. enterocolitica (Yen-HPI) and that in Y. pestis and Y. pseudotuberculosis revealed that, in contrast to genes of the variable region, genes of the core region (genes irp9 to fyuA) are highly homologous. In the present work the Yen-HPI core genes were rescued from the chromosome of Y. enterocolitica WA-C (BG 1B, serotype O:8) using the FRT-FLP recombinase system. Transfer of the resulting plasmid pCP1 into the siderophore-deficient strain Y. enterocolitica NF-O (BG 1A) led to no halo on siderophore indicator chrome azurol S (CAS) agar. Transfer of pCP1 into the Y. enterocolitica strain MRS40 (serotype O:9, BG 2; phenotype, CAS negative) led to a CAS halo larger than that of parental strain WA-C, indicating high Ybt production. pCP1 was highly unstable in iron-deficient medium, and no enhanced mouse virulence conferred by MRS40 carrying pCP1 could be detected. To overcome the problem of instability, pCP1 was integrated into the chromosome of MRS40, leading to the formation of a CAS halo comparable to that seen with WA-C and correspondingly to increased mouse virulence. Thus, the core genes of Yen-HPI are sufficient to confer a positive CAS phenotype and mouse virulence to Y. enterocolitica MRS40, BG 2, but are insufficient to confer this phenotype to Y. enterocolitica NF-O, BG 1A.
During the last decade it became evident that evolution of microbial pathogenicity can occur in so-called quantum leaps due to lateral exchange of large mobile genetic elements (e.g., plasmids, phages, or transposons) between different bacterial genera (11). This concept is now supported by the analysis of numerous complete bacterial genome sequences. Bacterial genomes appear to be composed of core regions with species-characteristic homogenous G+C content and of genomic islands with significant deviations in the overall G+C content and codon usage. For genomic islands which carry complex functional cassettes required for pathogenicity (e.g., encoding fimbrial adhesions, protein secretion systems, and high-affinity iron uptake systems) as well as genetic modules mediating site-specific recombination and/or horizontal transfer, the term pathogenicity island (PAI) has been introduced (2). A large number of PAIs have been described as being associated with tRNA genes, indicating a phage-like integration process. Frequently, the original structures of PAIs change because of rearrangements and deletions, leading to their stable integration in the host genome.
Acquisition of a PAI during evolution might also be the reason for the divergence of Yersinia species into three groups with respect to their mouse virulence (as seen in intravenous challenge of BALB/c mice): (i) a group with a high level of pathogenicity (HP group) (50% lethal dose [LD50] <103 bacteria), (ii) a group with a low level of pathogenicity (LP group) (LD50 > 106 bacteria), and (iii) a nonpathogenic group (NP group) (LD50 > 108 bacteria) (1, 6). Members of the NP group belong to Yersinia enterocolitica biogroup (BG) 1A, which is frequently isolated from the environment and from stool samples of healthy humans. The LP group comprises Y. enterocolitica strains of BGs 2 to 5, which are commonly isolated worldwide from enteric sites in humans, pigs, and hares.
Members of the HP and LP groups but not the NP group carry a conserved 70-kb plasmid (pYV, for Yersinia virulence) that enables yersiniae to overcome host defense mechanisms (8). Typical representatives of the HP group are Y. pestis, Y. pseudotuberculosis serotype O:I, and Y. enterocolitica BG 1B, which is commonly isolated in North America but not in Europe. Exclusively the HP group carries a PAI denoted as the “high PAI” (HPI) of Yersinia on the chromosome which is essential for systemic infection and mouse lethality and thus is believed to differentiate the HP from the LP group (5). The HPI is a striking example of a PAI which presumably still carries the characteristic elements of a functional PAI, as there are (i) two attachment sites (attR and attL), (ii) an integrase, and (iii) genes determining a proven virulence factor (namely, the yersiniabactin [Ybt]) siderophore system, which mediates high-affinity ferric iron uptake). Sequence analysis revealed the two-part composition of the HPI: (i) the 30.5-kb core region with a G+C content of 57.5 mol% and (ii) an A+T-rich part of variable size and unknown function (25). In contrast to the A+T-rich part, the core is highly homologous (up to 98% identity) between the two evolutionary HPI lineages (23) represented by Y. enterocolitica (Yen-HPI) (43 kb) and Y. pestis and Y. pseudotuberculosis (36 kb), suggesting that this part is the major virulence determinant of the HPI encoding the Ybt system. This is supported by the presence of mutants of core genes which are attenuated in mouse virulence (3, 4, 22). However, these results do not demonstrate the fact that, in particular, core genes carry the complete set of genes for biosynthesis, transport, and utilization of Ybt, enabling iron provision in the mammalian host and as a consequence enhancing strains' virulence.
Recently we have observed that transfer of the Ybt transport genes of the Yen-HPI core region (fyuA, which encodes the outer membrane receptor for Fe-Ybt and bacteriocin pesticin uptake and irp6-irp7, which encode the ABC transporter of the cytoplasmic membrane) confers Fe-Ybt growth support to Escherichia coli but not to a strain of Y. enterocolitica BG 1A (NP group), suggesting that these three genes do not form a complete set of transport genes sufficient for Fe-Ybt utilization in enterobacteria (3). Probably, a gene encoding a Ybt-specific periplasmatic binding protein is required; however, such a gene has not been identified on the HPI.
Moreover, it has been demonstrated in vitro that the Ybt synthetases HMWP1 (encoded by irp1), HMWP2 (encoded by irp2), and YbtE (encoded by ybtE of Y. pestis and irp5 of Y. enterocolitica) are produced as apo-forms and have to be posttranslationally activated to holo-forms by phosphopantetheinyl (P-pant) (10).
The putative complex function of Yen-HPI core genes could become obvious by transfer of these genes into Y. enterocolitica strains of the LP and NP group, respectively. Therefore, we used the FRT-FLP recombinase system of Posfai et al. (21) to excise and rescue the HPI core genes of Y. enterocolitica WA-C serotype O:8 on a plasmid (pCP1). The Yen-HPI core-carrying plasmid pCP1 was introduced into Y. enterocolitica strains of the LP (MRS40/serotype O:9) and NP (strain NF-O, BG 1A) groups to study Ybt production and mouse virulence.
Using the recently reported integration module of the HPI (26) we were also able to integrate the Yen-HPI core into the genome of Y. enterocolitica MRS40 serotype O:9, resulting in a significantly enhanced mouse virulence of the strain.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown in Luria-Bertani (LB) broth or on LB agar plates (Difco Laboratories, Detroit, Mich.) at 28°C (Yersinia) or 37°C (E. coli). Iron-limited medium (NBD medium) was made by adding 200 μM α-α′-dipyridyl (Sigma, St. Louis, Mo.) to NB medium (8 g of nutrient broth [Difco] and 5 g of NaCl per liter).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli S17-1λpir | Tpr SmrrecA thi pro hsdR− M+ [PR4-2-Tc::Mu:Kanr Tn7] λpir | 19 |
| Y. enterocolitica | ||
| WA-C | Plasmidless derivative of strain WA-314; spontaneous nalidixin-resistant mutant | 12 |
| WA-CS irp1::Kanr | irp1 yersiniabactin synthesis mutant, CAS negative; Nalr Smr Kanr | 20 |
| WA-C Δirp6,irp7 | irp6-irp7 mutant of WA-C; Nalr | 3 |
| WA-C(pYVO:8) | WA-C carrying the pYVO:8 plasmid | This study |
| WA-CH− | WA-C with deleted core region (genes irp9 to fyuA); Kanr Nalr | This study |
| WA-CH−(pYVO:8) | WA-CH− carrying the pYVO:8 plasmid; Nalr | This study |
| WA-CH−(pYVO:8 pCP1, pPIR-K) | WA-CH− carrying the pYVO:8 plasmid, pPIR-K, and pCP1; Nalr Kanr Cmr | This study |
| WA fyuA | fyuA mutant of WA-C; Nalr | 22 |
| MRS40 | BG2, O:9, carrying the pYV40 plasmid, HPI and CAS negative; Nalr | Cornelis |
| MRS40(pCP1 pPIR-K) | Derivative of MRS40 carrying pCP1 and pPIR-K; Nalr Kanr Cmr | This study |
| MRS40INTA | Derivative of MRS40 with integrated pKR527 construct in asn tRNA genes; Kanr | This study |
| MRS40INTAP | Derivative of MRS40 with chromosomally integrated pCP1; Nalr Kanr Cmr | This study |
| NF-O | BG 1A, pYV plasmid and HPI negative; Nalr | 12 |
| NF-O(pCP1, pPIR-K) | NF-O carrying pPIR-K and pCP1; Nalr Kanr Cmr | This study |
| Plasmids | ||
| pCP1 | Carrying irp1-9, fyuA, ybtA genes and 400 nucleotides of intB, R6Kγ ori; Cmr | This study |
| pFT-A | Helper plasmid, carrying the FLP recombinase; Ampr | 21 |
| pKR527 | Suicide vector pKAS32 carrying intB and attP of Y. pestis KIM; Kanr Ampr | 26 |
| pPIR-K | Helper plasmid, carrying the pir gene; Kanr | 21 |
| pRK290B8-5::pYVO:8 | Mobilizable pYVO:8 cointegrate; Tetr | 13 |
| pSG76-K | Suicide vector, carrying R6Kγ ori and FRT site; Kanr | 21 |
| pSG76K/7 | pSG76-K with PCR insert covering the region upstream of irp9; Kanr | This study |
| pST76-C | Suicide vector, carrying pSC-101 ori and FRT site; Cmr | 21 |
| pSV15 | pST76-C with PCR insert covering the region downstream of fyuA; Ori pSC-101 replaced by ori R6Kγ; Cmr | This study |
Ybt production assays.
Ybt production was tested by a cross-feeding assay using Ybt synthesis mutant WA-CS irp1::Kanr as an indicator strain.
Strains were cultivated aerobically to the logarithmic phase (optical density at 600 nm [OD600], 0.6) in NB medium, centrifuged (2,250 × g, 7 min), and washed (0.85% NaCl). The cells were resuspended in NBD medium and grown at 28°C for an additional 24 h (induction of Ybt production). After centrifugation the supernatants were sterilized by filtration. WA-CS irp1::Kanr (OD600, 0.1; 100 μl) was seeded in 100 ml of CDM-H agar (9). Filter papers soaked with 12 μl of the sterile-filtered supernatants were applied to the agar surface. A visible halo around the filter papers was indicative of growth support. Bacteria were stained using a 0.5% solution of TTC (2,3,5,triphenyltetrazoliumchloride).
Strains were also tested for Ybt production by incubation on siderophore indicator (chrome azurol S [CAS]) agar for 2 days at 28°C (31). A clear visible red-orange halo (desferrated CAS is orange) around bacterial colonies was indicative of siderophore production (phenotype, CAS positive).
Pesticin assay.
Pesticin-containing supernatant and the pesticin assay was as previously described (14, 15). LB plates were overlaid with 0.6% agar containing about 107 bacteria (double-layer technique). Pesticin-containing culture supernatant (5 μl) was dropped on these plates, and this was followed by incubation at 37°C for 1 day. Lysis zones were visualized by staining with TTC.
Southern blotting.
Restriction enzyme-digested DNA fragments were resolved through 1% agarose gel, and DNA was transferred to ZETA-Probe BT blotting membrane (Bio-Rad Laboratories) with a vacuum blotter (Pharmacia, Freiburg, Germany). Blots were incubated overnight at 68°C without addition of formamide. Probes and detection were performed according to a DNA labeling and detection kit (Roche, Mannheim, Germany).
Immunoblotting.
Bacteria were grown (OD600, 0.6) in NB medium, centrifuged, washed (0.85% NaCl), resuspended in NBD, and grown for an additional 24 h. Cell lysates were obtained by boiling in solubilization buffer (17), and equal amounts were separated by sodium dodecyl sulfate-7.5% polyacrylamide gel (SDS-7.5% PAGE). Proteins were transferred electrophoretically to nitrocellulose membranes (BA85; Schleicher & Schuell, Inc., Dasserl, Germany) and incubated with an anti-FyuA rabbit antibody (14) directed against the Ybt-pesticin receptor FyuA or an anti-HMWP mouse antiserum directed against Ybt synthesis proteins HMWP1 (encoded by irp1) and HMWP2 (encoded by irp2) (16), respectively (kindly provided from E. Carniel, Institut Pasteur, Paris, France). Secondary peroxidase-conjugated antibodies were used for chemiluminescence detection (ECL kit from Amersham).
PCR amplifications.
PCR was performed in an automated thermal cycler (GeneAmp PCR system 2400; Perkin-Elmer) with TaqI polymerase and different pairs of oligonucleotide sequencing primers (Metabion; Martinsried, Munich, Germany). The initial denaturation step (94°C, 7 min) was followed by 35 cycles of denaturing, annealing, and extension with one final extension step. Annealing and extension temperatures were set according to the primers utilized. PCR amplification products were separated by 1.5% agarose gels.
Rescue of the Yen-HPI core.
The Yen-HPI core, which comprises genes irp1 to irp9, ybtA, and fyuA and 400 bp of the intB gene of the Ybt system, was isolated from Y. enterocolitica WA-C using the method of Posfai et al. (21). Briefly, 520 nucleotides (PCR product using primers C15-303-C15-620) comprising the intergenic region in between irp9 and intB and 400 nucleotides of the 3′end of intB (Fig. 1a) were ligated into suicide vector pSG76-K carrying a pir-dependent replicon, an FRT target sequence, and a kanamycin resistance cassette. The resulting plasmid, p76K/7, was inserted by homologous recombination into the 5′ end of the HPI in WA-C, resulting in WA-C (::p76K/7) (Fig. 1a). As the second suicide vector pST76-C replicated well in strain WA-C (::p76K/7) at 42°C the temperature-sensitive (pSC101) origin of replication was replaced by the pir-dependent origin of pSG76-K. A 620-nucleotide portion of the fyuA gene (obtained using primers fyuA18-HPI1688) was ligated into this suicide vector that carries FRT target sequence and a chloramphenicol resistance cassette as well. The resulting plasmid pSV15 recombined into the right extremity of the HPI, resulting in WA-C (::p76K/7, ::pSV15) (Fig. 1). The expected insertion locations of both suicide plasmids were verified by PCR.
FIG. 1.
(a) Structure of the HPI in Y. pestis and Y. pseudotuberculosis and Yen-HPI. Arrows indicate genes and direction of transcription. The vertical line divides core and variable parts of the HPI. Crosses indicate recombination sites of suicide vectors p76K/7 and pSV15. (b) Position of p76K/7 and pSV15 after recombination into the chromosome of Y. enterocolitica WA-C (::76K/7, ::pSV15). Explanation of designations: oriV, origin of replication, R6Kγ; Kanr, kanamycin resistance cassette; Cmr, chloramphenicol resistance cassette; FRT, FRT site (recognition site for the FLP recombinase).
Excision of the functional core was mediated by site-specific (FRT) recombination using plasmid pFT-A (Ampr), which carries a chlorotetracycline-inducible FLP recombinase. The core genes were rescued from chlorotetracycline-induced WA-C (::p76K/7, ::pSV15, pFT-A) by plasmid isolation and transformed into S17-1λ pir. The resulting plasmid, pCP1, harbors irp1 to irp9, ybtA, and fyuA genes and 400 bases of intB as well as the R6Kγ origin and Cmr marker of plasmid pSV15 (Fig. 1b).
Integration of pCP1 into pKR527.
We retrieved strain MRS40INTA by site-specific integration of pKR527 (26) into the asn RNA genes of MRS40 (see Results). Homologous recombination of pCP1 into pKR527, which resulted in MRS40INTAP, was confirmed by PCR with primer combination 2328 (upstream of fyuA)-int1597 (integrase) (Fig. 2). A PCR product could be detected in the case of the circular pCP1 construct but not in the case of the chromosomal DNA of MRS40INTAP. The chromosomal DNA of MRS40INTAP was isolated, digested with NruI or HpaI, and religated. The ligation was electroporated into S17-1λ pir and selected on chloramphenicol- or kanamycin-containing LB plates. Whereas Nru subclones (derived from NruI digestion) were Cmr only, Hpa clones (derived from HpaI digestion) were found to be Kanr as well (Fig. 2). Both clones were Amps.
FIG. 2.
Integration and position of pCP1 in the chromosome of Y. enterocolitica MRS40 (serotype O:9). Construct pKR527 was inserted (step 1) into the asn tDNA site of MRS40 by integrase-mediated site-specific recombination, resulting in strain MRS40INTA. Homologous recombination (step 2) of pCP1 into the vector part of pKR527 resulted in strain MRS40INTAP. Gray arrows indicate genes and direction of transcription, and small black arrows indicate primer annealing sites. Probes for Southern blot analysis are depicted as boxes. Vertical bars indicate cutting sites for restriction enzymes NruI, HpaI, ClaI, and EcoRI. Not all possible cutting sites are indicated. Explanation of designations: intB, integrase gene of Y. pestis KIM; ΔintB, 400 bp of the integrase gene of Y. enterocolitica WA-C; Kanr, kanamycin resistance cassette; Cmr, chloramphenicol resistance cassette; Ampr, ampicillin resistance cassette; FRT= FRT site (recognition site for the FLP recombinase); oriV, origin of replication, R6Kγ.
PCR revealed that Hpa and Nru clones and chromosomal DNA of MRS40INTAP react positively with primers C15-1620-Frt, C15-1620-in2620, and C15-303-C15-1620, whereas plasmid pKR527 and chromosomal DNA of MRS40INTA did not. In contrast to Hpa clones Nru clones expressed no band with primers aph157-aph606 and primers ybt815-int1597. Due to the circularization of the DNA only Nru clones revealed a product of the expected size with primers S452-aph1233 (Table 2; Fig. 2).
TABLE 2.
PCR results with various primer pairs
| Primer pair | PCR result
|
||||
|---|---|---|---|---|---|
| Nru clone | Hpa clone | MRS40INTA | MRS40INTAP | pKR527 | |
| Frt-C15-1620 | + | + | − | + | − |
| in2620-C15-1620 | + | + | − | + | − |
| C15-303-C15-1620 | + | + | − | + | − |
| ybtA815-int1597 | − | + | + | + | + |
| aph157-aph606 | − | + | + | + | + |
| aph1233-aphS452 | + | − | − | − | − |
Virulence testing.
To test the mouse virulence of WA-C, WA-CH−, and WA-CH−(pCP1, pPIR-K) the mobilizable cointegrate pRK290B8-5::pO8 (13) containing the pYVO:8 virulence plasmid and Tetr was introduced. The three exconjugants were passaged through mice by intraperitoneal (i.p.) injection of 106 CFU in 0.5% physiological salt solution and reisolation of the yersiniae by i.p. lavage 24 h later. Mouse passage resulted in dissociation of the cointegrate (loss of the vector plasmid pKR290B8-5 and maintenance of pYVO:8). pYVO:8-carrying strains WA-C(pYVO:8), WA-CH−(pYVO:8), and WA-CH−(pYVO:8, pCP1, pPIR-K) were selected for challenging BALB/c mice (four per strain) i.p. with 3 × 106 CFU. Strains MRS40, MRS40(pCP1, pPIR-K), and MRS40INTAP (all carrying the virulence plasmid pYV40) were passaged in mice, and afterwards 106 bacteria were injected (i.p.) into BALB/c mice (four to eight mice). Three and four days postinfection mice were sacrificed. To monitor the number of CFU per gram of spleen and liver, the organs were homogenized in phosphate-buffered saline (containing 0.5% Tergitol and 0.5% bovine serum albumin), serially diluted, and plated on LB agar. The number of CFU was determined after incubation at 28°C for 24 h.
RESULTS
Rescue of the HPI core region of Y. enterocolitica HPI.
The G+C-rich region of Yen-HPI of 30.5 kb (designated HPI core [24, 25]) is believed to carry the essential genes of the Ybt system including biosynthesis (irp1 to irp5 and irp9), transport (irp6, irp7, and fyuA), regulation (ybtA), and integration (intB) genes. Whether these core genes or additional genes from the AT-rich 3′ arm are required for Ybt utilization or mouse virulence has not been demonstrated by transfer experiments to date. We therefore retrieved the Yen-HPI core of Y. enterocolitica WA-C serotype O:8 by in vivo cloning using the FRT-FLP system described by Posfai et al. (21). For this purpose two suicide vectors carrying FRT target sites for FLP recombinase were inserted into the chromosome of Y. enterocolitica WA-C serotype O:8. The former (p76K/7) was inserted between irp9 and intB, and the latter (pSV15) was inserted downstream of the fyuA gene (Fig. 1a and Materials and Methods). FLP-mediated recombination resulted in the Yen-HPI core-negative strain WA-CH− and plasmid pCP1. pCP1 is 32.5 kb and carries Yen-HPI core genes (irp1-irp9, ybtA, and fyuA), 400 nucleotides of the intB gene, a chloramphenicol resistance cassette, and a pir-dependent origin of replication (Fig. 1b).
Strain WA-CH− was verified by PCR (using the primer pairs shown in parentheses) to be negative for irp1 (primers i8531-i8730), irp8 and irp9 (primers C15-291-S156), and fyuA (primers f13-f176). The inability of WA-CH− to synthesize Ybt was confirmed using the CAS siderophore indicator agar (31). Colonies that synthesize siderophores form a halo on the agar (CAS-positive phenotype). As expected WA-CH− was incapable of forming such a halo, demonstrating deficiency in Ybt synthesis (CAS-negative phenotype). Moreover, when seeded in iron-restricted CDM-H agar (9) (in which Ybt synthesis and uptake mutants grow only poorly) WA-CH− was unable to be fed by filters soaked with Ybt-containing supernatant of parental strain WA-C, indicating loss of Ybt uptake function.
Yen-HPI core genes of pCP1 are functional.
To confirm the functionality of the core genes, pCP1 and helper plasmid pPIR-K (carrying the pir gene) were transferred into different mutant strains of WA-C.
Presence of pCP1 in WA-CH− or WA-CS irp1::Kanr (Ybt synthesis mutant [20]) restored the CAS-positive phenotype, demonstrating the functionality of Yen-HPI core genes with respect to Ybt synthesis. The CAS halo was even somewhat larger than that of parental strain WA-C (Fig. 3a), which might be due to a gene dosage effect of plasmid pCP1. To exclude a defect of Ybt transport genes (fyuA, irp6, and irp7) pCP1 was transferred into receptor mutant WA fyuA (22) and double-mutant WA-C Δirp6,irp7 (3). It has been demonstrated that these mutants cannot take up Ybt efficiently. In contrast, WA-C Δirp6,irp7 (pCP1, pPIR-K) and WA fyuA (pCP1, pPIR-K) showed enhanced growth in CDM-H agar after addition of filter papers soaked with Ybt-containing supernatant. The efficient uptake of Ybt could also be demonstrated for WA-CH−(pCP1, pPIR-K) seeded in CDM-H agar.
FIG. 3.
CAS halo production of Y. enterocolitica strains. (a) WA-C (BG 1B, serotype O:8), NF-O (BG 1A), NF-O(pCP1, pPIR-K), MRS40 (BG 2, serotype O:9), MRS40INTAP, WA-CH−(pCP1, pPIR-K). (b) MRS40, WA fyuA, MRS40(pCP1; pPIR-K). Strains were grown on siderophore indicator agar (CAS agar) at 28°C for 2 days.
In summary the Yen-HPI core genes of pCP1 are able to complement Ybt synthesis and uptake in different mutants of Y. enterocolitica WA-C demonstrating their functionality.
Mouse virulence of WA-CH−(pCP1, pPIR-K).
After transfer of the pYVO:8 virulence plasmid (see Materials and Methods) into strains WA-C, WA-CH−, and WA-CH−(pPIR-K, pCP1) four BALB/c mice per group were challenged i.p. with 3 × 106 bacteria of resulting strains WA-C(pYVO:8), WA-CH−(pYVO:8), and WA-CH−(pYVO:8, pPIR-K, pCP1), respectively. After 3 days mice were sacrificed and the numbers of CFU per gram of liver and spleen were analyzed. The virulence test was repeated twice, leading to similar results. pCP1 was able to partially restore virulence. The numbers of CFU per gram of liver and spleen of mice challenged with WA-CH−(pYVO:8, pCP1, pPIR-K) were 20 times (spleen) and 10 times (liver) higher than the corresponding numbers in mice infected with WA-CH−(pYVO:8) but did not approach those of mice infected with parental strain WA-C(pYVO:8) (Table 3). Thus, pCP1 contributed to mouse virulence, although not to the degree of the parental strain. As complementation of Ybt transport and synthesis mutants with pCP1 was successful, it is likely that replication dependency of pCP1 on plasmid pPIR-K (carrying a temperature-sensitive origin) or the gene dosage effect of pCP1 affects the full restoration of mouse virulence in WA-CH−(pYVO:8, pCP1, pPIR-K).
TABLE 3.
Mouse virulence assay resultsa
| Strainb | Dose (no. of bacteria) | Time (h postinfection) | Amt of bacteria (CFU/g) in:
|
|
|---|---|---|---|---|
| Spleen | Liver | |||
| WA-C | 3 × 106 | 96 | 4 × 105 | 4 × 104 |
| WA-CH− | 3 × 106 | 96 | 2.5 × 103 | 1.7 × 101 |
| WA-CH(pCP1) | 3 × 106 | 96 | 5 × 104 | 1.5 × 102 |
| MRS40 | 1 × 106 | 96 | 103 | 102 |
| MRS40(pCP1) | 1 × 106 | 96 | single colonies | |
| MRS40c | 1 × 106 | 36 | (3.6 ± 1.9) × 107 | (3.5 ± 1.9) × 104 |
| MRS40 | 1 × 106 | 72 | (3.7 ± 2.0) × 103 | (8.6 ± 5.9) × 101 |
| MRS40INTAPd | 1 × 106 | 72 | (2.1 ± 1.1) × 106 | (2.4 ± 1.8) × 105 |
| MRS40INTAP | 1 × 106 | 96 | (6.9 ± 2.9) × 106 | (6.3 ± 4.1) × 106 |
BALB/c mice received i.p. injection of the indicated number of bacteria. The numbers of CFU in spleen and liver were determined, and values are presented as means ± standard deviations for four mice except as noted.
All WA-C strains harbor the pYVO:8 plasmid. Strains carrying pCP1 also harbor pPIR-K.
DFO (5 mg) was administered 8 h before infection.
CFU determinations based on data from three mice.
Indeed, when WA-CH−(pYVO:8, pCP1, pPIR-K) colonies isolated from spleen and liver were plated on CAS agar, only 10 to 50% of the colonies showed a CAS-positive phenotype.
Transferable traits of pCP1. (i) Transfer of pCP1 into Y. enterocolitica NF-O (NP group) does not lead to a CAS-positive phenotype.
In a following experiment pCP1 was transferred into the nonpathogenic Y. enterocolitica NF-O, a naturally pYV-, HPI-, and CAS-negative strain belonging to BG 1A. The presence of pCP1 was verified by PCR with primers corresponding to genes irp1, irp8, irp9, and fyuA (Table 4). The resulting strain, NF-O(pCP1, pPIR-K), produced no halo when streaked on CAS agar (CAS-negative phenotype) (Fig. 3a). In order to analyze whether the strain is able to express Yen-HPI core genes, whole-cell lysates of NF-O and NF-O(pCP1, pPIR-K) grown in NBD medium were separated by SDS-7.5% PAGE. Western blotting performed with FyuA and HMWP antibodies revealed that NF-O(pCP1, pPIR-K) but not NF-O synthesized the Ybt outer membrane receptor FyuA (Fig. 4a) as well as HMWP1 and HMWP2, indicating expression of irp1 and irp2 (Fig. 4b). Functionality of the FyuA receptor was demonstrated by the pesticin sensitivity assay (see Materials and Methods), revealing NF-O(pCP1, pPIR-K) but not NF-O to be pesticin sensitive (Fig. 4c). A feeding test with WA-CS irp1::Kanr as the detector strain seeded in CDM-H agar and filters soaked with NBD supernatants of NF-O and NF-O(pCP1, pPIR-K), respectively, demonstrated that NF-O(pCP1, pPIR-K) supernatant weakly supported the growth of the irp1 mutant, whereas the supernatant of the parental strain did not (Fig. 4d).
TABLE 4.
Primers used in this study
| Primer | Sequence | Description |
|---|---|---|
| fyu18 | 5′-AGGCGACTGAACGGATAAACA-3′ | |
| HPI1688 | 5′-GTGGCGAATTGGTTTTACTG-3′ | PCR product downstream of fyuA gene |
| C15-303 | 5′-GCCGCATATCGTCCCCCTCTC-3′ | |
| C15-1620 | 5′-TCGCTCTTTGATTCCTCTGT-3′ | PCR product upstream of irp9 gene |
| S156 | 5′-TCTGCTGCTGCCGATTCTCC-3′ | |
| C15-291 | 5′-ACGGTGACATTTCCCTTTTC-3′ | For irp8 and irp9 genes |
| i8513 | 5′-TGAATCGCGGGTGTCTTATGC-3′ | |
| i8730 | 5′-TCCCTCAATAAAGCCCACGCT-3′ | For irp1 gene |
| f13 | 5′-AAAAAGCTTGACCGTTATCGCCATTCTGC-3′ | |
| f176 | 5′-CAGAGGATAAAGCCGTGTCAT-3′ | For fyuA gene |
| C15-205 | 5′-TACAGGCAGGTTCCCGATGAC-3′ | |
| int520 | 5′-ACATCCTTGCGAATCCTTATC-3′ | intB fragment as probe for Southern blotting |
| 2328 | 5′-CTTTCTCGTTCCTTTCACTCC-3′ | |
| aph157 | 5′-CTCACCGAGGCAGTTCCATAG-3′ | |
| aph606 | 5′-CGACCATCAAGCATTTTATCC-3′ | |
| aph1233 | 5′-ATCTGCAGGAAAGCCACGTTGTGTCTC-3′ | |
| asn468 | 5′-CCGTATGTCACTGGTTCG-3′ | |
| bla767 | 5′-AATGAAAGTGGTCGCAAAGAC-3′ | |
| C15-205 | 5′-TACAGGCAGGTTCCCGATGAC-3′ | |
| Frt | 5′-CCGAAGTTCCTATTCTCTAG-3′ | |
| in2620 | 5′-GAAAACGGGGGGCGAAGAAG-3′ | |
| int1597 | 5′-AAAGGTACCCCTGTGGAGGTGGTGGTAAT-3′ | |
| S452 | 5′-CCCCGGCGATGGTGGATAAAC-3′ | |
| ybtA815 | 5′-CATTGCGCTGGGTGCCTATC-3′ | Integration and recombination of pKR527 and pCP1 |
FIG. 4.
(a and b) Immunoblots of NF-O and NF-O(pCP1, pPIR-K) using a FyuA-specific antibody (a) and an HMWP1- and HMWP2-specific antibody (kindly provided by E. Carniel, Institut Pasteur) (b). Whole-cell lysates of the strains grown in NBD medium were subjected to SDS-7.5% PAGE and transferred to nitrocellulose. Arrows indicate FyuA (a) and HMWPs (b). (c) Pesticin assay. NF-O and NF-O(pCP1; pPIR-K) were seeded in an overlay agar, and 5 μl of a pesticin preparation was applied on the agar surface. Pesticin sensitivity (growth inhibition, clear halo) was indicative of a functional FyuA receptor. (d) Feeding test with reporter strain WA-CS irp1::Kanr seeded in iron-limited CDM-H agar. Filters soaked with sterilized NBD supernatants of NF-O and NF-O(pPIR-K, pCP1) were applied to the agar surface. Bacteria were stained using a 1% solution of TTC.
Thus, Yen-HPI core genes are expressed in NF-O(pCP1, pPIR-K) but result in low Ybt production at best.
(ii) Transfer of pCP1 into Y. enterocolitica MRS40 (LP group) leads to a CAS-positive phenotype.
Y. enterocolitica MRS40 serotype O:9 belongs to the LP group of strains of Yersinia (BG 2) that harbor the O:9 pYV-plasmid but not the HPI and therefore are CAS negative (12). Electroporation of pCP1 and helper plasmid pPIR-K into MRS40 led to MRS40(pCP1, pPIR-K). A large CAS halo around the colony was indicative of siderophore production in MRS40(pCP1, pPIR-K). The halo was even larger than that of WA-C or of receptor mutant WA fyuA (Fig. 3b). To confirm synthesis of Ybt in MRS40(pCP1, pPIR-K), the growth response of the irp1 mutant seeded in CDM-H agar was tested. As expected NBD supernatant of MRS40(pCP1, pPIR-K) but not of MRS40 supported growth of WA-CS irp1::Kanr significantly.
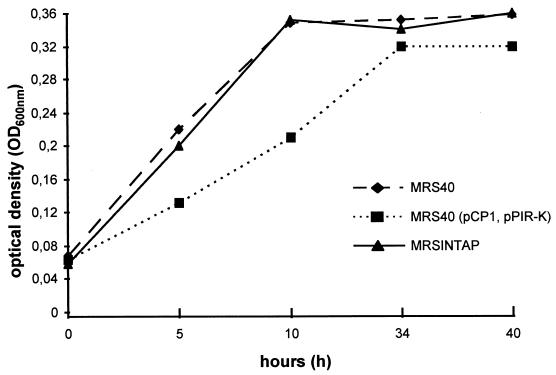
MRS40 and MRS40(pCP1, pPIR-K) were inoculated in NBD medium, and growth rates were measured at an OD600 for 40 h (28°C). Surprisingly MRS40(pCP1, pPIR-K) showed a decreased growth rate compared to MRS40, implying that even though pCP1 confers a Ybt synthesis phenotype, this is of no benefit to MRS40(pCP1, pPIR-K) under these growth conditions (Fig. 5). After growth in chloramphenicol-containing NBD broth and subsequent plating on CAS agar, more than 90% of the colonies were CAS negative. Interestingly, these colonies carried plasmids about 5 kb smaller than pCP1 (as estimated by agarose gel electrophoresis). Obviously deletions occurred in pCP1, resulting in the Ybt biosynthesis deficiency.
FIG. 5.
Growth rate of MRS40, MRS40(pPIR-K, pCP1), and MRS40INTAP in NBD medium. Strains were grown in NB medium (overnight) with addition of antibiotics. Antibiotics containing NBD medium were inoculated (1:100), and bacteria were shaken at 28°C (200 rpm). The OD600 of the growing bacteria was measured.
MRS40(pCP1, pPIR-K) does not express mouse virulence.
The HPI is thought to be one of the main factors conferring mouse virulence to Yersinia spp. Although Y. enterocolitica MRS40(pCP1, pPIR-K) did not show better growth than its parental strain under iron-limited conditions in vitro it might benefit from pCP1 under in vivo conditions during mouse infection. Four BALB/c mice were infected (i.p.) with 106 bacteria of mouse-passaged MRS40 and MRS40(pCP1, pPIR-K), respectively. At 54 h postinfection no mice showed any clinical symptoms. The spleen and liver were taken from two mice, and two mice were used for i.p. lavage. About 102 CFU/g could be detected in liver and spleen of mice challenged with MRS40 (Table 3). In the case of MRS40(pCP1, pPIR-K) only single colonies could be recovered from mice (2 CFU in liver, 11 CFU in spleen, and 4 CFU by i.p. lavage). To analyze the presence of pCP1 the colonies were plated on CAS agar. Both colonies from the liver and three out of four colonies from the lavage were found to be CAS positive. Colonies from spleen were all CAS negative.
This result suggests that pCP1 was not able to increase the pathogenicity of an O:9 strain but rather decreased it. As colonies from spleen were CAS negative, the result might reflect the instability of plasmid pCP1 in strain MRS40(pCP1, pPIR-K) as has been observed after growth in NBD broth.
Integration of pCP1 into the chromosome of MRS40.
As in vitro subcultivation of MRS40(pCP1, pPIR-K) in iron-limited medium revealed the rapid loss of pCP1, we developed a strategy to insert pCP1 adjacent to the asn tRNA gene of MRS40 according to the location of the HPI in strain WA-C.
First MRS40 was electroporated with plasmid pKR527 (Table 1), which carries the intB gene (encoding the integrase of the HPI) and the attP site of Y. pestis HPI (26).
Integrase-mediated site-specific recombination of pKR527 into the 3′ end of asn tRNA (attB) gene of MRS40 was confirmed using primers asn468 and C15-205 (Fig. 2). The resulting strain was designated MRS40INTA. As yersiniae carry three highly homologous asn tRNA genes, Southern blotting was performed using the first 300 bp of intB (primers int520-C15-205) as a probe and chromosomal DNA of MRS40INTA and MRS40 cut with ClaI (cutting inside the integrase) as a template. Whereas MRS40 revealed no band, three bands could be detected for MRS40INTA. This indicates that pKR527 is inserted into each of the three asn tRNA genes (data not shown). Homologous recombination of the 400-bp fragment of intB (ΔintB) present on pCP1 and intB of the chromosomally inserted pKR527 was expected to lead to integration of pCP1 into the chromosome of MRS40INTA. Therefore, pCP1 was electroporated into strain MRS40INTA. The resulting strain, MRS40INTAP, expressed a CAS-positive phenotype with a halo size equal to that of WA-C (Fig. 3a) as well as a Cmr fragment that is present on pCP1. PCR and Southern blotting revealed that no recombination via the intB fragment had occurred (data not shown). As both pKR527 and pCP1 carry the R6Kγ origin it was likely that insertion of pCP1 occurred via these vector sequences. This could be confirmed by PCR and DNA subcloning (see Materials and Methods). In addition Southern blotting was performed using ClaI-cut chromosomal DNA of MRS40, MRS40INTA, and MRS40INTAP as templates and suicide vector pKAS32 as a probe. Whereas MRS40 showed no visible band, MRS40INTA as well as MRS40INTAP expressed three bands (due to the integration of pKR527 into the three asn tRNA genes). Two of the bands were of the same size, and one was larger in MRS40INTAP than in MRS40INTA. The larger band not only reacted positively with the vector probe but also with an irp5 and fyuA probe (Fig. 2), confirming the recombination of pCP1 into the chromosomally integrated pKR527 (data not shown).
MRS40INTAP expresses mouse virulence.
In NBD medium strain MRS40INTAP showed growth rates similar to those of MRS40 (Fig. 5). As the strain was CAS positive, with a CAS halo similar in size to that of WA-C (Fig. 3a), mouse virulence was tested. Strains MRS40 and MRS40INTAP were first passaged through BALB/c mice. (i) Four BALB/c mice were treated i.p. with 5 mg of desferrioxamine (DFO) and (8 h later) challenged with 106 CFU of MRS40 (siderophore-mediated mouse virulence), (ii) four BALB/c mice were challenged (i.p.) with 106 CFU of MRS40 (negative control), and (iii) eight BALB/c mice were challenged (i.p.) with 106 CFU of MRS40INTAP.
Treatment with 5 mg of DFO provides MRS40 with iron sufficient for mouse virulence. This control experiment should demonstrate that MRS40, like other serotype O:9 strains (27), is able to become mouse virulent under these conditions. At 36 h after MRS40 injection DFO-treated mice were highly colonized (3.6 × 107 and 3.5 × 104 CFU/g of spleen and liver, respectively) (Table 3).
At 72 h postinfection mice challenged with MRS40 alone showed no clinical symptoms of disease, whereas mice challenged with MRS40INTAP displayed symptoms of severe infection (disheveled fur, apathy, and impaired mobility). One mouse died. Spleen and liver samples from the four mice infected with MRS40 and three mice infected with MRS40INTAP were analyzed for CFU. About 103 CFU/g (Table 3) could be detected in spleen and 8.6 × 101 CFU/g could be detected in liver of mice infected with the parental MRS40 strain.
In contrast mice challenged with MRS40INTAP developed up to 106 CFU/g of spleen and 105 CFU/g of liver 72 h postinfection. The remaining four mice infected with MRS40INTAP were sacrificed 96 h postinfection. About 106 CFU was determined per g of spleen and liver (Table 3). One hundred fourteen clones of MRS40INTAP isolated from spleen and liver of the infected mice were streaked on CAS agar, and all but one were found to be CAS positive. The CAS-negative strain was still positive in PCR for the gene irp2. Thus, the CAS-negative phenotype was not attributable to excision of the whole Yen-HPI core.
DISCUSSION
The main objective of this study was the isolation of the Yen-HPI core of Y. enterocolitica WA-C serotype O:8 for transfer experiments. This approach should elucidate the following issues. (i) Is Yen-HPI core sufficient to transfer the Ybt production phenotype to HPI-negative Y. enterocolitica strains of different BGs? (ii) Is Yen-HPI core sufficient to enhance mouse virulence in Y. enterocolitica of LP group? (iii) Is Ybt production affected by the extrachromosomal location of the Yen-HPI core region?
Using the FRT-FLP recombinase system (21) we succeeded in excising and rescuing in vivo the Yen-HPI core of Y. enterocolitica WA-C serotype O:8 as plasmid pCP1. The resulting strain, WA-CH−, carried the corresponding truncated island Yen-HPI-Δcore and was CAS negative. pCP1 was proven to carry functional genes of the Ybt system (see Results). Moreover, we could show that functional expression of Yen-HPI core genes is independent of chromosomal or extrachromosomal location. This is similar to the LEE island of enteropathogenic E. coli strains but in contrast to several virulence plasmids regulated by H-NS or H-NS-like proteins such as pINV of enteroinvasive E. coli or pYV of Y. pestis (7, 18, 33), which are shut down when integrated into the chromosome because of disturbed regulation or insertional gene inactivation.
The CAS halo of WA-CH−(pPIR-K, pCP1) was larger than that of parental strain WA-C, which may be assigned to the gene dosage effect of the plasmid. However, introduction of pCP1 into WA-CH−(pYVO:8) enhanced mouse virulence but not to the level of the parental strain [in spleen, 5 × 104 CFU of WA-CH−(pYVO:8, pCP1, pPIR-K) per g versus 4 × 105 CFU of WA-C(pYVO:8) per g (Table 3)]. This might be explained by progressive loss of pCP1 or inactivation of HPI core genes in WA-CH−(pYVO:8, pCP1, pPIR-K) during the course of infection. On the other hand it is also conceivable that hyperproduction of Ybt by WA-CH−(pPIR-K, pCP1) as observed in vitro (Fig. 3a) may be not beneficial for the infecting strain.
Introduction of pCP1 into LP group Y. enterocolitica MRS40 also resulted in Ybt hyperproduction as demonstrated by CAS halo size (Fig. 3b). Surprisingly, strain MRS40(pCP1, pPIR-K) did not benefit from the Ybt system when growing under iron-restricted in vitro conditions in comparison to the parental strain MRS40 (Fig. 5). As Y. enterocolitica is endowed with diverse iron uptake systems (28, 30) which are sufficient for iron provision during growth in NBD medium, the presence of pCP1 is evidently not favorable for growth of MRS40(pCP1, pPIR-K) under these conditions. This seems also to be the case for MRS40(pCP1, pPIR-K) when injected into mice, as the exconjugant was attenuated. For this reason we developed a technique to integrate the Yen-HPI core adjacent to asn tRNA gene in the genome of MRS40 (which corresponds to the integration site of the HPI in parental strain WA-C), resulting in strain MRS40INTAP. Phenotypical analysis of MRS40INTAP revealed a CAS halo similar to that of strain WA-C. More strikingly, strain MRS40INTAP was shown to be more virulent to mice than MRS40. Comparison of CFU per gram of spleen and liver 72 h after mouse infection indicated that MRS40INTAP grew to an even higher bacterial concentration than HPI donor strain WA-C(pYVO:8) (Table 3). At 96 h after infection with MRS40INTAP, mice developed severe clinical symptoms, with about 106 CFU/g of spleen and liver. In conclusion Y. enterocolitica strain MRS40 expresses higher virulence after chromosomal integration of the HPI core of Y. enterocolitica serotype O:8. Moreover, these transfer experiments also show that Yen-HPI is rather a PAI than a fitness island. However, we cannot exclude the possibility that Yen-HPI core genes such as transcriptional regulator YbtA are also involved in regulation of additional virulence genes on the MRS40 genome. The provocative question remains why the HPI is restricted naturally to Y. enterocolitica BG 1B and is absent in other BGs although it is widely distributed within the family of Enterobacteriaceae. In general, evolution of bacterial pathogenicity is not driving towards increasing mouse virulence but towards host-specific adaptation allowing coexistence of host and pathogen. In the case of the LP group (Y. enterocolitica BGs 2 to 4) swine and wild boar are the natural hosts, whereas the HP group (Y. enterocolitica BG 1B) naturally infects a broad host spectrum, ranging from rodents to carnivores (29, 32). It appears to be attractive to compare the outcome of infection of swine with strain MRS40 versus MRS40INTAP.
A striking result is that Yen-HPI core transfer into strain NF-O (NP group, Y. enterocolitica BG 1A) is not sufficient to confer the CAS-positive phenotype in spite of irp1, irp2, and fyuA gene expression. This deficiency in Ybt production in strain NF-O could have different reasons. (i) A possibility which is in line with the HMWP and FyuA synthesis of strain NF-O(pCP1, pPIR-K) (Fig. 4a and b) is an insufficient precursor production (e.g., chorismate and isochorismate) for yersiniabactin to enable a CAS-positive phenotype. (ii) Strain NF-O might not carry a gene encoding an appropriate P-pant transferase (e.g., EntD-like enzyme) and thus is unable to activate posttranscriptionally the Ybt synthases (10). Interestingly, transfer of pCP1 into an E. coli entD mutant did not lead to a CAS-positive phenotype of the exconjugant (unpublished data), which supports this suggestion.
In summary we have demonstrated that transfer of Yen-HPI core genes of Y. enterocolitica WA-C serotype O:8 is sufficient to confer a CAS-positive phenotype and a mouse virulence trait to strain MRS40 of Y. enterocolitica (LP group). Intriguingly, Y. enterocolitica NF-O (NP group) remains CAS negative after reception of pCP1, indicating that Yen-HPI core is not able on its own to confer the CAS-positive phenotype because of lack of essential genes.
Acknowledgments
We thank A. Rakin, E. Carniel, and G. Cornelis for kindly providing plasmid pKR527, HMWP antibody, and strain MRS40 as well as C. Nölting, D. Brem, W.-D. Hardt, and S. Schubert for critical reading of the manuscript.
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (He 1297/8-2).
Editor: D. L. Burns
REFERENCES
- 1.Autenrieth, I. B., R. Reissbrodt, E. Saken, R. Berner, U. Vogel, W. Rabsch, and J. Heesemann. 1994. Desferrioxamine-promoted virulence of Yersinia enterocolitica in mice depends on both desferrioxamine type and mouse strain. Infect. Immun. 169:562-567. [DOI] [PubMed] [Google Scholar]
- 2.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschäpe, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brem, D., C. Pelludat, A. Rakin, C. A. Jacobi, and J. Heesemann. 2001. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147:1115-1127. [DOI] [PubMed] [Google Scholar]
- 4.Carniel, E., A. Guiyoule, I. Guilvout, and O. Mercereau Puijalon. 1992. Molecular cloning, iron-regulation and mutagenesis of the irp2 gene encoding HMWP2, a protein specific for the highly pathogenic Yersinia. Mol. Microbiol. 6:379-388. [DOI] [PubMed] [Google Scholar]
- 5.Carniel, E., I. Guilvout, and M. Prentice. 1996. Characterization of a large chromosomal high-pathogenicity island in biotype 1B Yersinia enterocolitica. J. Bacteriol. 178:6743-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, P. B. 1975. Pathogenicity of Yersinia enterocolitica for mice. Infect. Immun. 11:164-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colonna, B., M. Casalino, P. A. Fradiani, C. Zagaglia, S. Naitza, L. Leoni, G. Prosseda, A. Coppo, P. Ghelardini, and M. Nicoletti. 1995. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J. Bacteriol. 177:4703-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R. 1998. The Yersinia deadly kiss. J. Bacteriol. 21:5495-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flossmann, K. D., C. Grajetzki, and H. Rosner. 1985. Nachweis von Eisen-Transport-Aktivität in Pasteurella multocida-Kulturen. J. Basic Microbiol. 25:559-567. [DOI] [PubMed] [Google Scholar]
- 10.Gehring, A. M., E. DeMoll, J. D., Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron aquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 11.Groisman, E. A., and H. Ochman. 1996. Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791-794. [DOI] [PubMed] [Google Scholar]
- 12.Heesemann, J. 1987. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol. Lett. 48:229-233. [Google Scholar]
- 13.Heesemann, J., and R. Laufs. 1983. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J. Bacteriol. 155:761-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heesemann, J., K. Hantke, T. Vocke, E. Saken, A. Rakin, I. Stojiljkovic, and R. Berner. 1993. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol. Microbiol. 8:397-408. [DOI] [PubMed] [Google Scholar]
- 15.Hu, P. C., G. C. Yang, and R. R. Brubaker. 1972. Specificity, induction, and absorption of pesticin. J. Bacteriol. 112:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kooi, C., and P. A. Sokol. 1995. Characterization of monoclonal antibodies of Yersinia enterocolitica iron regulated proteins. Can. J. Microbiol. 41:562-571. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 19.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rakin, A., E. Saken, D. Harmsen, and J. Heesemann. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13:253-263. [DOI] [PubMed] [Google Scholar]
- 23.Rakin, A., P. Urbitsch, and J. Heesemann. 1995. Evidence for two evolutionary lineages of highly pathogenic Yersinia species. J. Bacteriol. 177:2292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakin, A., S. Schubert, C. Pelludat, D. Brem, and J. Heesemann. 1999. The high-pathogenicity island of yersiniae, p. 77-90. In J. B. Kaper and J. Hacker (ed.), Pathogenicity islands and other mobile virulence factors, 1st ed. American Society for Microbiology, Washington, D.C.
- 25.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakin, A., P. Schropp, C. Noelting, and J. Heesemann. 2001. Integrative module of the high-pathogenicity island of Yersinia. Mol. Microbiol. 39:407-415. [DOI] [PubMed] [Google Scholar]
- 27.Robins Browne, R. M., and J. K. Prpic. 1985. Effects of iron and desferrioxamine on infections with Yersinia enterocolitica. Infect. Immun. 47:774-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saken, E., A. Rakin, and J. Heesemann. 2000. Molecular characterization of a novel siderophore-independent iron transport system in Yersinia. Int. J. Med. Microbiol. 290:51-60. [DOI] [PubMed] [Google Scholar]
- 29.Schiemann, D. A., and C. A. Fleming. 1981. Yersinia enterocolitica isolated from the throats of swine in eastern and western Canada. Can. J. Microbiol. 27:1326-1333. [DOI] [PubMed] [Google Scholar]
- 30.Schubert, S., D. Fischer, and J. Heesemann. 1999. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J. Bacteriol. 181:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 32.Shayegani, M., and L. M. Parson. 1987. Epidemiology and Pathogenicity of Yersinia enterocolitica in New York State. Contr. Microbiol. Immunol. 9:41-47. [PubMed] [Google Scholar]
- 33.Zsigray, R. M., J. B. Hopper, K. Zugowski, and W. R. Chesbro. 1985. Repression of the virulence of Yersinia pestis by an F′ plasmid. Infect. Immun. 47:670-673. [DOI] [PMC free article] [PubMed] [Google Scholar]