Abstract
Attenuated Salmonella enterica serovar Typhimurium expressing recombinant antigens from other pathogens elicits primarily a Th1-type dominant immune response to both recombinant and Salmonella antigens. The immunogenicity and appropriate subcellular location of the recombinant antigen in the Salmonella vaccine strain may contribute to augmenting immune responses by facilitating adequate exposure of recombinant antigen to antigen-presenting cells for processing. To allow for secretion from gram-negative bacteria and overexpression of antigen, a DNA fragment encoding a highly antigenic α-helical region of PspA (pneumococcal surface protein A) was subcloned downstream from the β-lactamase signal sequence in the multicopy Asd+ pYA3493 vector to create pYA3494. pYA3493 was derived from a class of Asd+ vectors with reduced expression of Asd to minimize selective disadvantage and enhance immunization of expressed recombinant antigens. The S. enterica serovar Typhimurium vaccine strain was constructed by the introduction of deletion mutations Δcrp-28 and ΔasdA16. Approximately 50% of the recombinant PspA (rPspA) expressed in a Salmonella strain harboring pYA3494 was detected in the combined supernatant and periplasmic fractions of broth-grown recombinant Salmonella. After a single oral immunization in BALB/c mice with 109 CFU of the recombinant Salmonella vaccine strain carrying pYA3494, immunoglobulin G (IgG) antibody responses were stimulated to both the heterologous antigen rPspA and Salmonella lipopolysaccharide (LPS) and outer membrane proteins (OMPs). About half, and even more at later times after immunization, of the antibodies induced to rPspA were IgG1 (indicating a Th2-type response), whereas 60 to 70% of the antibodies to LPS and 80 to 90% of those to OMPs were IgG2a (indicating a Th1-type response). A sublethal infection with Streptococcus pneumoniae WU2 boosted PspA antibody levels and maintained IgG2a/IgG1 ratios similar to those seen before the challenge. Oral immunization with Salmonella-PspA vaccine protected 60% of immunized mice from death after intraperitoneal challenge with 50 times the 50% lethal dose of virulent S. pneumoniae WU2.
Orally administered Salmonella enterica serovar Typhimurium colonizes the gut-associated lymphoid tissue (Peyer's patches) and the secondary lymphatic tissues, including the liver and spleen, to elicit anti-Salmonella immune responses during infection of the mouse. (17). The immune responsiveness to orally administered Salmonella has been applied to develop live attenuated oral Salmonella vaccines (13). Attenuated Salmonella vaccines have been constructed by introduction of mutations in the genes required for virulence, including the cyclic AMP receptor protein gene (crp) (12). Crp is a global regulator involved in a variety of biological functions, including carbohydrate utilization (4). Attenuated Salmonella vaccine strains have been genetically modified to express another pathogen's antigen (s) specified by multicopy plasmids. These recombinant vaccines induce immunity to the pathogen whose antigen gene is expressed as well as to Salmonella. It is essential that the antigen-specifying plasmids in Salmonella vaccines are stably maintained during the in vivo colonization process. A balanced-lethal host-vector system based on the essential bacterial gene for aspartate β-semialdehyde dehydrogenase (asd) has been used to specify recombinant antigens from Asd+ plasmids that are retained in vivo in Salmonella vaccine strains with the asd gene deleted (16, 36).
Streptococcus pneumoniae is a human pathogen that causes life-threatening diseases, including community-acquired pneumonia, otitis media, meningitis, and bacteremia, in persons of all ages (35). S. pneumoniae is the leading cause of childhood pneumonia worldwide, resulting in about 3 million deaths per year (21). The recent emergence of antibiotic-resistant strains has the potential to threaten the treatment of pneumococcal disease in the near future (5). Thus, the development of a safe, effective, and lower-cost antipneumococcal vaccine is urgently needed. Capsular polysaccharide-based pneumococcal vaccines are currently available and are moderately effective. A 23-valent pneumococcal polysaccharide vaccine is recommended for the prevention of infection in adults (48), and a 7-valent conjugated polysaccharide vaccine is licensed for use in children (49). However, vaccination with the pneumococcal polysaccharide vaccine does not reduce the frequency of hospitalization, costs, and mortality caused by pneumococcal pneumonia (23), which reinforces the need for effective new vaccines.
Studies on the protective efficacy of subunit vaccines may further the development of a more protective pneumococcal vaccine. The pneumococcal PspA (pneumococcal surface protein A) protein has been evaluated and considered to be a pneumococcal vaccine candidate because of its immunogenicity and protection of mice against challenge with virulent S. pneumoniae (6, 8, 9, 25). Native PspARx1 (PspA originating from S. pneumoniae strain Rx1) contains several functional domains: an N-terminal signal sequence, an α-helical region, a proline-rich domain, 10 tandem-repeat choline-binding regions, and a 17-amino-acid residue carboxy terminus. Pneumococcal protection assays with mice immunized with various recombinant PspARx1 oligopeptides showed that the α-helical domain contains the protective epitopes (7). In a previous study, mice orally immunized with an S. enterica serovar Typhimurium vaccine strain expressing a recombinant PspARx1 (from the ATG start codon specifying the native signal sequence, the entire α-helical domain, and up to the fifth tandem repeat) showed PspA-specific immune responses and were protected against challenge with virulent S. pneumoniae (37). Expression of recombinant PspA (rPspA) in this recombinant Salmonella vaccine strain was somewhat toxic, such that the high-copy-number plasmid pYA3193 (pUC ori) was relatively unstable. Thus, approximately 50% of cells lost the plasmid after 24 h of growth as a standing culture in the presence of diaminopimelic acid (DAP). This phenomenon forced us to construct an improved plasmid vector to enable stable expression of rPspA in attenuated Salmonella. An additional goal of our research is to construct recombinant attenuated Salmonella vaccines that induce higher immune responses to the foreign expressed antigen than to Salmonella antigens.
In this work, we constructed a stable multicopy Asd+ antigen expression vector encoding the β-lactamase signal sequence fused in frame to the immunogenic α-helical region of PspA. This plasmid was designed to translocate PspA into the periplasmic space of the Salmonella vaccine strain, although about 25% of the synthesized PspA reached the supernatant fluid without cell lysis. We report the immunogenicity, type of immune responses, and protection against both Salmonella and S. pneumoniae in mice immunized with a Salmonella vaccine expressing rPspA by an improved antigen expression system.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteriophage P22HTint (46) was used for generalized transduction. Escherichia coli and S. enterica serovar Typhimurium cultures were grown at 37°C in Lennox broth (29) or Luria-Bertani (LB) broth or on LB agar (1). MacConkey agar (Difco, Detroit, Mich.) supplemented with 1% sugar was used for fermentation assays. When required, antibiotics were added to culture media at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 15 μg/ml. DAP was added (50 μg/ml) for the growth of Asd− strains (36). LB agar containing 5% sucrose was used for sacB gene-based counterselection in allelic exchange experiments (18). S. pneumoniae WU2 was cultured on brain heart infusion agar containing 5% sheep blood or in Todd-Hewitt broth plus 0.5% yeast extract (6).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Derivation, source, or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| χ6212 | F− λ− φ80 Δ(lacZYA-argF) endA1 recA1 hsdR17 deoR thi-1 glnV44 gyrA96 relA1 ΔasdA4 | 36 |
| MGN-617 | thi-1 thr-1 leuB6 fhuA21 lacY1 glnV44 ΔasdA4 recA1 RP4 2-Tc::Mu [λpir]; Kmr | 44 |
| S. enterica serovar Typhimurium | ||
| χ3339 | SL1344 hisG | 22 |
| χ4550 | SR-11 gyrA1816 Δcrp-1 ΔasdA1 Δ(zhf-4::Tn10) Δcya-1 | 47 |
| χ4746 | χ3339 nadA540::Tn10 Δ(galE-chl-uvrB)-1005; Tetr | Lab collection |
| χ8499 | hisG Δcrp-28 | χ3339 |
| χ8501 | hisG Δcrp-28 ΔasdA16 | χ8499 |
| χ8554 | hisG ΔasdA16 | χ3339 |
| χ8599 | hisG ΔasdA16 atrB13::MudJ | χ8554 |
| JF2430 | LT2 atrB13::MudJ | 15 |
| S. pneumoniae WU2 | Wild-type virulent, encapsulated type 3 | 6 |
| Plasmids | ||
| pYA3193 | Asd+ vector harboring 1.5-kb C-terminally truncated pspA gene; pUCori | 37 |
| pYA3333 | Asd+; pBRori | Lab collection |
| pYA3334 | Asd+; pUCori | Lab collection |
| pYA3341 | Asd+; pUCori | This study |
| pYA3342 | Asd+; pBRori | This study |
| pYA3493 | pYA3342 derivative β-lactamase signal sequence-based periplasmic secretion plasmid | This study |
| pYA3494 | 0.7-kb DNA encoding the α-helical region of PspA in pYA3493 | This study |
| pYA3496 | 0.7-kb DNA encoding the α-helical region of PspA in pYA3342 for the expression of His-tagged PspA | This study |
| pBR322 | Cloning vector; Apr Tcr | 3 |
| pMEG-443 | Recombinant suicide plasmid to generate Salmonella ΔasdA16 mutant; Apr Cmr | 27 |
| pMEG-493 | Recombinant suicide plasmid to generate Salmonella Δcrp-28 mutant; Apr Cmr | Megan Health Inc. |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
General DNA procedures.
DNA manipulations were carried out as described by Sambrook et al. (45). Transformation of E. coli and Salmonella was done by electroporation (Bio-Rad, Hercules, Calif.). Transformants containing Asd+ plasmids were selected on L agar plates without DAP. Only clones containing the recombinant plasmids were able to grow under these conditions. Transfer of recombinant suicide plasmids to Salmonella was accomplished by conjugation using E. coli MGN-617 (Asd−) (44) as the plasmid donor. Bacteriophage P22HT int-mediated general transduction was performed by standard methods (52). PCR amplification was employed to obtain DNA fragments for cloning and for verification of chromosomal deletion mutations. The PCR conditions were as follows: denaturation at 95°C for 20 s, primer annealing at 55°C for 20 s, polymerization at 72°C for 2 min, and a final extension at 72°C for 10 min. Nucleotide sequencing reactions were performed using ABI Prism fluorescent Big Dye terminators according to the instructions of the manufacturer (PE Biosystems, Norwalk, Conn.).
Characterization of phenotype.
MacConkey agar supplemented with 1% maltose was used to detect the phenotypes of Salmonella crp mutants. Motility was evaluated by observing Salmonella spread on a semisolid medium composed of 1% casein enzyme hydrolysate, 0.5% NaCl, and 0.5% agar. Triphenyltetrazolium chloride (50 μg/ml) was added to motility medium to observe Salmonella as red cells. The presence of the ΔasdA16 mutation in Salmonella was confirmed by inability of the strain to grow on media without DAP (36). Lipopolysaccharide (LPS) profiles of Salmonella strains were examined by previously described methods (24).
SDS-PAGE and immunoblot analyses.
Protein samples were boiled for 5 min and then separated by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein bands were visualized by Coomassie brilliant blue R250 (Sigma, St. Louis, Mo.) staining. For immunoblotting, proteins separated by SDS-PAGE were transferred eletrophoretically to nitrocellulose membranes. The membranes were blocked with 3% bovine serum albumin in 10 mM Tris-0.9% NaCl (pH 7.4) and incubated with mouse monoclonal antibodies specific for PspA (Xi126) (32), OmpC (50), or β-galactosidase (Sigma) and then with a peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Bio-Rad). Immunoreactive bands were detected by the addition of 4-chloro-1-naphthol (Sigma) in the presence of H2O2. The reaction was stopped after 2 min by washing with several large volumes of deionized water.
Purification of rPspA.
For overexpression of His6-tagged PspA, a fragment of the pspA gene specifying the α-helical region from amino acid residue 3 to 257 of the mature PspA protein of S. pneumoniae Rx1 was PCR amplified from pYA3193 (37) template DNA using a pair of primers (N terminal, 5′CCGGAATTCATCACCATCACCATCACTCTCCCGTAGCCAGTCAGT3′; C terminal, 5′GGGAAGCTTCTATTATTCTACATTATTGTT3′). The 0.8-kb amplified fragment was then cloned into the pYA3342 vector, resulting in pYA3496 (Table 1). The N-terminal primer contains an EcoRI site (underlined) and six consecutive histidine codons (alternate use of CAT and CAC; boldface) for His6 tagging at the N terminus. The C-terminal primer specifies two consecutive stop codons (TAA TAG; boldface) followed by a HindIII site (underlined). In-frame cloning was confirmed by nucleotide sequencing. E. coli χ6212 harboring pYA3496 expressed a large amount of soluble His6-tagged rPspA in its cytoplasm. According to the protocol of the manufacturer (Qiagen, Valencia, Calif.), rPspA protein was purified by an affinity purification process with Ni2+-nitrilotriacetic acid-agarose support. The protein purity was verified by Coomassie blue staining of SDS-polyacrylamide gels, and the total amount of purified protein was determined by using the Pierce (Rockford, Ill.) protein assay kit with bovine serum albumin as a standard. Immunoblotting with the Xi126 PspA monoclonal antibody (32) was performed to confirm the purified protein.
Salmonella subcellular fractionation.
The periplasmic fraction was prepared by a modification of the lysozyme-osmotic shock method (56). Cultures grown in LB broth to an optical density at 600 nm (OD600) of 0.8 were centrifuged at 7,000 × g for 10 min, and the supernatant fluid was saved for analysis of secreted proteins. The cell pellets were resuspended in 800 μl of 100 mM Tris-HCl buffer (pH 8.6) containing 500 mM sucrose and 0.5 mM EDTA. Hen egg white lysozyme (40 μl of a 4-mg/ml stock solution) was added, followed immediately by the addition of 3.2 ml of 50 mM Tris-HCl buffer (pH 8.6) containing 250 mM sucrose, 0.25 mM EDTA, and 2.5 mM MgCl2. After gentle agitation, the suspension was incubated for 15 min in an ice bath. Cells were removed by centrifugation at 7,000 × g for 6 min followed by filtration of the supernatant through a 0.45 μm-pore-size filter. The filtered supernatant fluid served as the periplasmic fraction. Cells resuspended in 4 ml of 20 mM Tris-HCl (pH 8.6) were disrupted by two passages through a French pressure cell (American Instrument Company, Silver Spring, Md.). Cell lysates were centrifuged at 7,000 × g at 4°C for 6 min to remove unbroken cells. The supernatant fluid was then centrifuged at 132,000 × g at 4°C for 1 h to separate the soluble fraction and insoluble cell envelopes. The soluble fraction contained the cytoplasmic proteins. To isolate the outer membrane fraction, total envelope pellets were suspended in 4 ml of 20 mM Tris-HCl (pH 8.6) containing 1% Sarkosyl and incubated for 30 min on ice. The outer membrane fraction was obtained as a pellet after centrifugation at 132,000 × g at 4°C for 1 h. The pellet was resuspended in 4 ml of 20 mM Tris-HCl buffer (pH 8.6). The original culture supernatant was filtered (0.22 μm-pore-size filter), and secreted proteins were precipitated with 10% trichloroacetic acid (1 h, 4°C). An equal volume of each fraction sample was separated by SDS-PAGE for Western blot analysis. By using the outer membrane protein preparation procedure described above, Salmonella outer membrane proteins (SOMPs) were prepared from S. enterica serovar Typhimurium χ4746 cells grown in LB broth without galactose for analysis by enzyme-linked immunosorbent assay (ELISA). The use of SOMPs obtained from χ4746 precludes LPS O-antigen contamination.
Construction of an S. enterica serovar Typhimurium vaccine strain.
The Δcrp mutation was introduced into S. enterica serovar Typhimurium χ3339 by allelic exchange using the suicide vector pMEG-493 to yield χ8499. The presence of the 680-bp deletion was confirmed by PCR with a primer set flanking crp (5′-AAAGTCGCAATGGAAGGC-3′ and 5′-CGTAGACGACGATGGTCTTG-3′) and a strain phenotype of Mal− and nonmotility. The ΔasdA16 mutation was then introduced into χ8499 by P22HTint transduction from χ8554 with the suicide vector pMEG-443 integrated into a strain with the ΔasdA16 mutation, followed by sucrose selection to eliminate the suicide vector to yield χ8501 (27). The presence of the 1,242-bp asd deletion in χ8501 was confirmed by PCR using flanking a asd primer set (5′-CGGAAATGATTCCCTTCCTAACG-3′ and 5′-TATCTGCGTCGTCCTACCTTCAG-3′) (27).
Immunization of mice.
Two groups of five inbred 7-week-old female BALB/c mice were deprived of food and water for 4 h before infection. The recombinant S. enterica serovar Typhimurium χ8501(pYA3494) vaccine (1.9 × 109 CFU in 20 μl of phosphate-buffered saline containing 0.01% gelatin [BSG]) grown in LB broth to an OD600 of 0.8 was orally administered to BALB/c mice. The recombinant S. enterica serovar Typhimurium χ8501(pYA3493) vaccine (2 × 109 CFU in 20 μl of BSG) was used as a vector control. Food and water were returned to the immunized mice 30 min after immunization. Blood was obtained by retro-orbital puncture with heparinized capillary tubes at biweekly intervals. Following centrifugation at 4,000 × g for 5 min, the serum was removed from the whole blood and stored at −20°C. Vaginal secretion specimens were collected in a 50-μl BSG wash and stored at −20°C (59).
Pneumococcal challenge.
To observe the immune response caused by pneumococcal infection after vaccination, a sublethal dose of 3.8 × 105 S. pneumoniae WU2 CFU in 200 μl of BSG was administered by intravenous (i.v.) injection to S. enterica serovar Typhimurium vaccine-immunized BALB/c mice at 16 weeks after primary immunization. The ability of the Salmonella-PspA vaccine to protect the immunized mice against S. pneumoniae was assessed by intraperitoneal challenge with 4.8 × 103 CFU of S. pneumoniae in 100 μl of BSG. The 50% lethal dose (LD50) of S. pneumoniae WU2 in BALB/c mice was >106 CFU by i.v. administration (Larry McDaniel, personal communication) and <102 CFU by intraperitoneal administration (37). Significant differences in mortality were determined by chi-square analysis at day 21 after challenge. A difference was considered significant at a P value of <0.05.
ELISA.
ELISA was used to assay antibodies in vaginal secretions and serum to S. enterica serovar Typhimurium LPS and SOMPs and to rPspA. Polystyrene 96-well flat-bottom microtiter plates (Dynatech Laboratories Inc., Chantilly, Va.) were coated with S. enterica serovar Typhimurium LPS (100 ng/well; Sigma), SOMPs (100 ng/well), or purified rPspA (100 ng/well). Antigens suspended in sodium carbonate-bicarbonate coating buffer (pH 9.6) were applied with 100-μl volumes in each well. The coated plates were incubated at 37°C for 1 h, followed by an overnight incubation at 4°C. Free binding sites were blocked with a blocking buffer (phosphate-buffered saline [pH 7.4], 0.1% Tween 20, and 1% bovine serum albumin). Vaginal secretions and sera obtained from the same experimental group (five mice per group) were pooled and diluted 1:10 and 1:600, respectively. A 100-μl volume of diluted sample was added to individual wells in duplicate and incubated for 2 h at 37°C. Plates were treated with biotinylated goat anti-mouse IgG, IgG1, or IgG2a (Southern Biotechnology Inc., Birmingham, Ala.) for sera and IgA for vaginal secretions. Wells were developed with streptavidin-alkaline phosphatase conjugate (Southern Biotechnology) followed by p-nitrophenylphosphate substrate (Sigma) in diethanolamine buffer (pH 9.8). Color development (absorbance) was recorded at 405 nm using an automated ELISA plate (model EL311SX; Biotek, Winooski, Vt.). Absorbance readings two times higher than preimmune serum baseline values were considered positive reactions.
RESULTS
Construction of Asd+ vectors to use for antigen expression.
Vaccine strains harboring mulicopy Asd+ vectors (pBR ori or pUC ori) containing the entire asd gene with its promoter synthesized the Asd protein at a much higher level than necessary to complement the chromosomal asd mutation in a balanced-lethal host-vector system. In fact, the 200- to 300-fold excess production of Asd in a strain such as χ8554 (Δasd16) with the pYA3334 Asd+ vector (pUC ori) increases the generation time slightly and the LD50 10-fold compared to the same strain with an Asd+ vector with the pSC101 ori or p15A ori. In an attempt to reduce the level of Asd, the asd promoter region was deleted to determine whether there would be sufficient transcription to permit a promoterless asd gene to complement the chromosomal ΔasdA16 mutation. The asd gene sequence was amplified by PCR starting at bp 286 and ending at bp 1421 of the S. enterica serovar Typhimurium asd sequence (accession number AF015781) with an N-terminal BglII site and a C-terminal XbaI site. This sequence contains the Shine-Dalgarno (SD) sequence for ribosome recognition but lacks the −35 and −10 promoter sequence and ends just after the asd gene TAG stop codon. The BglII-XbaI DNA fragment was used to construct Asd+ vectors pYA3342 (pBR ori) and pYA3341 (pUC ori) (Table 1). It was possible to clone this fragment onto pSC101 ori or p15A ori vectors, but this did not result in sufficient Asd to permit construction of balanced-lethal systems with strains such as χ8554 which could grow in the absence of DAP. Both pYA3342 and pYA3341 complemented the asd mutations of E. coli χ6212 and S. enterica serovar Typhimurium χ4550. Salmonella strains possessing pYA3342 and pYA3341 produced significantly reduced amounts of Asd protein (39 kDa) compared to strains containing plasmids that had asd genes with the asd native promoter (Fig. 1). pYA3342 and pYA3341 in a Δasd S. enterica serovar Typhimurium strain such as χ8554 yielded recombinants that had wild-type LD50s following oral inoculation of BALB/c mice. Plasmid pYA3342 (Fig. 2A) was used for further construction.
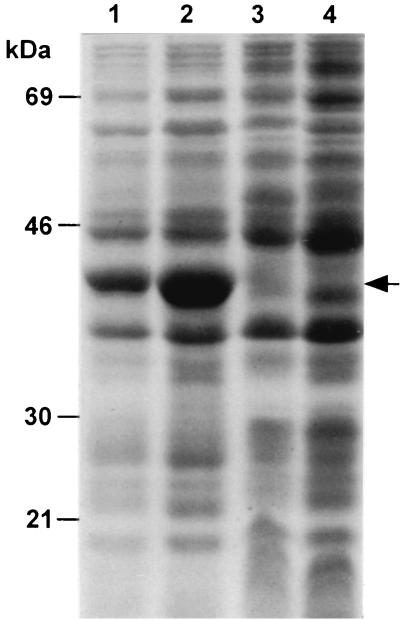
FIG. 1.
Reduced expression of Asd protein by deletion of the asd gene promoter region. SDS-PAGE was performed with cell lysates of S. enterica serovar Typhimurium χ4550 with pYA3333 (entire asd gene with Pasd, pBR ori), pYA3334 (entire asd gene with Pasd, pUC ori), pYA3342 (promoterless SD-asd gene, pBR ori), and pYA3341 (promoterless SD-asd gene, pUC ori). Standards are indicated to the left, and Asd protein (39 kDa) is indicated by an arrow. Lanes; 1, pYA3333; 2, pYA3334; 3, pYA3342; 4, pYA3341.
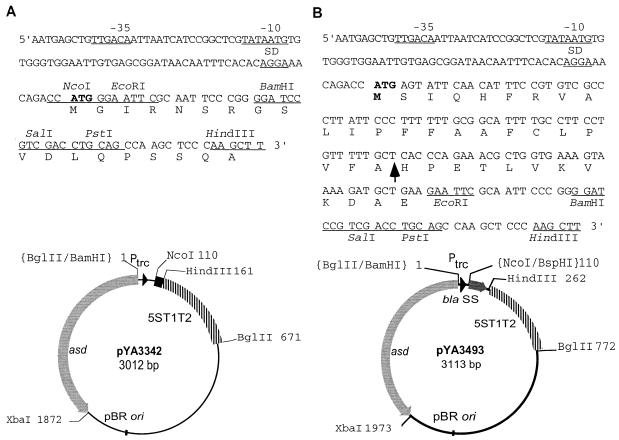
FIG. 2.
Asd+ antigen expression vectors. (A) Asd+ vector pYA3342. The map of pYA3342 and the nucleotide sequences of the Ptrc promoter region and multicloning sites are shown. (B) Periplasmic secretion Asd+ vector pYA3493. A DNA fragment encoding the β-lactamase signal sequence and 12 amino acid residues of the N terminus of mature β-lactamase of plasmid pBR322 was positioned under the control of the Ptrc promoter of the Asd+ vector pYA3342 (pBR ori). The map of pYA3493 and the nucleotide sequences of the Ptrc promoter region, β-lactamase signal sequence (bla SS), and multicloning sites are shown. The Ptrc sequences for −35, −10, and SD are indicated, and the translation start codon is in boldface. An arrow within the sequence indicates the signal peptidase cleavage site. Unique restriction enzyme sites in the multicloning site are indicated. 5ST1T2 is a transcriptional terminator.
Since export of PspA into the periplasmic space of Salmonella was inefficient and caused toxicity when the export depended on the signal sequence for PspA (37), we constructed a recombinant plasmid by cloning a DNA fragment specifying the signal sequence of β-lactamase which is efficiently transported into the periplasmic space in Salmonella. A 105-bp DNA fragment (nucleotides 4049 to 4153 of the sequence under accession number J01749) of the β-lactamase gene was PCR amplified from the pBR322 DNA template by using a pair of primers (N-terminal, 5′GCATTCATGAGTATTCAACATTTCC3′-BspHI [underlined]; C-terminal, 5′CCGGAATTCTTCAGCATCTTTTACT3′-EcoRI [underlined]). The PCR-amplified fragment included the N terminus of β-lactamase from the ATG start codon through the signal sequence (23 amino acids) plus 12 amino acids of the N terminus of the mature β-lactamase. These additional 12 amino acid residues were included to increase the efficiency of secretion of the recombinant protein (53). The 105-bp PCR product was digested with BspHI and EcoRI enzymes and cloned into the NcoI site (compatible with the BspHI site) and EcoRI site of the Asd+ vector pYA3342, resulting in plasmid pYA3493 (Fig. 2B). The in-frame position of the β-lactamase signal sequence was confirmed by nucleotide sequencing. Transcription promoted by Ptrc can be stopped by the 5ST1T2 transcriptional terminator located following the multicloning sites. pYA3493 was stably maintained for 50 or more generations in E. coli χ6212 and S. enterica serovar Typhimurium (Δasd) hosts grown in the presence or absence of DAP.
Construction of the rPspA-expressing plasmid.
A highly immunogenic α-helical region of PspA from amino acid residue 3 to 257 (765 bp; 255 amino acids) of the mature PspARx1 protein (588 amino acids) was selected to use as a test antigen in antigen delivery by a Salmonella carrier. The 765-bp DNA fragment of the pspA gene of S. pneumoniae Rx1 was PCR amplified from the pYA3193 DNA template with a pair of primers (N-terminal, 5′CCGGAATTCTCTCCCGTAGCCAGTCAGTCT3′; C-terminal, same as used in the construction of His6-tagged PspA, which introduces the TAA TAG stop codons after the pspA coding sequences). The PCR product, digested with EcoRI and HindIII enzymes, was cloned into the EcoRI and HindIII sites of pYA3493, resulting in pYA3494 (Fig. 3). The in-frame fusion of the rPspA with the β-lactamase signal sequence was confirmed by nucleotide sequencing. E. coli χ6212 harboring pYA3494 expressed rPspA as approximately 1% of the total cell protein (data not shown).
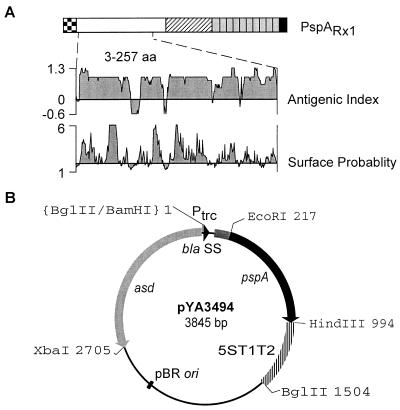
FIG. 3.
Recombinant plasmid pYA3494 for PspA expression. (A) The PspA region used in this study. Functional domains of native PspA from S. pneumoniae (PspARx1) are diagramed. Dotted box, leader sequence (31 amino acids [aa]); open box, immunodominant α-helical region (aa 1 to 288); hatched box, proline-rich region (aa 289 to 370); 10 gray boxes, choline-binding repeats (aa 371 to 571); black box, C terminus (aa 572 to 588). Dotted lines represent the limit of the rPspA region used in this study. Bioinformatical analyses of the rPspA for antigenic index and surface probability are presented. Analyses were performed with the Protean module of the Lasergene sequence analysis software. (B) Map of recombinant plasmid pYA3494. A 0.7-kb EcoRI-HindIII fragment of PCR-amplified DNA of pspARx1 was cloned into the EcoRI and HindIII sites of pYA3493 (Fig. 2B). The cloned fragment included the immunogenic α-helical region of PspA including amino acids 3 through 257 of mature PspA (255 amino acids).
Expression and subcellular localization of rPspA in Salmonella.
An S. enterica serovar Typhimurium strain was constructed to examine expression and subcellular localization of rPspA. The atrB13::MudJ allele (15), causing constitutive expression of β-galactosidase, in S. enterica serovar Typhimurium JF2430 was transduced into S. enterica serovar Typhimurium χ8554 by P22HTint-mediated generalized transduction (52), resulting in χ8599 (hisG ΔasdA16 atrB13::MudJ). χ8599 was Lac+ on MacConkey agar plus lactose and DAP. β-Galactosidase production from the atrB13::MudJ allele in χ8599 was used as a cytoplasmic protein marker and as an indicator of membrane leaking in the examination of subcellular fractionations. No periplasmic protein marker was used, since the use of the β-lactamase signal sequence in the pYA3494 construct precluded use of β-lactamase and the commercially available monoclonal antibody to the E. coli maltose binding protein (Sigma) did not react with this protein from S. enterica serovar Typhimurium. To observe rPspA expression, plasmid pYA3494 was introduced into S. enterica serovar Typhimurium χ8599. χ8599 harboring pYA3493 (vector alone) was used as the control (data not shown).
With the expectation of the periplasmic secretion of the rPspA, various subcellular fractions, including cytoplasm, periplasm, outer membrane, and culture supernatant of χ8599(pYA3494), were prepared to examine the location of rPspA. Although the calculated size of rPspA was approximately 30 kDa, PspA-specific monoclonal antibody Xi126 reacted with an approximately 35-kDa protein (Fig. 4). Aberrant migration of a PspA protein has been seen in previous studies (37, 54). Although a large amount of the rPspA resided in the cytoplasmic fraction, half of the rPspA was detected in the periplasmic fraction and the culture supernatant fluid. Little or no rPspA was detected in the outer membrane fraction. Densitometry analyses of immunoreactive bands showed that approximately 50% of the rPspA was located in the combined periplasm (25%) and culture supernatants (25%). In the immunoblot analyses of subcellular fractions with anti-β-galactosidase and -OmpC monoclonal antibodies, the β-galactosidase and OmpC proteins were detected in the cytoplasm and outer membrane fractions, respectively, suggesting that the rPspA detected in the periplasmic fraction and culture supernatant fluid was actively secreted instead of resulting from nonspecific membrane leaking or cell death by lysis.
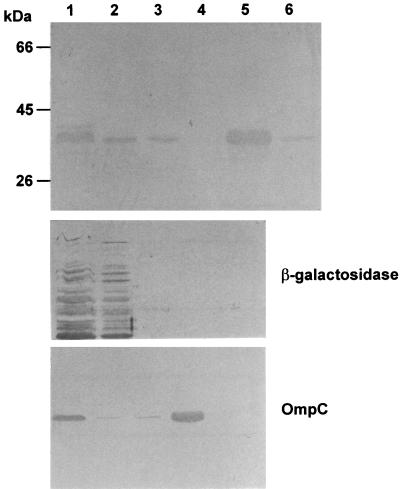
FIG. 4.
Subcellular location of expressed rPspA in S. enterica serovar Typhimurium. Subcellular fractions were prepared from S. enterica serovar Typhimurium χ8599(pYA3494) cells grown in LB broth at 37°C by the procedures described in Materials and Methods. Fractions equivalent to 30-μl volumes of the culture at an OD600 of 0.8, except for supernatant fluids, were analyzed by SDS-PAGE, and the rPspA was detected by immunoblotting with PspA-specific monoclonal antibody Xi126 (33). β-Galactosidase and OmpC were used as fractionation controls for cytoplasmic and outer membrane fractions, respectively. Standards are indicated to the left. Lanes: 1, total cell lysate; 2, cytoplasm; 3, periplasm; 4, outer membrane; 5, concentrated supernatant (750 μl); 6, supernatant (10 μl).
Recombinant S. enterica serovar Typhimurium Δcrp-28 vaccine expressing rPspA antigen.
pYA3493 (vector control) and pYA3494 (encoding rPspA) were electroporated into the Δcrp-28 ΔasdA16 strain χ8501. The S. enterica serovar Typhimurium χ8501 (Δcrp-28 ΔasdA16) vaccine strain containing pYA3494 expressed the rPspA protein at an approximate molecular mass of 35 kDa. In the analyses of Coomassie blue-stained SDS-polyacrylamide gels, the amount of rPspA protein was as much as approximately 1 to 2% of the total χ8501(pYA3494) proteins (Fig. 5). With results consistent with those seen in the rPspA localization analysis (75% of rPspA cell associated [50% in cytoplasm and 25% in periplasm] and 25% of rPspA secreted), the rPspA expressed in the χ8501 vaccine strain was secreted into the culture supernatant along with other secreted proteins. To examine the stability of plasmids pYA3493 and pYA3494 in Salmonella χ8501 in vitro, χ8501 cells containing pYA3493 and pYA3494 were cultured with daily passage of 1:1,000 dilutions for five consecutive days in L broth containing DAP. All χ8501 clones examined (300 clones/day) kept the Asd+ plasmid pYA3493 and pYA3494, indicating that pYA3493 and pYA3494 were very stable in the χ8501 vaccine strain. Cells obtained from the last-day culture of the stability test expressed amounts of the 35-kDa rPspA similar to those from the first day (data not shown), suggesting stable expression of rPspA without rearrangements.
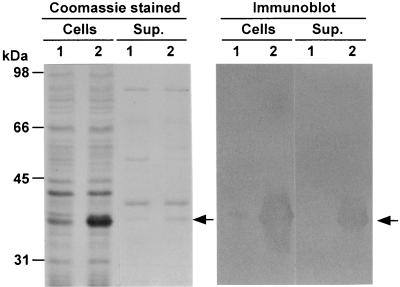
FIG. 5.
Expression of rPspA in the S. enterica serovar Typhimurium vaccine strain. χ8501 harboring pYA3494 (specifying rPspA) or pYA3493 (vector control) was cultured in LB broth at 37°C. Total cells (equivalent to 7.5 × 108 cells) and concentrated culture supernatants (Sup.) (equivalent to 750 μl of supernatant of cultures at an OD600 of 0.8) were subjected to SDS-PAGE analysis. Left panel, Coomassie brilliant blue-stained gel. Right panel, immunoblot of the duplicated gel with PspA-specific monoclonal antibody Xi126. Molecular markers are indicated to the left. PspA proteins are indicated by arrows. Lanes 1 and 2, protein profiles of χ8501(pYA3493) and χ8501(pYA3494), respectively.
Immune responses in mice after oral immunization with the recombinant S. enterica serovar Typhimurium vaccines.
All mice orally administered a single dose of 1.3 × 109 CFU S. enterica serovar Typhimurium χ8501(pYA3493) (vector control) survived for the 30-day monitoring period. Those mice were protected for a 30-day observation period against wild-type χ3339 (LD50, <106 CFU) challenge (1.7 × 109 CFU) 30 days after the initial immunization. There were no survivors in a group of unimmunized mice challenged with 1.7 × 107 CFU of χ3339. These results indicate that χ8501 with an Asd+ vector is avirulent for mice and elicits a protective immune response against challenge with S. enterica serovar Typhimurium.
A single dose of S. enterica serovar Typhimurium χ8501(pYA3494) (1.9 × 109 CFU) or χ8501(pYA3493) (control, 2 × 109 CFU) was orally administered to 7-week-old female BALB/c mice. All immunized mice survived, and we did not observe any signs of disease in the immunized mice during the entire experimental period. The antibody responses to Salmonella LPS and SOMPs and to the foreign antigen rPspA in the sera and the vaginal secretions of the immunized mice were measured. The serum IgG responses to LPS, SOMPs, and rPspA are presented in Fig. 6. At 2 weeks after administration, little IgG response to the antigens was observed. Maximal anti-LPS, -SOMP, and -rPspA IgG levels without boost immunization were detected at 6 weeks postimmunization.
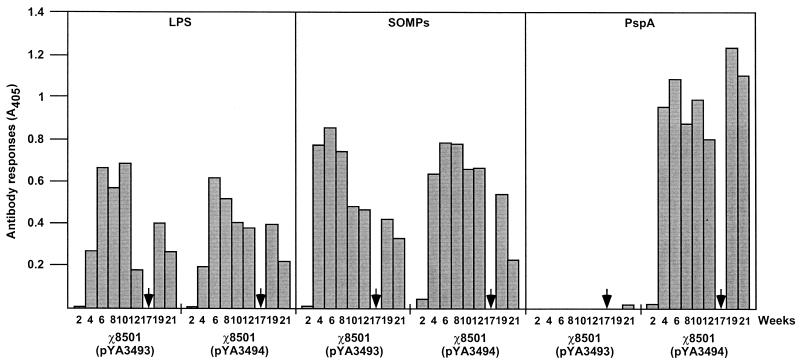
FIG. 6.
Serum IgG responses to S. enterica serovar Typhimurium LPS and SOMPs and to rPspA. The data represent IgG antibody levels induced in mice orally immunized with χ8501(pYA3493) (vector control) and χ8501(pYA3494) (expressing rPspA) at the indicated weeks after immunization. ELISA and data analysis are described in Materials and Methods. Arrows indicate sublethal i.v. infection with S. pneumoniae WU2.
At 17 weeks postimmunization, we infected mice i.v. with a sublethal dose (3.8 × 105 CFU) of the virulent S. pneumoniae WU2 strain to monitor the changes of anti-rPspA antibody titers. Sublethal i.v. infection with S. pneumoniae did not kill mice immunized with the χ8501(pYA3493) or χ8501(pYA3494) vaccine. Because native PspA is a highly immunogenic pneumococcal surface protein, the pneumococcal challenge boosted rPspA-specific immune responses in χ8501(pYA3494)-immunized mice (Fig. 6). In comparison to the anti-rPspA IgG level (A405, 0.81) at 12 weeks after S. enterica serovar Typhimurium χ8501(pYA3494) immunization, the pneumococcal challenge boosted 53% more anti-rPspA IgG (A405, 1.24) 1 week later. This suggests that the S. enterica serovar Typhimurium χ8501-rPspA vaccine induces immunological memory for a rapid responsiveness to subsequently administered PspA antigen. Anti-rPspA IgG was not detected in sera obtained from mice immunized with χ8501(pYA3493), the vector control vaccine. The χ8501(pYA3493) vaccine elicited anti-LPS and -SOMP IgG responses with kinetics and levels similar to those induced by χ8501(pYA3494). These results suggest that Salmonella-delivered rPspA antigen had minimal influence on the immune response to Salmonella itself.
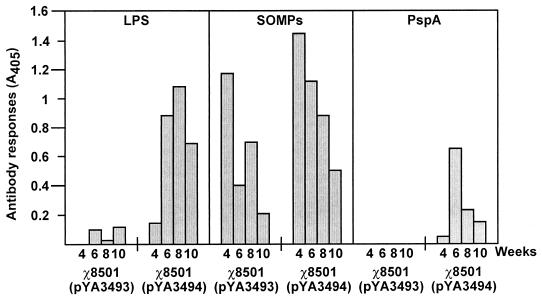
IgA levels, mostly secretory IgA, for LPS, SOMPs, and rPspA in the vaginal fluids of immunized mice were measured. The χ8501(pYA3493) and χ8501(pYA3494) vaccines elicited anti-LPS and anti-SOMP IgA. rPspA-specific IgA was detected in the vaginal fluids from mice immunized with χ8501(pYA3494) but not in those from mice immunized with the χ8501(pYA3493) vector-only control (Fig. 7).
FIG. 7.
Secretory IgA responses to S. enterica serovar Typhimurium LPS and SOMPs and to rPspA. The data represent anti-LPS, -SOMP, and -rPspA IgA antibody levels in vaginal secretions of BALB/c mice orally immunized with χ8501(pYA3493) (vector control) and χ8501(pYA3494) (expressing rPspA) at weeks 4, 6, 8, and 10 after immunization.
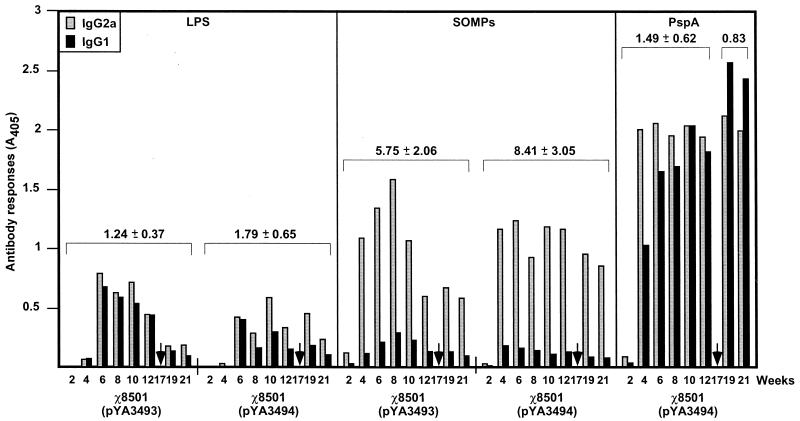
IgG isotype analyses.
The types of immune responses to Salmonella LPS and SOMPs and to the rPspA were further examined by measuring the levels of IgG isotype subclasses IgG2a and IgG1. The Th1-helper cells direct cell-mediated immunity and promote class switching to IgG2a, and Th2 cells provide potent help for B-cell antibody production and promote class switching to IgG1 (39, 51). IgG2a isotype dominant responses were observed for the Salmonella LPS and SOMP antigens (Fig. 8). The IgG2a/IgG1 ratios for anti-LPS and -SOMPs in sera obtained from mice immunized with χ8501(pYA3493) were not significantly different from those from mice immunized with χ8501(pYA3494) (LPS, P = 0.07; SOMPs, P = 0.054). The IgG2a/IgG1 ratios for anti-SOMPs (ranging from 6.4 to 11.5) are higher than those for LPS (ranging from 1.1 to 2.5), with statistical significance [χ8501(pYA3493), P = 0.0004; χ8501(pYA3494), P = 0.0003]. Th1-type dominant immune responses are frequently observed after immunization with attenuated Salmonella (30, 40). In contrast to the type of immune responses to LPS and SOMPs, a Th1- and Th2-type mixed response was observed for the rPspA antigen. Although the IgG2a levels were higher than IgG1 levels in the early phase (up to 8 weeks postimmunization), the level of anti-rPspA IgG1 isotype antibodies gradually increased. After 10 weeks postimmunization, a 1:1 ratio of IgG2a to IgG1 or IgG1 dominant responses was detected (Fig. 8). The pneumococcal i.v. challenge stimulated more IgG1 responses (mean IgG2a/IgG1 ratio, 0.83) to PspA than seen before challenge (mean IgG2a/IgG1 ratio, 1.49), with statistical significance (P = 0.023).
FIG. 8.
Serum IgG2a and IgG1 responses to S. enterica serovar Typhimurium LPS and SOMPs and to rPspA. The data represent IgG2a and IgG1 subclass antibody levels to Salmonella LPS and SOMPs and to rPspA in sera of BALB/c mice orally immunized with χ8501(pYA3493) (vector control) and χ8501(pYA3494) (expressing rPspA) at the indicated weeks after immunization. Arrows indicate sublethal i.v. infection with S. pneumoniae WU2. Anti-rPspA IgG2a and IgG1 responses of χ8501(pYA3493) (negative control) are not shown. The overall IgG2a/IgG1 ratios (means ± standard deviations) for each antigen are shown above the bars. Statistical analyses were performed with a paired Student t test to compare the IgG2a/IgG1 ratios for antigens. P values of <0.05 were considered significant. The results were as follows: (i) LPS for χ8501(pY3493) versus χ8501(pYA3494), P > 0.05; (ii) SOMPs for χ8501(pY3493) versus χ8501(pYA3494), P > 0.05; (iii) LPS for χ8501(pYA3493) versus SOMPs for χ8501(pYA3493), P < 0.05; (iv) LPS for χ8501(pYA3494) versus SOMPs for χ8501(pYA3494), P < 0.05; (v) PspA before versus after sublethal challenge of S. pneumoniae, P < 0.05; (vi) LPS for χ8501(pYA3494) versus PspA for χ8501(pYA3494), P > 0.05 (before challenge) and P < 0.05 (after challenge); (vii) SOMPs for χ8501(pYA3494) versus PspA for χ8501(pYA3494), P < 0.05 (before and after challenge).
Evaluation of protective immunity.
To examine the ability of Salmonella-rPspA vaccines to protect against pneumococcal infection, BALB/c mice were immunized with either S. enterica serovar Typhimurium χ8501(pYA3493) (dose of 1.3 × 109 CFU) or χ8501(pYA3494) (dose of 1.7 × 109 CFU). At 10 weeks after the initial immunization, a second dose of 109 CFU of each vaccine was administered. We did not detect weakness or disease signs in vaccinated mice during the immunization periods. At 5 weeks after the second immunization, mice were challenged intraperitoneally with 4.8 × 103 CFU (50 times the LD50) of S. pneumoniae WU2. Sixty percent of the mice immunized with χ8501(pYA3494) were protected from pneumococcal challenge, with statistical significance (P < 0.05). This challenge dose killed 100% of unimmunized and χ8501(pYA3493)-immunized mice (Table 2). Following challenge, mice unimmunized or immunized with χ8501(pYA3493) died much faster, with death at a mean of day 2, than mice immunized with χ8501(pYA3494), which died at a mean of day 5.
TABLE 2.
Oral immunization with rPspA-expressing S. enterica serovar Typhimurium χ8501(pYA3494) vaccine protects BALB/c mice against challenge with virulent S. pneumoniae strain WU2
| Vaccinea | rPspA expressionb | Protectionc (% alive) | Days to death |
|---|---|---|---|
| 5, 5 | |||
| χ8501(pYA3494) | + | 60* | |
| >21, >21, >21 | |||
| χ8501(pYA3493) | − | 0 | 1, 2, 2, 3, 3 |
| None (unimmunized) | NA | 0 | 1, 2, 2, 2, 3 |
Mice were orally immunized a total of two times at 10-week intervals with ∼109 CFU of indicated vaccine strain per dose.
+, rPspA expressed; −, rPspA not expressed; NA, not applicable.
Five weeks after the last immunization, mice (five mice per group) were challenged intraperitoneally with approximately 4.8 × 103 CFU of virulent S. pneumoniae WU2. The LD50 of WU2 by intraperitoneal infection in unimmunized BALB/c mice was <102 (data not shown). Mortality was monitored for 3 weeks after pneumococcal challenge. Significance was determined by chi-square analysis. ∗, P < 0.05.
DISCUSSION
Attenuated Salmonella strains constructed by recombinant DNA technology have been developed as live vaccines for humans and animals to prevent disease caused by Salmonella infections. The ability of live recombinant Salmonella vaccines to colonize the gut-associated lymphoid tissue (Peyer's patches) and visceral lymphoid tissues following oral administration is beneficial in that it stimulates all arms of the immune response, including mucosal, humoral, and cellular immunities (13, 17, 34). Recombinant Salmonella vaccines have also been developed as multivalent vaccines to deliver recombinant antigens originating from viruses, bacteria and parasites (13, 34).
Analysis of convalescent-phase sera from patients or animals infected with bacterial pathogens reveals that the proteins located in the envelopes of or secreted by the bacterial pathogens act as dominant immunogens for the immune responses (31, 38, 58). These observations indicate that envelope and secreted proteins are highly immunogenic and/or more readily interact with antigen-presenting cells due to their subcellular location. Translocation of such highly immunogenic antigens into the cell envelope or secretion from the cell should increase the strength of the immune response elicited by vaccine strains expressing foreign antigens. In the development of attenuated Salmonella-based multivalent vaccines, a preferable system would have a recombinant antigen secreted from the cytoplasm of Salmonella vaccines (19, 20). β-Lactamase, encoded by the ampicillin resistance gene, is a well-characterized periplasmic secreted protein in gram-negative bacteria (43). The β-lactamase gene contained in plasmid pBR322 was originally obtained from an S. enterica serovar Paratyphi B isolate (3, 14). It is well known that β-lactamase produced from pBR322 is secreted to the periplasmic space of gram-negative bacteria, and its translocation depends upon the presence of a signal sequence consisting of 23 amino acid residues at the N terminus (26, 43). Evidence obtained from previous studies confirms that the signal sequence plus an additional 12 amino acids of the mature β-lactamase are required to translocate β-lactamase through the cytoplasmic membrane of gram-negative bacteria (28, 53). Fusion of a protein to the β-lactamase signal peptide promotes the secretion of the fusion protein into the periplasm of E. coli (42, 53). We reasoned that a protein antigen attached to the β-lactamase signal peptide should be secreted into the periplasm of Salmonella vaccine strains. The pYA3493 plasmid (Fig. 2B) constructed in this study was designed to use for the periplasmic secretion of recombinant antigens for antigen delivery by Salmonella vaccines.
The recombinant plasmid pYA3494 (pBR ori) (Fig. 3) was constructed for the periplasmic secretion of the α-helical region of the PspARx1. In contrast to the previously described PspA-specifying pYA3193 (37), pYA3494 was maintained stably (100%) over 50 generations in the S. enterica serovar Typhimurium vaccine strain grown in the presence of DAP. Both E. coli and Salmonella containing pYA3494 expressed the rPspA protein with an apparent molecular mass of 35 kDa, and the rPspA proteins were detected in the periplasm (25%) as well as in the culture supernatant (25%) (Fig. 4). The N-terminal His6-tagged rPspA protein (no apparent signal peptide) expressed in S. enterica serovar Typhimurium χ8599 containing pYA3496 was detected only in the cytoplasm and not in the periplasm of the Salmonella host (data not shown). These results suggest that the signal peptide and 12 residues of the N terminus of β-lactamase (present in pYA3494) promote the periplasmic secretion of rPspA. The mechanism to explain how rPspA was translocated from the periplasm outside cells (Fig. 4 and 5) remains unknown. The secondary and tertiary structures of rPspA may permit it to traverse through the Salmonella outer membrane. Alternatively, accumulated rPspA in the periplasm may be encapsulated in membrane vesicles which are discharged from the cells. Membrane vesicles have been identified in many gram-negative bacteria (2), and Ciofu et al. (10) reported that β-lactamase was packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. Approximately 50% of the rPspA remained in the cytoplasm, perhaps because the amount of endogenous signal peptidase necessary to process all of the overexpressed rPspA is limiting. Alternatively, it has been reported that the C-terminal sequence of mature β-lactamase is important for the efficient periplasmic secretion of β-lactamase, along with the signal sequence (28).
The immunogenicity and appropriate subcellular location of rPspA in the Salmonella vaccine strain may contribute to the augmented immune responses by facilitating adequate exposure of rPspA antigen to antigen-presenting cells for processing. BALB/c mice orally immunized with a single dose of S. enterica serovar Typhimurium χ8501 expressing rPspA showed immune responses to both Salmonella antigens and rPspA. Although there was variation in antibody levels between samples due to mucus materials in vaginal washings, mucosal anti-rPspA IgA was detected in vaginal washings only of mice immunized with Salmonella-rPspA vaccine (Fig. 7). It is likely that mucosal immunity will act as a primary immune defense system against natural infection by S. pneumoniae. Detection of anti-rPspA IgG with a typical antibody response in sera from Salmonella-rPspA vaccine-immunized mice (Fig. 6) indicates that the χ8501(pYA3494) vaccine likely reached appropriate lymphoid tissues to stimulate a systemic immune response. Similar levels of anti-LPS and -SOMP IgG were induced by strains χ8501(pYA3493) (vector) and χ8501(pYA3494), suggesting that rPspA-specific immunity did not interfere with immunity against Salmonella itself.
In T-cell-dependent, antigen-specific immune responses, Th1 cells direct cell-mediated immunity and promote class switching to IgG2a. Th2 cells direct humoral immunity and promote class switching to IgG1 and IgA (39, 51). To date, the mechanism determining Th1- or Th2-type immunity to a given antigen is not well understood. Th2-type immune responses are rare in the immune response elicited by attenuated Salmonella vaccines (11, 41). While Th1-type dominant responses were observed for the LPS and SOMPs in this study, mixed Th1- and Th2-type immune responses were observed for rPspA (Fig. 8). After 10 weeks postimmunization, the Th2-type response became dominant. The mechanisms stimulating these types of immune responses by the Salmonella-rPspA vaccine remain unknown. It has been reported that oral immunization of purified PspA with the mucosal adjuvant cholera toxin in mice elicited Th2-type immune responses (57). In the development of pneumococcal vaccines, the anti-rPspA IgG isotype switching to a mixed or IgG1-dominant type along with a mucosal IgA response, indicating induction of a strong Th2-type humoral immune response, is preferred to prevent colonization of S. pneumoniae in the respiratory tract (pneumonia) or ear mucosa (otitis media) (33, 55). A sublethal challenge of S. pneumoniae boosted the rPspA immune responses with maintenance of the IgG1 isotype dominant response observed before infection.
Mice orally vaccinated with Salmonella-rPspA vaccine were significantly protected (60%) (P < 0.05) against virulent S. pneumoniae WU2 intraperitoneal challenge at 10 weeks postimmunization. These results indicate that the Salmonella χ8501 vaccine, which expresses the rPspA by an improved antigen expression system, induces protective immunity against pneumococcal infection, and they are consistent with the results demonstrating that the α-helical portion of PspA represents a protective domain (7).
In conclusion, the results of this study demonstrate that a recombinant attenuated Salmonella vaccine expressing rPspA antigen elicited mixed Th1- and Th2-type immune responses to rPspA and protected mice against pneumococcal challenge. Recombinant attenuated Salmonella-rPspA vaccines may have great potential for use for inducing effective protection against pneumococci which establish infection via a mucosal surface. However, there are some issues that need to be evaluated in future studies. The factors causing augmented Th2-type immune responses will be investigated. It remains to be examined whether rPspA secretion could be associated with the formation of membrane vesicles. Further modifications of the rPspA antigen expression construct may be required to enhance the rPspA secretion and to induce antibodies which can opsonize diverse pneumococcal strains with high avidity. Answers to these questions will subsequently improve the recombinant attenuated Salmonella-based pneumococcal vaccine.
Acknowledgments
We thank Larry S. McDaniel for providing S. pneumoniae strain WU2, Steven A. Tinge for providing pMEG-443, and Xin Zhang for technical assistance with animal experiments. We also thank Josephine Clark-Curtiss and Richard Groger for critical comments on the manuscript.
This research project was supported by a grant (DE06669) from the Public Health Service through the National Institutes of Health.
Editor: E. I. Tuomanen
REFERENCES
- 1.Bertani, G. 1952. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beveridge, T. J. 1999. Structure of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breiman, R., J. C. Butler, F. C. Tenover, J. Elliot, and R. R. Facklam. 1994. Emergence of drug-resistant pneumococcal infections in the United States. JAMA 271:1831-1835. [PubMed] [Google Scholar]
- 6.Briles, D. E., J. D. King, M. A. Gray, L. S. McDaniel, E. Swiatlo, and K. A. Benton. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858-867. [DOI] [PubMed] [Google Scholar]
- 7.Briles, D. E., R. C. Tart, E. Swiatlo, J. P. Dillard, P. Smith, K. A. Benton, B. A. Ralph, A. Brooks-Walter, M. J. Crain, S. K. Hollingshead, and L. S. McDaniel. 1998. Pneumococcal diversity: considerations for new vaccine strategies with emphasis on pneumococcal surface protein A (PspA). Clin. Microbiol. Rev. 11:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briles, D. E., S. K. Hollingshead, G. S. Nabors, J. C. Paton, and A. Brooks-Walter. 2001. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine 19:S87-S95. [DOI] [PubMed] [Google Scholar]
- 9.Briles, D. E., S. K. Hollingshead, J. King, A. Swift, P. A. Braun, M. K. Park, L. M. Ferguson, M. H. Nahm, and G. S. Nabors. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J. Infect. Dis. 182:1694-1701. [DOI] [PubMed] [Google Scholar]
- 10.Ciofu, O., T. J. Beveridge, J. Kadurugamuwa, J. Walther-Rasmussen, and N. Hoiby. 2000. Chromosomal β-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:9-13. [DOI] [PubMed] [Google Scholar]
- 11.Corthésy-Theulaz, I. E., S. Hopkins, D. Bachmann, P. F. Saldinger, N. Porta, R. Haas, Y. Zheng-Xin, T. Meyer, H. Bouzourène, A. L. Blum, and J. Kraehenbuhl. 1998. Mice are protected from Helicobacter pylori infection by nasal immunization with attenuated Salmonella typhimurium phoPc expressing urease A and B subunits. Infect. Immun. 66:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtiss, R., III, and S. M. Kelly. 1987. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect. Immun. 55:3035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtiss, R., III, T. Doggett, A. Nayak, and J. Srinivasan. 1996. Strategies for the use of live recombinant avirulent bacterial vaccines for mucosal immunization, p. 499-511. In H. Kiyono and M. F. Kagnoff (ed.), Essentials of mucosal immunology. Academic Press, San Diego, Calif.
- 14.Datta, N., and P. Kontomichalou. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239-241. [DOI] [PubMed] [Google Scholar]
- 15.Foster, J. W., and B. Bearson. 1994. Acid-sensitive mutants of Salmonella typhimurium identified through a dinitrophenol lethal screening strategy. J. Bacteriol. 176:2596-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gálan, J. E., K. Nakayama, and, R. Curtiss III. 1990. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene 94:29-35. [DOI] [PubMed] [Google Scholar]
- 17.Gálan, J. E., and P. J. Sansonetti. 1996. Molecular and cellular bases of Salmonella and Shigella interaction with host cells, p. 2757-2773. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 18.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentschev, I., H. Mollenkopf, Z. Sololovic, J. Hess, S. H. Kaufmann, and W. Goebel. 1996. Development of antigen-delivery systems, based on the Escherichia coli hemolysin secretion pathway. Gene 179:133-140. [DOI] [PubMed] [Google Scholar]
- 20.Gentschev, I., I. Glaser, W. Goebel, D. J. McKeever, A. Musoke, and V. T. Heussler. 1998. Delivery of the p67 sporozoite antigen of Theileria parva by using recombinant Salmonella dublin: secretion of the product enhances specific antibody responses in cattle. Infect. Immun. 66:2060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenwood, B. 1999. The epidemiology of pneumococcal infection in children in the developing world. Phil. Trans. R. Soc. London B 354:777-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulig, P. A., and R. Curtiss III. 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55:2891-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirschmann, J. V. 2000. Use of the pneumococcal polysaccharide vaccine is unwarranted in the United States. ASM News 66:326-327. [Google Scholar]
- 24.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollingshead, S. K., R. Becker, and D. E. Briles. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect. Immun. 68:5889-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadonaga, J. T., A. Plückthun, and J. R. Knowles. 1985. Signal sequence mutants of β-lactamase. J. Biol. Chem. 260:16192-16199. [PubMed] [Google Scholar]
- 27.Kang, H. Y., C. M. Dozois, S. A. Tinge, T. H. Lee, and R. Curtiss III. 2002. Transduction-mediated transfer of unmarked deletions and point mutations through the use of counterselectable suicide vectors. J. Bacteriol. 184:307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koshland, D., and D. Botstein. 1980. Secretion of beta-lactamase requires the carboxy end of the protein. Cell 20:749-760. [DOI] [PubMed] [Google Scholar]
- 29.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 30.Lo-Man, R., J. P. M. Langeveld, E. Dériaud, M. Jehanno, M. Rojas, J.-M. Clément, R. H. Meloen, M. Hofnung, and C. Leclerc. 2000. Extending the CD4+ T-cell epitope specificity of the Th1 immune response to an antigen using a Salmonella enterica serovar Typhimurium delivery vehicle. Infect. Immun. 68:3079-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathers, K., M. Leinonen, and D. Goldblatt. 1999. Antibody response to outer membrane proteins of Moraxella catarrhalis in children with otitis media. Pediatr. Infect. Dis. J. 18:982-988. [DOI] [PubMed] [Google Scholar]
- 32.McDaniel, L. S., G. Scott, J. F. Kearney, and D. E. Briles. 1984. Monoclonal antibodies against protease sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J. Exp. Med. 160:386-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGhee, J. R., and H. Kiyono. 1993. New perspectives in vaccine development: mucosal immunity to infections. Infect. Agents Dis. 2:55-73. [PubMed] [Google Scholar]
- 34.Medina, E., and C. A. Guzman. 2001. Use of live bacterial vaccine vectors for antigen delivery: potential and limitations. Vaccine 19:1573-1580. [DOI] [PubMed] [Google Scholar]
- 35.Mufson, M. A. 1990. Streptococcus pneumoniae, p. 1539-1550. In G. L. Mandell, R. G. Douglas, Jr., and J. E. Bennett (ed.), Principles and practice of infectious disease. Churchill Livingstone, Inc., New York, N.Y.
- 36.Nakayama, K., S. M. Kelly, and R. Curtiss III. 1988. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Bio/Technology 6:693-697. [Google Scholar]
- 37.Nayak, A. R., S. A. Tinge, R. C. Tart, L. S. McDaniel, D. E. Briles, and R. Curtiss III. 1998. A live recombinant avirulent oral Salmonella vaccine expressing pneumococcal surface protein A induces protective responses against Streptococcus pneumoniae. Infect. Immun. 66:3744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicolle, L. E., E. Ujack, J. Brunka, and L. E. Bryan. 1988. Immunoblot analysis of serologic response to outer membrane proteins of Escherichia coli in elderly individuals with urinary tract infections. J. Clin. Microbiol. 26:2087-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Garra, A., and N. Arai. 2000. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell. Biol. 10:542-550. [DOI] [PubMed] [Google Scholar]
- 40.Okahashi, N. M. Yamamoto, J. L. VanCott, S. N. Chatfield, M. Roberts, H. Bluethmann, T. Hiroi, H. Kiyono, and J. R. McGhee. 1996. Oral immunization of interleukin-4 (IL-4) knockout mice with a recombinant Salmonella strain or cholera toxin reveals that CD4+ Th2 cells producing IL-6 and IL-10 are associated with mucosal immunoglobulin A responses. Infect. Immun. 64:1516-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pascual, D. W., D. M. Hone, S. Hall, F. W. van Ginkel, M. Yamamoto, N. Walters, K. Fujihashi, R. J. Powell, S. Wu, J. L. Vancott, H. Kiyono, and J. R. McGhee. 1999. Expression of recombinant enterotoxigenic Escherichia coli colonization factor antigen I by Salmonella typhimurium elicits a biphasic T helper cell response. Infect. Immun. 67:6249-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pawelek, J. M., K. B. Low, and D. Bermudes. 1997. Tumor-targeted Salmonella as a novel anticancer vector. Cancer Res. 57:4537-4544. [PubMed] [Google Scholar]
- 43.Plückthun, A., and J. R. Knowles. 1987. The consequences of stepwise deletion from the signal-processing site of β-lactamase. J. Biol. Chem. 262:3951-3957. [PubMed] [Google Scholar]
- 44.Roland, K., R. Curtiss III., and D. Sizemore. 1999. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 43:429-441. [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Schmieger, H., and H. Backhaus. 1976. Altered cotransduction frequencies exhibited by HT-mutants of Salmonella-phage P22. Mol. Gen. Genet. 143:307-309. [DOI] [PubMed] [Google Scholar]
- 47.Schödel, F., S. M. Kelly, D. L. Peterson, D. R. Milich, and R. Curtiss III. 1994. Hybrid hepatitis B virus core-pre-S proteins synthesized in avirulent Salmonella typhimurium and Salmonella typhi for oral vaccination. Infect. Immun. 62:1669-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro, E. D., A. T. Berg, R. Austrian, D. Schroeder, V. Parcells, A. Margolis, R. K. Adair, and J. D. Clemmens. 1991. Protective efficacy of polyvalent pneumococcal polysaccharide vaccine. N. Engl. J. Med. 325:1453-1460. [DOI] [PubMed] [Google Scholar]
- 49.Shinefield, H. R., S. Black, P. Ray, I. Chang, N. Lewis, B. Fireman, J. Hackell, P. R. Paradiso, G. Siber, R. Kohberger, D. V. Madore, F. J. Malinowski, A. Kimura, C. Le, I. Landaw, J. Aguilar, and J. Hansen. 1999. Safety and immunogenicity of heptavalent pneumococcal CRM197 conjugate vaccine in infants and toddlers. Pediatr. Infect. Dis. J. 18:757-763. [DOI] [PubMed] [Google Scholar]
- 50.Singh, S. P., Y. Upshaw, T. Abdullah, S. R. Singh, and P. E. Klebba. 1992. Structural relatedness of enteric bacterial porins assessed with monoclonal antibodies to Salmonella typhimurium OmpD and OmpC. J. Bacteriol. 174:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spellberg, B., and J. E. Edwards, Jr. 2001. Type 1/type 2 immunity in infectious diseases. Clin. Infect. Dis. 32:76-102. [DOI] [PubMed] [Google Scholar]
- 52.Sternberg, N. L., and R. Maurer. 1991. Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204:18-43. [DOI] [PubMed] [Google Scholar]
- 53.Summers, R. G., and J. R. Knowles. 1989. Illicit secretion of a cytoplasmic protein into the periplasm of Escherichia coli requires a signal peptide plus a portion of the cognate secreted protein. J. Biol. Chem. 264:20074-20081. [PubMed] [Google Scholar]
- 54.Talkington, D. F., D. C. Voellinger, L. S. McDaniel, and D. E. Briles. 1992. Analysis of pneumococcal PspA microheterogeneity in SDS polyacrylamide gels and the association of PspA with the cell membrane. Microb. Pathog. 13:343-355. [DOI] [PubMed] [Google Scholar]
- 55.van den Dobbelsteen, G. P. J. M., and E. P. van Rees. 1995. Mucosal immune responses to pneumococcal polysaccharides: implications for vaccination. Trends Microbiol. 3:155-159. [DOI] [PubMed] [Google Scholar]
- 56.Witholt, B., M. Boekhout, M. Brock, J. Kingma, H. van Heerikhuizen, and L. de Leij. 1976. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal. Biochem. 74:160-170. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto, M., L. S. McDaniel, K. Kawabata, D. E. Briles, R. J. Jackson, J. R. McGhee, and H. Kiyono. 1997. Oral immunization with PspA elicits protective humoral immunity against Streptococcus pneumoniae infection. Infect. Immun. 65:640-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yerdugo-Rodriguez, A., L. H. Gam, C. L. Koh, S. D. Puthucheary, E. Calva, and T. Pang. 1993. Detection of antibodies against Salmonella typhi outer membrane protein (OMP) preparation in typhoid fever patients. Asian Pac. J. Allergy Immunol. 11:45-52. [PubMed] [Google Scholar]
- 59.Zhang, X., S. M. Kelly, W. S. Bollen, and R. Curtiss III. 1997. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 Δcrp and Δcdt deletion mutants. Infect. Immun. 65:5381-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]