Abstract
Clostridium perfringens iota-toxin is a binary toxin consisting of two individual proteins, the binding component (Ib) and the enzyme component (Ia). Wild-type Ib bound to Vero cells at 4 and 37°C and formed oligomers at 37°C but not at 4°C. The Ib-induced K+ release from the cells was dependent on the oligomer formation of Ib in the cells, but the oligomer formation did not induce rounding activity or cytotoxicity. After incubation of the cells with recombinant Ib (rIb) at 37°C, the Ib oligomer in the cell became resistant to pronase treatment with time, but the Ib monomer was sensitive to the treatment. Furthermore, treatment of Vero cells with rIb in the presence of bafilomycin, methylamine, or ethylamine resulted in accumulation of the oligomer in the cells but had no effect on K+ release. Moreover, incubation with Ib plus Ia in the presence of these agents caused no rounding in the cells. These observations suggest that Ib binds to Vero cells, itself oligomerizing to form ion-permeable channels and that the formation of oligomer then induces endocytosis.
Clostridium perfringens iota-toxin consists of an enzyme component (Ia) and binding component (Ib) (31). Each protein has been reported to lack toxic activity when injected alone (25, 31, 32). These proteins act in binary combinations to produce toxic activities, cytotoxic, lethal, and dermonecrotic activities (24, 26). Ib binds to a receptor and transfers Ia into the cytosol, where Ia ADP-ribosylates actin (7, 20, 29, 36).
Clostridium botulinum and Clostridium spiroforme have been reported to produce binary toxins (the light-chain component and the heavy-chain component), each of which is similar in molecular weight and action (1, 23). It has been reported that these light-chain components ADP-ribosylate synthetic substrates, such as homo-poly-l-arginine and actin-like proteins in platelets (27, 28), and that the heavy-chain component (C2II) of C. botulinum C2 toxin is responsible for the binding of the light-chain component to the eucaryotic cell surface (2, 3, 9).
Ia belongs to a family of bacterial ADP-ribosylating toxins, along with the light-chain components of C. botulinum C2 toxin and C. spiroforme toxin, as well as pertussis toxin, diphtheria toxin, cholera toxin, and Escherichia coli heat-labile enterotoxin (8). ADP-ribosylating toxins are known to contain three conserved regions, aromatic residues R and E-X-E and hydrophobic residue S-T-S (8). Perelle et al. (21) reported that Arg-295, Glu-378, and Glu-380 are involved in the ADP-ribosylation of Ia. We reported that Glu-378 within E378-X-E380 in Ia plays an important role in NAD+-glycohydrolase activity but not in ADP-ribosyltransferase activity and that Glu-380 within E378-X-E380 and Arg-295 within aromatic residue-R295 in Ia are essential for the catalytic mechanism of ADP-ribosyltransferase activity (19).
Recent cloning and sequencing of the gene encoding Ib revealed significant sequence similarities with the genes of binding components of other actin-ADP-ribosylating toxins (11) and the gene of the protective antigen (PA) of Bacillus anthracis (20). Crystallography of PA showed that PA is characterized by a four-domain structure (22). Milne et al. (14) reported that PA inserted into membranes oligomerized to form ion-conductive channels in the plasma membrane. Popoff et al. (23) showed that iota-like toxin from C. spiroforme also entered cells by receptor-mediated endocytosis. Considine and Simpson (7) reported that C2 toxin enters cells by receptor-mediated endocytosis. Furthermore, the Aktories group has reported that C2II oligomers bind to carbohydrate receptor, assemble C2I, enter cells by endocytosis, and release C2I into the cytosol after acidification of the endosomal compartment (3, 9). Recently, the Stiles group (12, 33) has reported that Ib strongly binds to the cell surface receptor of Vero and MDCK cells, which are sensitive to iota-toxin, but not that of HeLa, NIH 3T3, and MIC-5 cells, which are highly resistant to the toxin and that the N- and C-terminal domains of Ib are important for Ia docking and cell surface binding, respectively. Furthermore, Blocker et al. (4) reported that acidification of endosomal compartments is required for iota-toxin uptake. However, little is known about the biological activity of Ib and the role of Ib in the cell entry of iota-toxin. Here we present evidence for K+ release induced by Ib and the oligomerization of Ib in Vero cells.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and DNA-modifying enzymes were obtained from Takara Shuzo Co. Ltd. (Kyoto, Japan) and Toyobo Co. Ltd. (Osaka, Japan). Bovine serum albumin was purchased from Sigma (St. Louis, Mo.). pT-7 blue was obtained from Novagen (Madison, Wis.), and pGEX-4T-1 from Amersham Pharmacia Biotech (Tokyo, Japan). All other chemicals were of analytical grade.
Bacterial strains and plasmids.
C. perfringens type E strain NCIB 10748 (donated by M. Popoff) was grown in brain heart infusion broth under anaerobic conditions. Plasmid DNA was extracted and purified as described by Perelle et al. (20).
E. coli JM109 or C600 was the host for the plasmid used. B. subtilis ISW1214 was used for production of the toxin (16).
DNA cloning.
C. perfringens type E strain NCIB 10748 encoding the entire iota-toxin gene was amplified by PCR using a pair of primers, Ib 1 ((5′-GATGGAACTGTAACAAAACT-3′) and primer Ib 2 (5′-AGGTAGCCAGTATGAATAAC-3′), designed from the published iota-toxin sequence (20). The PCR was carried out for 25 cycles under standard reaction conditions with a GeneAmp DNA amplification reagent kit (Perkin-Elmer Cetus, Norwalk, Conn.). A 2.8-kbp fragment obtained after the amplification was gel purified and cloned into pT-7 blue (pTIB).
Preparation of Ib expression vector.
Plasmid pTIB was used as a template for PCR. To obtain the Ib gene, the forward primer (5′-GGAGATCTGAAGAAATAACAAATGA-3′) was used to introduce a 5′ BglII site and the reverse primer (5′-CCCTCGAGCTAGCTTTATTAATTTT-3′) was used to introduce a 3′ XhoI site. The product was isolated, digested with BglII and XhoI, and ligated into a pGEX vector previously digested with BamHI and XhoI so that the correct reading frame was maintained with the thrombin cleavage site under the glutathione S-transferase (GST) gene (pGEX-Ib).
Expression and purification of rIb.
Recombinant Ib (rIb) was expressed as a fusion protein with GST in E. coli BL21 as described previously (18). After growth (at 30°C) and induction (with isopropyl-β-d-thiogalactopyranoside to 1 mM) of a large culture, the cells were centrifuged and disrupted by a sonicator on ice in a short burst. Centrifugation of the lysate and passage of the soluble fraction over a glutathione-Sepharose (Amersham Pharmacia Biotech) column yielded the GST-Ib fusion protein at about 2 mg/liter. After cleavage of the purified GST-Ib with chymotrypsin as described by Gibert et al. (10), the cleaved protein was passed through a glutathione-Sepharose column and then subjected to anion-exchange chromatography (Mono-Q HR10/10; Amersham Pharmacia Biotech). Native Ib was purified from culture supernatant fluid of C. perfringens type E NCTC 8084 as described previously (25). Protein was eluted with a 0 to 1.0 M NaCl gradient. Fractions containing Ib were collected. The protein assay was performed by the method of Bradford (6).
Determination of molecular weight by SDS-3.5% PAGE.
To determine the molecular weight of Ib, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 3.5% gel was performed according to the method of Weber and Osborn (37). High-molecular-mass standards for SDS-PAGE were phosphorylase b cross-linked SDS molecular weight markers (Sigma): monomer (97 kDa), dimer (194 kDa), trimer (292 kDa), tetramer (390 kDa), pentamer (487 kDa), and hexamer (584 kDa). Low-molecular-mass standards containing phosphorylase b (94 kDa), albumin (67 kDa), and ovalbumin (43 kDa) were obtained from Amersham Pharmacia Biotech.
Determination of cytotoxic activity.
The test for cytotoxicity was done on Vero cells. The cells were cultivated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. For cytotoxicity assays, cells were inoculated in 48-well tissue culture plates (Falcon, Oxnard, Calif.). Various concentrations of Ib and 50 ng of Ia/ml purified as reported previously (19) were mixed in DMEM containing 10% fetal bovine serum and were inoculated onto cell monolayers. The cells were observed for morphological alterations 8 h after inoculation as described previously (19).
To measure the effect of methylamine, ethylamine, or bafilomycin A1 (Sigma) on the cytotoxicity of iota-toxin, Vero cells were preincubated with the agent at 37°C for 1 h and then incubated with various concentrations of Ia and Ib (150 ng/ml) at 37°C for 8 h.
Determination of ADP-ribosylating activity.
Vero cells were washed with cold phosphate-buffered saline (PBS) and detached from their support with a rubber policeman. They were resuspended in 100 μl of 10 mM Tris-HCl buffer (pH 7.2)-50% sucrose containing 10 μM leupeptin and 1 μM pepstatin. Cells were lysed by three cycles of freezing and thawing. The protein content was determined by the Bradford method (6). ADP-ribosylation was performed as described previously (19). In brief, 10 mg of cell lysate was incubated with [32P]NAD (0.1 mM) and 50 ng of Ia for 60 min at 37°C. The reaction was stopped by SDS-sample buffer, and the samples were boiled for 3 min and subjected to SDS-PAGE. [32P]ADP-ribosylated actin was detected by autoradiography with a BAS 2000 system (Fuji Photo Film Co., Ltd., Tokyo, Japan).
Iodination of Ib.
125I-labeled rIb and oligomer were prepared according to the method of Bolton and Hunter (5) as described previously (15). rIb or the oligomer (25 μg) was incubated with 250 μCi of 125I-labeled Bolton-Hunter reagent (2,000 Ci/mmole; Amersham Pharmacia Biotech). The labeled Ib plus Ia retained over 90% of the original toxicity (lethality) of Ia plus rIb.
Binding of labeled Ib to Vero cells.
Vero cells were seeded in 48-well plates at a concentration of 5 × 104 cells/well. The labeled Ib (10 ng, representing 12,000 cpm) was added to cells in 0.2 ml of DMEM containing 10% fetal bovine serum at 4°C for 2 h. Unbound Ib was removed by chilled PBS, and then the cells were incubated in prewarmed DMEM containing 10% fetal bovine serum at 37°C in a 5% CO2 incubator. The cells were washed twice with 1 ml of PBS, lysed in SDS-sample buffer, and analyzed by SDS-PAGE (7.5% polyacrylamide gel). The gel was dried and autoradiographed. For quantification of radioactivity in the gel, a Fuji BAS 2000 system was used.
To measure the cellular uptake of Ib, the Ib remaining on the cell surface was digested by incubation with a prewarmed pronase solution (5 mg/ml in PBS) at 37°C for 5 min (35). After the addition of chilled calf serum to the plate, cells were harvested to a tube and were washed once with chilled DMEM containing 10% fetal bovine serum. Detection of labeled Ib in the cells was performed using a bioimage analyzer.
To measure the effect of methylamine or bafilomycin A1 on uptake of Ib, Vero cells were preincubated with these agents at 37°C for 1 h and were incubated with labeled Ib at 37°C for various periods.
Measurement of K+ flux.
Vero cells were seeded in 9-cm2 dishes (Sumitomo Bakelite Co., Tokyo, Japan) at a concentration of 2 × 105 cells/dish, incubated for 18 h at 37°C in a 5% CO2 incubator, and washed twice in Hanks balanced salt solution (HBSS). The cells were incubated with or without rIb or the oligomer in 1 ml of HBSS at 37°C for 18 h. K+ concentrations of the supernatants were determined at given times by atomic absorption spectrometer (Hitachi Z-8200) as described previously (17).
RESULTS
Purification of rIb from E. coli transformant containing the Ib gene.
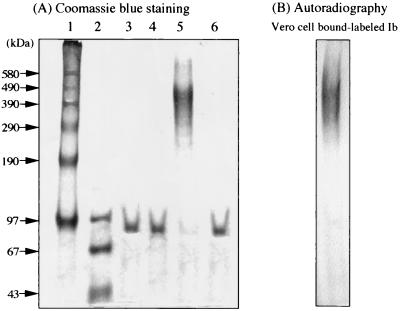
Sonicated lysates of E. coli transformants containing the Ib gene were centrifuged at 40,000 × g and 4°C for 30 min. The GST-Ib which was purified on a glutathione-Sepharose column was digested with chymotrypsin. To obtain purified rIb, the chymotrypsin-treated fraction was loaded onto a Pharmacia fast-performance liquid chromatography Mono-Q HR55 column equilibrated with 0.02 M Tris-HCl buffer (pH 8.0). The column was eluted with a linear gradient of 0 to 1.0 M NaCl. Two protein peaks which were eluted with 0.2 M NaCl (first fraction) and 0.5 M NaCl (second fraction) and reacted on anti-Ib antiserum were observed. These fractions were dissolved in SDS sample solution, analyzed by SDS-3.5% PAGE with or without prior heating for 3 min at 95°C, and stained with Coomassie blue. Figure 1 shows two bands, one at about 76 kDa which corresponds to the expected size of Ib (Fig. 1A, lane 3) and one at about 500 kDa (Fig. 1B, lane 5), a protein band of a high molecular mass. However, when these fractions were heated for 3 min at 95°C and analyzed by SDS-PAGE, one band at about 76 kDa only was detected in each fraction (Fig. 1A, lanes 4 and 6). These observations showed that monomeric Ib and the oligomer were isolated by this procedure. It therefore is apparent that Ib spontaneously forms an oligomer. Incubation of Vero cells with the purified rIb (50 ng/ml) in the presence of Ia (50 ng/ml) induced rounding of the cells but that with the purified oligomer (5 mg/ml) in the presence of Ia (50 ng/ml) did not. Furthermore, the cytotoxicity induced by Ia plus the rIb was identical to that induced by Ia plus native Ib purified from culture of the C. perfringens type E strain (data not shown).
FIG. 1.
Purification of rIb from GST-Ib. (A) Chymotrypsin-treated GST-Ib purified by glutathione-Sepharose column chromatography was applied to a Mono-Q column and was eluted with a 0 to 1.0 M NaCl gradient in 20 mM Tris-HCl buffer (pH 8). The first and second fractions were analyzed by SDS-3.5% PAGE with incubation at 37°C for 15 min or with heating at 95°C for 3 min. Protein (10 μg each) was stained with Coomassie blue. Lane 1, high-molecular-mass standard (phosphorylase b cross-linked; Sigma); lane 2, low-molecular-mass standard (Pharmacia); lane 3, first fraction (37°C, 15 min); lane 4, first fraction (95°C, 3 min); lane 5, second fraction (37°C, 15 min); and lane 6, second fraction (95°C, 3 min). (B) Vero cells were incubated with 125I-labeled Ib at 37°C for 60 min, washed, dissolved in SDS-sample buffer, and subjected to SDS-3.5% PAGE prior to incubation at 37°C for 15 min. After electrophoresis, the gel was dried and autoradiographed. The radiolabeled band was analyzed with a Fuji BAS 2000 system.
Biological action of Ib.
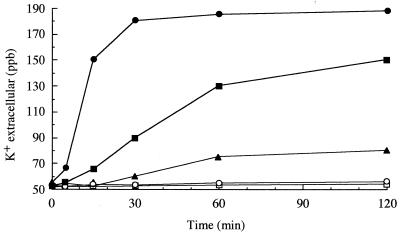
It has been reported that Ib alone has no activity for living cells (7, 24, 26). Vero cells were still alive, even when the cells were incubated with large amounts of Ib (10 μg/ml) at 37°C for 12 h. To clarify the biological activity of Ib, Vero cells were incubated with various concentrations of rIb in HBSS (pH 7.0) at 37°C. At concentrations at over 28 ng/ml, Ib induced release of K+ from the cells in a dose- and time-dependent manner, as shown in Fig. 2. However, the oligomer at a concentration of 5.6 μg/ml did not cause K+ release from the cells. Incubation of the cells in the absence of rIb at 37°C (Fig. 2) or in the presence of rIb at 4°C (data not shown) induced no release of K+. Furthermore, when the cells were incubated with rIb at 4°C for 120 min, washed, and incubated at 37°C for 60 min, release of K+ was observed (data not shown). In addition, the Ib-induced K+ release was completely inhibited by anti-Ib antiserum (data not shown).
FIG. 2.
K+ release from Vero cells treated with rIb. Vero cells were incubated with rIb or the oligomer at 37°C. K+ concentrations in the medium were determined at given times by atomic absorption spectrometer. Symbols: •, Ib at 280 ng/ml; ▪, Ib at 56 ng/ml; ▴, Ib at 28 ng/ml; ○, Ib oligomer at 5.6 mg/ml; and □, without Ib (control)
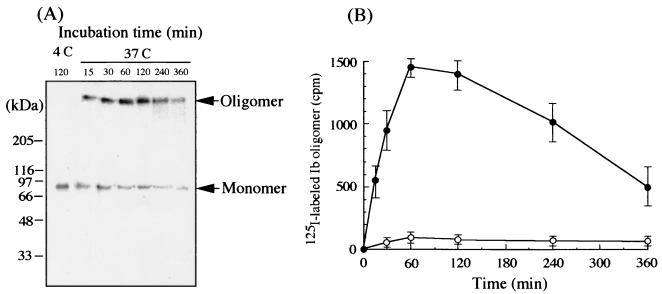
Next, to test if Ib forms an oligomer on Vero cells, the cells were preincubated with 100 ng of 125I-labeled rIb/ml in DMEM containing 10% fetal bovine serum (pH 7.0) at 4°C for 120 min, washed, and incubated in the same medium at 37°C for various periods. The treated cells were dissolved in SDS sample solution, analyzed by SDS-7.5% PAGE without heating at 95°C, and autoradiographed (Fig. 3A). The result showed two bands, a 76-kDa band that corresponds to the expected size of Ib cleaved by chymotrypsin (10) and a second band of 500 kDa (Fig. 3A). The migration of the 500-kDa band was identical to that of the oligomer of rIb (Fig. 2B). Furthermore, heating of the Ib-treated cells in SDS sample solution at 95°C abolished the 500-kDa band and induced the appearance of the 76-kDa band (data not shown). In addition, even in cells treated with rIb at 4°C for 120 min, washed, and incubated at 4°C for 120 min, no oligomer formation was observed (data not shown). On the other hand, when Vero cells were incubated with the labeled Ib in the presence of 50 mol of excess cold rIb at 4°C for 120 min, washed, and incubated at 37°C for 120 min, the 76- and 500-kDa bands were below the detection limit (Fig. 3B), suggesting that the binding of the labeled Ib to the cells is specific. These observations suggest that Ib formed a hexameric or heptameric oligomer on the cell membrane. The formation of the 500-kDa band, the oligomer, reached a maximum after 60 min of incubation and later decreased in a time-dependent manner, as shown in Fig. 3B. When Vero cells were incubated with labeled rIb at 4°C for 120 min, only the monomer band was detected (Fig. 3A), suggesting that the oligomer formation of Ib is dependent on temperature. No release of the labeled oligomer was detected from Vero cells that were incubated with the labeled Ib at 37°C for 60 min and then washed (data not shown). It therefore appears that the oligomer is not released from the cells.
FIG. 3.
Oligomer formation of rIb on Vero cells. (A) Vero cells were incubated with 125I-labeled Ib at 4°C for 120 min, washed, and incubated at 37°C for the time (in minutes) indicated. The cells were dissolved in SDS-sample buffer and subjected to SDS-7.5% PAGE without heating followed by autoradiography. (B) Vero cells were incubated with 125I-labeled Ib at 4°C for 120 min, washed, and incubated in the presence (○) or the absence (•) of a 50-mol excess of cold rIb at 37°C for the time indicated. The radioactivity of labeled Ib oligomer (a 500-kDa band) was determined using a Fuji BAS 2000 system.
The fate of Ib bound to Vero cells.
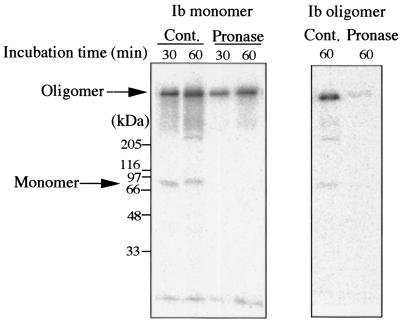
To clarify the disappearance of the oligomer bound to Vero cells, the cells were incubated with labeled rIb at 4°C for 120 min, washed, and then incubated at 37°C for 30 or 60 min. The treated cells were incubated with pronase at 37°C for 5 min, dissolved in SDS sample solution without heating, analyzed by SDS-PAGE, and autoradiographed (Fig. 4). The monomer band (about 76 kDa) was detected from the untreated cells (Cont.) but not from the cells treated with pronase. However, treatment of the cells with pronase led to no reduction in the oligomer band (about 500 kDa). On the other hand, when the cells were incubated with the labeled oligomer (the second fraction) at 37°C for 60 min, washed, and treated with or without pronase at 37°C for 5 min, SDS-PAGE analysis indicated that the pronase-untreated cells showed the oligomer band but that the pronase-treated cells did not (Fig. 4). Pronase completely hydrolyzed rIb bound to the cells at 4°C for 120 min under experimental conditions (data not shown).
FIG. 4.
Effect of pronase treatment on Vero cell-bound oligomer. Vero cells were incubated with 125I-labeled Ib or labeled oligomer at 4°C for 120 min, washed, and incubated at 37°C for the time indicated. The cells were washed with PBS and were again incubated with or without pronase at 37°C for 5 min. Cell samples were dissolved in SDS-sample buffer and subjected to SDS-7.5% PAGE without heating followed by autoradiography. Cont., control.
As shown in Fig. 3, the oligomer band decreased in a time-dependent manner after 60 min of the incubation, but any release of radioactivity was not detected on the gel. To determine the fate of the oligomer in the cells, the cells were incubated with the lableled Ib at 37°C for 60 min, washed with the fresh medium three times, and further incubated in the fresh medium at 37°C for 60 min. The cells were dissolved in SDS sample solution without heating and analyzed by SDS-7.5% PAGE and autoradiographed. The monomer and the oligomer mainly were detected from the cells, and further faint bands such as a tailing of the oligomer were detected, as shown in Fig. 5. However, the tailing phenomenon of the monomer was not observed under experimental conditions (Fig. 5). The monomer band only was detected from the culture supernatants on the gel, but any other band was not detected (data not shown).
FIG. 5.
Change of rIb bound to Vero cells. Vero cells were incubated with 125I-labeled Ib at 37°C for 60 min, washed, and incubated at 37°C for the time indicated. Lane 1, 0 min; lane 2, 60 min; lane 3, 120 min; lane 4, 240 min; and lane 5, 360 min. The cells were dissolved in SDS-sample buffer and subjected to SDS-7.5% PAGE without heating followed by autoradiography.
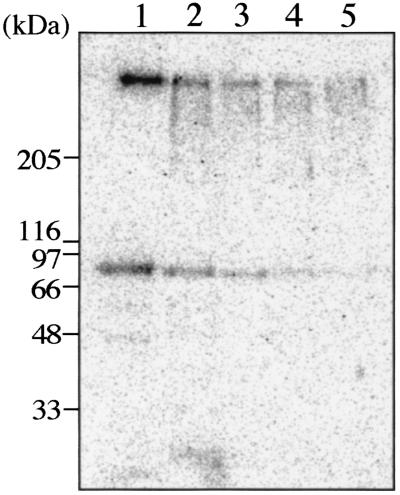
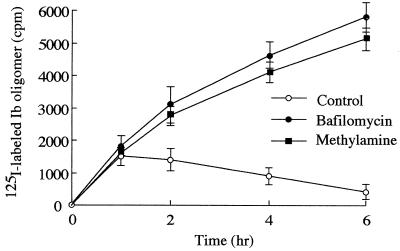
Barth et al. (3) reported that C2 toxin enters the cell via receptor-mediated endocytosis. Our observation also suggests that Ib enters Vero cells. Thus, we investigated whether Ib uptake into the cell is related to an acidic cellular compartment. It has been reported that bafilomycin A1 inhibits the vacuole-type H+-ATPase (34) and that methylamine and ethylamine neutralize pH in the vacuole (13). K+ release from Vero cells treated with rIb in the presence of these inhibitors was not different from that of the cells treated with rIb only (data not shown). However, when Vero cells were incubated with labeled rIb, the oligomer formation profile of rIb was changed in the presence of bafilomycin or methylamine (Fig. 6). In the absence of these inhibitors, the amount of oligomer associated with cells was maximum at about 1 h, as shown in Fig. 3, and then decreased, while in the presence of these inhibitors, the amount of radioactivity increased with time, reaching a maximum in about 6 h. SDS-PAGE analysis indicated that the oligomer remarkably increased in the presence of these inhibitors (data not shown). These observations suggested that the treatment of the cells with rIb in the presence of bafilomycin or methylamine induced accumulation of the oligomer in the cell. These inhibitors did not affect Ib binding to the cells under experimental conditions (data not shown). Furthermore, bafilomycin, methylamine, or ethylamine inhibited rounding of the cells induced by rIb plus Ia (Table 1). In addition, these inhibitors blocked ADP-ribosylation of actin in cytosol induced by rIb plus Ia (data not shown). These inhibitors themselves did not have any morphological effects under our experimental conditions.
FIG. 6.
Effect of bafilomycin A1 or methylamine on uptake of Ib oligomer in Vero cells. Vero cells were preincubated with or without 500 nM bafilomycin A1 or 20 mM methylamine for 60 min. 125I-labeled Ib was added to the cell suspension, and the cells were incubated at 37°C for the time indicated. The cells were dissolved in SDS-sample buffer and subjected to SDS-7.5% PAGE without heating followed by autoradiography. The radioactivity of the oligomer was determined using a Fuji BAS 2000 system.
TABLE 1.
Effect of methylamine, ethylamine, and bafilomycin A1 on rounding activity of iota-toxinlegend
| Treatment | Effect of dose of Ia (ng/ml)
|
||||
|---|---|---|---|---|---|
| 1 | 5 | 10 | 50 | 100 | |
| None | − | + | ++ | +++ | +++ |
| Methylamine | − | − | − | − | + |
| Ethylamine | − | − | − | − | + |
| Bafilomycin A1 | − | − | − | − | − |
Vero cells were preincubated with methylamine (20 mM), ethylamine (20 mM), or bafilomycin A1 (500 nM) at 37°C for 30 min. The cells were then incubated with various amounts of Ia in the presence of Ib (50 ng/ml). Cell rounding was recorded after 8 h. Cell rounding in a Vero cell assay (cytotoxic activity) was scored as follows: +++, 100% rounding; ++, 50 to 80% rounding; +, 20 to 40% rounding and −, no rounding.
DISCUSSION
The genes encoding iota-toxin revealed that Ib shares significant sequence homology with PA (about 34% identity and 54% similarity) (20). The receptor-bound PA is reported to be internalized by receptor-mediated endocytosis (30). Milne et al. (14) reported that PA formed oligomers during intoxication of mammalian cells. The Aktories group has reported that C2II toxin, which resembles PA, forms an oligomer during intoxication (3). It has been reported that Ib possesses no activity (2, 7, 31). However, Ib has been reported to specifically bind to Vero cells (4). In the present study, we examined the role of Ib in the biological activities induced by iota-toxin. rIb at the concentration of 10 mg/ml showed no cytotoxicity for Vero cells under our experimental conditions. However, here we showed that rIb of over 28 ng/ml induced release of K+ from the cell in a dose- and time-dependent manner. The result shows that Ib forms ion-permeable channels in Vero cells. The SDS-PAGE analysis revealed that the migration of rIb complex formed by incubation of Vero cells with rIb at 37°C was identical to that of rIb oligomer isolated from the Ib fraction, suggesting that Ib forms an oligomer on the cell membrane. The high-molecular-mass complex of about 500 kDa exceeds the monomer molecular mass by about six times. It is likely that hexamers or heptamers of Ib as the oligomer are present on membrane. The time course of K+ release from the cells treated with rIb was coincidental with that of oligomer formation, suggesting that the release is dependent on the formation. On the other hand, when Vero cells were incubated with the oligomers isolated from the rIb fraction, release of K+ from the cells was not detected, rounding was not observed in the presence of Ia, and the oligomers which bound to the cells were hydrolyzed by treatment with pronase. These observations imply that the monomer molecule of Ib binds to the cell and then forms a functional oligomer on the membrane.
The monomer and the oligomer mainly were detected from the cells incubated with rIb at 37°C; these bands decreased in a incubation time-dependent manner, and faint bands (>76 kDa) such as a tailing of the oligomer were detected. It therefore is likely that the oligomer was gradually degraded in the cells. In addition, the monomer band was detected in the culture supernatants, but no other band was detected. We cannot explain the observations. However, from these results, it is speculated that the oligomers degraded in the cell accumulated in cytosol under our experimental conditions. It is possible that the monomer is secreted from the cells after degradation of the oligomer to the monomer in the cells. On the other hand, the result does not exclude the possibility that the monomer bound to the surface of the cells was released to the buffer.
The oligomer formed on the cell surface by incubation of Vero cells with rIb was resistant to pronase treatment, but the oligomer isolated from the rIb fraction was sensitive to enzyme treatment, even if incubated with the cell membranes. It therefore is possible that the oligomer formed by incubation of the cells with rIb inserts the cell membrane. PA and diphtheria toxin were shown to translocate into the cytosol from an endosomal compartment. C2 toxin was reported to enter cells by receptor-mediated endocytosis (7). Furthermore, Barth et al. (3) reported that C2 toxin is translocated and released into the cytosol after acidification of the endosomal compartment. Considine and Simpson (7) suggested that internalization of Ib by receptor-mediated endocytosis is essential for toxicity of iota-toxin. We obtained evidence of the cellular routing of Ib as follows. Treatment of Vero cells with Ib in the presence of bafilomycin, methylamine, or ethylamine, which prevents acidification of endosomal vesicles, had no effect on Ib-induced K+ release but resulted in an accumulation of Ib in the cells. Furthermore, treatment of the cells with Ib plus Ia in the presence of these inhibitors had no effect on the cells and, in addition, did not cause ADP-ribosylation of actin in the cytosol, as reported by Blocker et al. (4). These observations suggest that Ib forms ion-permeable channels in the membrane (functional oligomer formation) and that, after acidification of the endosomal compartment, the oligomeric Ib is inserted into the endosomal membrane. Endocytosis seems to occur with Ib in the presence of Ia. It therefore is likely that Ib alone is endocytosed after binding to the cell. We cannot explain why the formation of an oligomer does not lead to cytotoxicity. However, the Ib-induced release of K+ and accumulation of Ib in the cells reached a maximum within 30 and 60 min, respectively. Thus, it is speculated that the oligomers formed in the cell membranes are inserted into the endosome within 30 min and that Ib is translocated into the cytosol from the endosomal compartment within 60 min and is degraded in cytosol.
Acknowledgments
We thank Yumi Yoshida, Yusuke Emoto, and Noriko Teramoto for competent technical assistance.
This research was supported in part by a Grant-in-Aid for Scientific Research (12670270) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Editor: J. T. Barbieri
REFERENCES
- 1.Aktories, K., M. Barmann, I. Ohishi, S. Tsuyama, K. H. Jakobs, and E. Habermann. 1986. Botulinum C2 toxin ADP-ribosylates actin. Nature 322:390-392. [DOI] [PubMed] [Google Scholar]
- 2.Aktories, K., and A. Wegner. 1989. ADP-ribosylation of actin by clostridial toxins. J. Cell Biol. 109:1385-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, H., D. Blocker, J. Behlke, W. Bergsma-Schutter, A. Brisson, R. Benz, and K. Aktories. 2000. Cellular uptake of Clostridium botulinum C2 toxin requires oligomerization and acidification. J. Biol. Chem. 275:16478-16483. [DOI] [PubMed] [Google Scholar]
- 4.Blocker, D., J. Behlke, K. Aktories, and H. Barth. 2001. Cellular uptake of Clostridium perfringens binary iota-toxin. Infect. Immun. 69:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolton, A. E., and M. M. Hunter. 1973. The labeling of proteins to high specific radioactivities by conjugation to a 125I-containing acylating agent. Biochem. J. 133:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Considine, R. V., and L. L. Simpson. 1991. Cellular and molecular actions of binary toxins possessing ADP-ribosyltransferase activity. Toxicon 29:913-936. [DOI] [PubMed] [Google Scholar]
- 8.Domenighini, M., and R. Rappuoli. 1996. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol. Microbiol. 21:667-674. [DOI] [PubMed] [Google Scholar]
- 9.Eckhardt, M., H. Barth, D. Blocker, and K. Aktories. 2000. Binding of Clostridium botulinum C2 toxin to asparagine-linked complex and hybrid carbohydrates. J. Biol. Chem. 275:2328-2334. [DOI] [PubMed] [Google Scholar]
- 10.Gibert, M., L. Petit, S. Raffestin, A. Okabe, and M. R. Popoff. 2000. Clostridium perfringens iota-toxin requires activation of both binding and enzymatic components for cytopathic activity. Infect. Immun. 68:3848-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura, K., T. Kubota, I. Ohishi, E. Isogai, H. Isogai, and N. Fujii. 1998. The gene for component-II of botulinum C2 toxin. Vet. Microbiol. 62:27-34. [DOI] [PubMed] [Google Scholar]
- 12.Marvaud, J. C., T. Sith, M. L. Hale, M. R. Popoff, L. A. Smith, and B. G. Stiles. 2001. Clostridium perfringens iota-toxin: mapping of receptor binding and Ia docking domains on Ib. Infect. Immun. 69:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekada, E., T. Uchida, and Y. Okada. 1981. Methylamine stimulates the action of ricin toxin but inhibits that of diphtheria toxin. J. Biol. Chem. 256:1225-1228. [PubMed] [Google Scholar]
- 14.Milne, J. C., D. Furlong, P. C. Hanna, J. S. Wall, and R. J. Collier. 1994. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J. Biol. Chem. 269:20607-20612. [PubMed] [Google Scholar]
- 15.Nagahama, M., and J. Sakurai. 1992. High-affinity binding of Clostridium perfringens epsilon-toxin to rat brain. Infect. Immun. 60:1237-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagahama, M., Y. Okagawa, T. Nakayama, E. Nishioka, and J. Sakurai. 1995. Site-directed mutagenesis of histidine residues in Clostridium perfringens alpha-toxin. J. Bacteriol. 177:1179-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagahama, M., S. Ochi, and J. Sakurai. 1998. Assembly of Clostridium perfringens epsilon-toxin on MDCK cell membrane. J. Nat. Toxins 7:291-302. [PubMed] [Google Scholar]
- 18.Nagahama, M., A. Kihara, T. Miyawaki, M. Mukai, Y. Sakaguchi, S. Ochi, and J. Sakurai. 1999. Clostridium perfringens beta-toxin is sensitive to thiol-group modification but does not require a thiol group for lethal activity. Biochim. Biophys. Acta 1454:97-105. [DOI] [PubMed] [Google Scholar]
- 19.Nagahama, M., Y. Sakaguchi, K. Kobayashi, S. Ochi, and J. Sakurai. 2000. Characterization of the enzymatic component of Clostridium perfringens iota-toxin. J. Bacteriol. 182:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perelle, S., M. Gibert, P. Boquet, and M. R. Popoff. 1993. Characterization of Clostridium perfringens iota-toxin genes and expression in Escherichia coli. Infect. Immun. 61:5147-5156. (Erratum, 63:4967, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perelle, S., M. Domenighini, and M. R. Popoff. 1996. Evidence that Arg-295, Glu-378, and Glu-380 are active-site residues of the ADP-ribosyltransferase activity of iota toxin. FEBS Lett. 395:191-194. [DOI] [PubMed] [Google Scholar]
- 22.Petosa, C., R. J. Collier, K. R. Klimpel, S. H. Leppla, and R. C. Liddington. 1997. Crystal structure of the anthrax toxin protective antigen. Nature 385:833-838. [DOI] [PubMed] [Google Scholar]
- 23.Popoff, M. R., F. W. Milward, B. Bancillon, and P. Boquet. 1989. Purification of Clostridium spiroforme binary toxin and activity of the toxin on HEp-2 cells. Infect. Immun. 57:2462-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakurai, J. 1995. Toxins of Clostridium perfringens. Rev. Med. Microbiol. 6:175-185. [Google Scholar]
- 25.Sakurai, J., and K. Kobayashi. 1995. Lethal and dermonecrotic activities of Clostridium perfringens iota toxin: biological activities induced by cooperation of two nonlinked components. Microbiol. Immunol. 39:249-253. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai, J., M. Nagahama, and S. Ochi. 1997. Major toxins of Clostridium perfringens. J. Toxicol.-Toxin Rev. 16:195-214. [Google Scholar]
- 27.Schering, B., M. Barmann, G. S. Chhatwal, U. Geipel, and K. Aktories. 1988. ADP-ribosylation of skeletal muscle and non-muscle actin by Clostridium perfringens iota toxin. Eur. J. Biochem. 171:225-229. [DOI] [PubMed] [Google Scholar]
- 28.Simpson, L. L. 1984. Molecular basis for the pharmacological actions of Clostridium botulinum type C2 toxin. J. Pharmacol. Exp. Ther. 230:665-669. [PubMed] [Google Scholar]
- 29.Simpson, L. L., B. G. Stiles, H. H. Zepeda, and T. D. Wilkins. 1987. Molecular basis for the pathological actions of Clostridium perfringens iota toxin. Infect. Immun. 55:118-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh, Y., K. R. Klimpel, N. Arora, M. Sharma, and S. H. Leppla. 1994. The chymotrypsin-sensitive site, FFD315, in anthrax toxin protective antigen is required for translocation of lethal factor. J. Biol. Chem. 269:29039-29046. [PubMed] [Google Scholar]
- 31.Stiles, B. G., and T. D. Wilkins. 1986. Purification and characterization of Clostridium perfringens iota toxin: dependence on two nonlinked proteins for biological activity. Infect. Immun. 54:683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiles, B. G., and T. D. Wilkins. 1986. Clostridium perfringens iota toxin: synergism between two proteins. Toxicon 24:767-773. [DOI] [PubMed] [Google Scholar]
- 33.Stiles, B. G., M. L. Hale, J. C. Marvaud, and M. R. Popoff. 2000. Clostridium perfringens iota-toxin: binding studies and characterization of cell surface receptor by fluorescence-activated cytometry. Infect. Immun. 68:3475-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umata, T., Y. Moriyama, M. Futai, and E. Mekada. 1990. The cytotoxic action of diphtheria toxin and its degradation in intact Vero cells are inhibited by bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPase. J. Biol. Chem. 265:21940-21945. [PubMed] [Google Scholar]
- 35.Umata, T., and E. Mekada. 1998. Diphtheria toxin translocation across endosome membranes. A novel cell permeabilization assay reveals new diphtheria toxin fragments in endocytic vesicles. J. Biol. Chem. 273:8351-8359. [DOI] [PubMed] [Google Scholar]
- 36.Vandekerckhove, J., B. Schering, M. Barmann, and K. Aktories. 1987. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 225:48-52. [DOI] [PubMed] [Google Scholar]
- 37.Weber, K., and M. Osborn. 1967. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]