Abstract
Babesia bovis rhoptry-associated protein 1 (RAP-1), which confers partial protection against B. bovis challenge, is recognized by antibodies and T lymphocytes from cattle that have recovered from infection and are immune to subsequent challenge. RAP-1 is a 60-kDa protein with an N-terminal (NT) region that contains four cysteine residues conserved among all Babesia RAP-1 family members and a C-terminal (CT) region that contains multiple, degenerate, tandem 23-amino-acid (aa) repeats. To define the location of CD4+-T-cell epitopes for vaccine development using a recombinant protein or minigene construct, a series of truncated recombinant RAP-1 proteins and peptides were tested for stimulation of T-cell lines derived from B. bovis-immune cattle. CD4+-T-cell lines from three B. bovis-immune cattle with different DRB3 haplotypes responded to the NT region of RAP-1, whereas T cells from only one animal responded weakly to the CT region. T-cell lines from the three individuals recognized two to six NT-region peptides spanning aa 134 to 316 and representing at least four dominant epitopes. Using RAP-1-specific CD4+-T-cell clones, two NT-region epitopes, EYLVNKVLYMATMNYKT (aa 187 to 203) and EAPWYKRWIKKFR (aa 295 to 307), and one CT-region repeat epitope, FREAPQATKHFL, which is present twice at aa positions 391 to 402 and 414 to 425, were identified. Several peptides representing degenerate repeats of the agonist CT-region peptide FREAPQATKHFL neither stimulated responses of T-cell clones specific for this peptide nor inhibited responses to the agonist peptide. Upon stimulation with specific antigen, T-cell clones specific for NT or CT epitopes produced gamma interferon. The presence of T-helper-cell epitopes in the NT domain of RAP-1, which is highly conserved among otherwise antigenically different strains of B. bovis, supports the inclusion of this region in vaccine constructs to be tested in cattle.
Babesia bovis is an intraerythrocytic protozoan parasite of cattle transmitted by Boophilus ticks. B. bovis infection causes a significant disease characterized by fever, anemia, cachexia, and, in severe cases, cerebral babesiosis or respiratory distress syndrome (12, 45). Similar to what occurs with malaria, control measures are largely inadequate and the lack of a safe and effective vaccine for babesiosis results in large economic losses in tropical countries where babesiosis is endemic (23).
Protective acquired immune mechanisms against apicomplexan parasites, including B. bovis, are thought to involve both humoral and cell-mediated responses which result in macrophage activation and parasite killing (12, 21, 24, 25, 32, 44). It was recently demonstrated that age-related resistance to acute infection correlated with earlier induction of interleukin-12 (IL-12), gamma interferon (IFN-γ), and nitric oxide responses in the spleens of infected calves than in more susceptible adults (22). Effective vaccines against these complex organisms will likely require the inclusion of multiple B- and T-lymphocyte epitopes representing different antigens and perhaps different parasite stages. In fact, better protection against the development of parasitemia in cattle challenged with B. bovis was achieved by a combination of two recombinant parasite proteins than by either protein alone (44). Identification of T-cell epitopes on candidate vaccine antigens conserved among parasite strains is therefore needed to develop a multiple-antigen, peptide-based vaccine or minigene construct. A study has characterized merozoite antigens that induce memory IFN-γ-producing CD4+-T-cell responses in cattle recovered from infection and subsequently shown them to be protected against B. bovis challenge (7, 34). Several immunostimulatory proteins, including rhoptry-associated protein 1 (RAP-1), cysteine-rich protein 12D3, spherical body protein SBP1, major surface antigen 1 (MSA-1), and the small heat shock protein Hsp-20, were identified (12, 14, 34). Of these, B. bovis RAP-1 is one of the leading candidate antigens for vaccine development. Immunization of cattle with recombinant RAP-1 antigen was shown to significantly reduce parasitemia upon challenge infection (44). RAP-1 is recognized by sera from B. bovis-infected cattle that are immune to challenge (38). It is an immunodominant antigen for T lymphocytes derived from such cattle and is recognized by memory CD4+-T-helper cells that produced IFN-γ but little or no IL-4 (10, 12).
B. bovis RAP-1 is a member of a family of 58- to 60-kDa proteins of multiple genes that have also been identified in Babesia bigemina, Babesia caballi, Babesia canis, Babesia divergens, and Babesia ovis parasites (17, 18, 27, 33, 37, 40). In B. bovis, RAP-1 is encoded by two nearly identical genes and consists of a unique N-terminal (NT) region (amino acids [aa] 1 to 316) and a C-terminal (CT) region (aa 317 to 565) that contains seven tandem repeats of a degenerate 23-aa sequence (Fig. 1A) (37-40). The NT region of RAP-1 is conserved in the B. bigemina RAP-1a orthologues , with an approximately 45% amino acid identity and a 14-aa sequence (PLSLPNPYQLDAAF) that is completely conserved despite changes in codon usage (37). Furthermore, four cysteine residues between aa 80 and 105 are conserved in all Babesia RAP-1 orthologues, consistent with structural conservation.
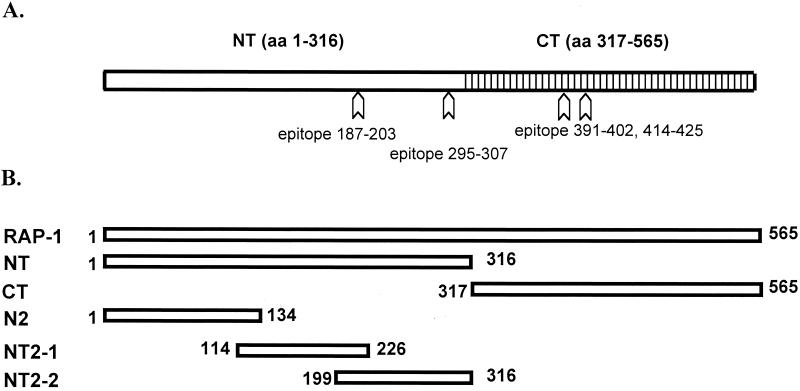
FIG. 1.
Schematic structure of B. bovis RAP-1. (A) RAP-1 consists of an NT region (aa 1 to 316) and a CT region (aa 317 to 565) containing tandem repeats of degenerate amino acid sequences. Arrows indicate locations of Th-cell epitopes identified in this study. (B) Truncated recombinant B. bovis RAP-1 proteins were used to identify CD4+-T-cell epitopes.
In contrast to those of other babesial proteins such as MSA-1 (13, 36), B. bovis RAP-1 T-cell epitopes are conserved among different strains of B. bovis (10). However, the T-cell epitopes in RAP-1 have not been mapped. Since antigens containing degenerative repetitive sequences may act as altered peptide ligand antagonists and compete for agonist peptide binding or induce T-cell anergy (20), it is important to determine whether T-cell epitopes are located in the unique NT region or the repeat-rich CT domain. In addition, it is important to identify “universal” T-cell epitopes that are recognized by individuals with multiple major histocompatibility complex (MHC) haplotypes to achieve the broadest possible protection at the population level. To achieve these objectives, T-cell responses against recombinant RAP-1 NT- and RAP-1 CT-region proteins (herein designated RAP-1 NT and RAP-1 CT, respectively) were determined using oligoclonal T-cell lines derived from three B. bovis-immune cattle with different MHC class II haplotypes. Weak CT-region-specific responses were observed in only one animal, whereas a predominant response against aa 114 to 316 in the NT region was observed in all the cattle. Synthetic peptides and cloned CD4+ T cells were used to further define epitopes in the NT region. These results support the use of selected B. bovis RAP-1 NT-region epitopes to prime for strong, anamnestic, interstrain-cross-reactive CD4+-T-cell responses upon exposure to B. bovis.
MATERIALS AND METHODS
Animals.
Crossbred Brahman × Angus cow C97 was infected by infestation with Boophilus microplus tick larvae infected with the Mexico strain of B. bovis. Crossbred Hereford cow C15 was inoculated intravenously with merozoites of the same strain cultured in autologous erythrocytes. Charolais cow G3 was infected with the Texas strain of B. bovis. All animals were confirmed to be resistant to homologous strain challenge (6, 15). The MHC class II DRB3 alleles were determined by the PCR-restriction fragment length polymorphism method developed by van Eijk et al. (42). The DRB3 haplotypes used were as follows: for cow C97, DRB3 *15/*34; for cow C15, DRB3 *24/*18; and for cow G3, DRB3 *18/*23. Because T-cell epitopes were precisely mapped using T-cell clones from cow C97, DRB3 exon 2 of cow C97 was further cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, Calif.) and sequenced to determine both alleles (DRB3 *3001/*4501) according to an established protocol (46). Nomenclature of the alleles was as described in the BoLA nomenclature website (http://www2.ri.bbscr.ac.uk.bola/).
B. bovis strains and antigen preparation.
For use in all T-cell experiments, B. bovis merozoite antigen was prepared from the cultured Mexico strain by homogenization of merozoites with a French pressure cell (SLM Instruments, Inc., Urbana, Ill.) and then ultracentrifugation to yield a fraction enriched in parasite cell membranes (CM) (6). Antigen from uninfected red blood cells (URBC) was similarly prepared for use as a negative control.
GenBank accession numbers (shown in parentheses) were obtained for the rap-1 gene sequences from the following strains of B. bovis: Texas (AF030054), Mexico (AF027149, L77326), Brazil (AF030057, AF030058), Argentina S2P (AF030053, AF030056), and Argentina R1A (AF030055, AF030062). Recombinant antigens spanning RAP-1 were prepared from the Mexico strain, and for one experiment, full-length RAP-1 from the Argentina R1A strain was made. Recombinant RAP-1 antigens were expressed as histidine-tagged thioredoxin fusion proteins. Each RAP-1 insert was amplified by reverse transcription-PCR using specific primer sets (Table 1) and was ligated into the pBAD/Thio-TOPO vector (Invitrogen) as recommended by the manufacturer. The direction and frame of each clone were confirmed by sequencing. For the expression of each recombinant antigen, a single recombinant Escherichia coli colony was inoculated in 2 ml of Luria broth containing 50 μg of ampicillin per ml and incubated overnight at 37°C with shaking. The culture was inoculated into 150 ml of Luria broth and incubated for 2 h at 37°C with shaking. Protein expression was induced with 0.2% L-(+)-arabinose for 4 h. E. coli cells were pelleted by centrifugation, lysed with 5 ml of PNLB (50 mM K2HPO4, 400 mM NaCl, 100 mM KCl, 10% glycerol, 0.5% Triton X-100, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride [pH 7.8]), and sonicated for 30 s. After centrifugation at 4°C at 10,000 × g for 15 min, the pellet was further lysed with 5 ml of guanidine buffer (6 M guanidine HCl, 100 mM K2HP4, 10 mM Tris [pH 8.0]) and sonicated for 15 s. The solubilized protein was centrifuged at 4°C at 10,000 × g for 15 min, and supernatant was collected for antigen purification. Recombinant protein was purified by affinity to a Ni2+ column with ProBond resin (Invitrogen) as recommended by the manufacturer. Briefly, a column containing a 1-ml bed volume of ProBond resin was equilibrated with guanidine buffer. The supernatant containing solubilized protein was applied to the column and mixed on a rotator at 4°C for 30 min. After incubation, the column was washed once with urea buffer 1 (8 M urea, 100 mM K2HPO4, 10 mM Tris [pH 8.0]), once with urea buffer 2 (8 M urea, 100 mM K2HPO4, 10 mM Tris [pH 6.3]), and twice with PNB (50 mM K2HPO4, 400 mM NaCl, 100 mM KCl, 10% glycerol, 10 mM imidazole [pH 7.8]). Protein was eluted with a series of imidazole solutions (50, 100, 200, and 500 mM and 1 M imidazole in PNB), and eluates were collected into separate tubes. The yields and expected sizes of the products were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Recombinant histidine-tagged major surface protein 5 (MSP-5) of Anaplasma marginale was expressed in the same vector and prepared in an identical manner for use as a negative control protein. Proteins were analyzed for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining. For use in T-cell proliferation assays, proteins were dialyzed extensively against phosphate-buffered saline and quantified using a Micro bicinchoninic acid protein assay reagent kit (Pierce, Rockford, Ill.).
TABLE 1.
Primer sequences used for amplification of recombinant RAP-1 proteins
| Protein | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| RAP-1 | ATGAGAATCATTAGCGGCGTTG | GAGGTATCCGGCGGTGTCTTCAC |
| RAP-1 NT | ATGAGAATCATTAGCGGCGTTG | TTGGGTAACGTTTTTAGAG |
| RAP-1 CT | CCTACAAAGAAGTTCATCGAGG | GAGGTATCCGGCGGTGTCTTCAC |
| RAP-1 N2 | ATGAGAATCATTAGCGGCGTTG | ACGCAGCATCCAACTGGTAA |
| RAP-1 NT2-1 | CCGTTGTATCAAGAGTACCAAC | ACTGAATATCTTTGTAGTGAAG |
| RAP-1 NT2-2 | ATGAACTACAAGACTTATTTG | TTGGGTAACGTTTTTAGAG |
Dialyzed recombinant proteins were also tested for the presence of endotoxin by using the Limulus amebocyte lysate assay according to the manufacturer's instructions (BioWhittaker, Inc., Walkersville, Md.). In samples containing 25 μg of protein per ml (the highest concentration of protein used in proliferation assays), only trace amounts (0.06 or 0.12 endotoxin unit per ml of sample) were detected. A rate of 0.06 endotoxin unit per ml is the limit of sensitivity of the assay, and this corresponds to approximately 6 pg of endotoxin per ml. For individual recombinant proteins, endotoxin levels were as follows: 12 pg/ml for RAP-1, 6 pg/ml for RAP-1 NT, 12 pg/ml for RAP-1 N2, 12 pg/ml for RAP-1 NT2-1, 12 pg/ml for RAP-1 NT2-2, 6 pg/ml for RAP-1 CT, and 6 pg/ml for MSP-5.
Peptides were synthesized by Gerhardt Munske, Laboratory for Biotechnology and Bioanalysis I, Washington State University, Pullman. A 30-mer peptide (P1) derived from A. marginale MSP-2 was used as a negative control peptide (10). Peptides were dissolved in phosphate-buffered saline. All proteins and peptides were stored at −20 or −80°C.
B. bovis-specific T-cell lines and clones.
To establish short-term T-cell lines, 4 × 106 peripheral blood mononuclear cells (PBMC) derived from B. bovis-immune cattle were cultured in 24-well plates (Costar, Cambridge, Mass.) in 1.5 ml of complete RPMI 1640 medium (6) with 25 μg of B. bovis CM antigen per ml for 1 to 6 weeks as described previously (10). T-cell lines were maintained by weekly stimulation of 7 × 105 T lymphocytes with B. bovis antigen and 2 × 106 irradiated (3,000 rads), autologous PBMC per well as antigen-presenting cells (APC). T-cell lines were often allowed to rest for 1 week after stimulation with B. bovis antigen to avoid high background proliferation.
RAP-1-specific clones 1B9, 1E7, 1G12, and 4D10 from cow C97 were obtained previously (10) and were shown in the present study to be specific for the NT2-2 region. Eight RAP-1 NT2-1-specific clones, including 3B3, 3C12, and 3E1, were obtained by limiting-dilution cloning of a 6-week-old cell line established by sequentially culturing PBMC from cow C97 with B. bovis CM antigen and then recombinant RAP-1 NT2-1 antigen. RAP-1 CT-specific T-cell clones 2C3, 2D11, 2G8, 2H1, and 3E5 were similarly obtained from a 3-week-old C97 cell line stimulated with B. bovis CM and then recombinant RAP-1 CT antigen. T-cell clones were maintained in 24-well plates by stimulating 7 × 105 cells with 10 μg of specific recombinant RAP-1 antigen and autologous APC per ml in a volume of 1.5 ml of complete RPMI 1640 medium containing 10% bovine T-cell growth factor (5). Cell surface phenotypes of the clones were determined by flow cytometry using monoclonal antibodies (MAbs) specific for bovine CD3 (MAb MM1A), CD4 (MAb CACT138A), CD8 (MAb CACT80C), and γδ T-cell receptor 1 (MAb CACT61A) purchased from the Washington State University Monoclonal Antibody Center, Pullman. All clones expressed the CD3+ CD4+ CD8− γδ T-cell receptor-negative phenotype.
T-cell proliferation assays.
Proliferation assays were performed for 3 days in duplicate or triplicate wells of round-bottom 96-well plates at 37°C in a humidified atmosphere of 5% CO2 in air (6, 10). Briefly, 3 × 104 T cells and 2 × 105 autologous APC were cultured with antigen in a total volume of 100 μl of complete RPMI 1640 medium (6). To measure proliferation, cells were radiolabeled for the last 7 to 18 h of culture with 0.25 μCi of [3H]thymidine (Dupont, New England Nuclear, Boston, Mass.) and radiolabeled nucleic acids were harvested onto glass filters and counted in a beta counter.
To determine whether T-cell clones were DRB3 restricted or DQ restricted, 2 × 105 autologous APC were preincubated in 96-well plates for 1 to 2 h with a 4-μg/ml concentration of MAb against the bovine MHC class II molecule (1, 2, 19) DRα (MAb TH14B) or DQα (MAb TH22A). These immunoglobulin G2a (IgG2a) MAbs and an isotype control MAb (Colis205D) were obtained from the Washington State University Monoclonal Antibody Center and purified by affinity chromatography to protein G using an Equilibrate Hi Trap protein G column (Pharmacia Biotech, Piscataway, N.J.) according to the manufacturer's protocol. This amount of MAb was determined to provide optimal blocking of proliferation without nonspecific effects.
Student's one-tailed t test was used to determine statistically significant differences in levels of proliferation induced by using different antigens or anti-MHC class II or control MAb.
Detection of IFN-γ in supernatants of T-cell clones.
To measure IFN-γ production, T-cell clones were cultured with specific antigen under the same conditions as for proliferation assays in 96-well plates and 50-μl supernatants were harvested from each well before being pulsed with [3H]thymidine. The bovine IFN-γ assay was performed with an enzyme-linked immunosorbent assay kit (BOVIGAM; CSL Limited, Parkville, Victoria, Australia) according to the manufacturer's protocol. The IFN-γ activities in culture supernatants diluted 1:4 to 1:20 were determined by comparison with a standard curve obtained with a supernatant from a Mycobacterium bovis purified protein derivative-specific Th-lymphocyte clone that contained 440 U of IFN-γ/ml (previously determined by the neutralization of vesicular stomatitis virus [9]). In our assay, 1 U corresponds to 1.7 ng of IFN-γ (3).
RESULTS
T-cell responses to RAP-1 were predominantly directed against the NT region.
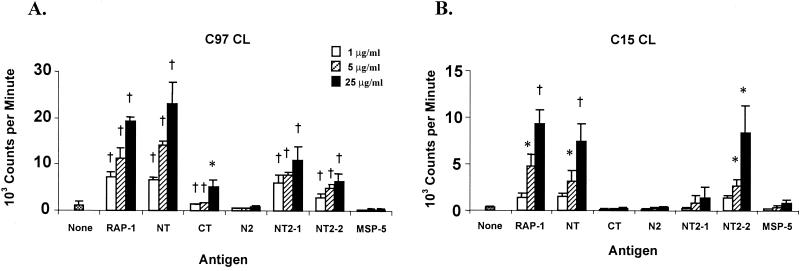
It was previously shown that B. bovis RAP-1 is strongly immunogenic for T-helper cells from the B. bovis-immune cattle C97, C15, and G3 and that Th-cell epitopes are conserved in different B. bovis strains (10, 34). To localize the T-cell epitopes, a full-length recombinant RAP-1 and several truncated RAP-1 proteins spanning the NT and CT regions were expressed (Fig. 1) and tested for stimulation of T-lymphocyte proliferation from B. bovis-immune cattle. T-cell lines consisting predominantly of CD4+ T cells established by stimulating PBMC from cow C97 or C15 with B. bovis CM responded significantly to RAP-1 NT at a level comparable to that induced by full-length RAP-1. In contrast, RAP-1 CT elicited a weak but significant proliferative response from C97 T cells and did not elicit proliferation of C15 T cells (Fig. 2).
FIG. 2.
Proliferative responses of T-cell lines (CL) from the B. bovis-immune cattle C97 and C15 against recombinant B. bovis RAP-1 NT and RAP-1 CT antigens. Short-term T-cell lines from cows C97 and C15 were stimulated with 1, 5, and 25 μg of full-length RAP-1, RAP-1 NT, and RAP-1 CT/ml and proteins RAP-1 N2, RAP-1 NT2-1, and RAP-1 NT2-2 spanning the RAP-1 NT region (Fig. 1). Results are the mean counts per minute of triplicate cultures of T cells stimulated with antigen for 3 days ± 1 standard deviation and are representative of at least three experiments. Responses significantly higher than those for A. marginale MSP-5 control antigen are indicated with a † (P < 0.005) or ∗ (P < 0.05).
Because the predominant T-cell response was directed against the NT region of RAP-1, additional truncated recombinant proteins, RAP-1 N2, RAP-1 NT2-1, and RAP-1 NT2-2 spanning the NT region (Fig. 1B), were used to further define T-cell epitopes in this region (Fig. 2). T-cell lines responded to the NT2-1 and/or NT2-2 region but not to the N2 region. Immune animal G3 also responded to the NT2-1 and NT2-2 regions of RAP-1 but not to the CT region, although high background responses to all E. coli-expressed proteins prevented us from obtaining statistically significant results in multiple assays (data not shown).
To rule out any possibility that differential T-lymphocyte responses to different fusion proteins could be explained by contaminating endotoxin (26), all samples were tested for endotoxin by the sensitive Limulus amebocyte lysate assay. The amount of endotoxin detected, which was 6 to 12 pg per ml of protein sample (25 μg per ml of protein used in proliferation assays), did not correlate with induction of T-lymphocyte proliferation. Furthermore, this trace amount of endotoxin is 10,000-fold less than the amount of 100 to 10,000 ng of endotoxin per ml reported to optimally stimulate proliferation of human T lymphocytes (26).
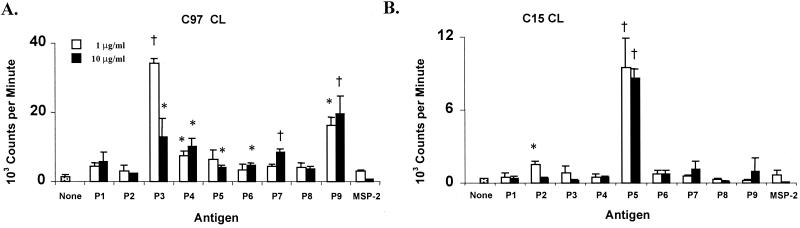
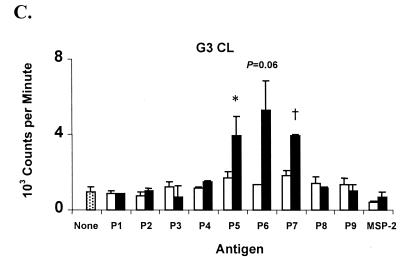
Since the T-cell epitopes were concentrated within the second half of the NT region of RAP-1, overlapping 30-mer peptides spanning this region were synthesized for use in epitope mapping (Table 2). Comparison of B. bovis antigen-stimulated T-cell lines from B. bovis-immune cattle revealed differences in levels of epitope recognition (Fig. 3), and these results were repeated in three or more assays. Cow C97 responded strongly to peptides P3 and P9 (P < 0.005). Although responses to peptides P4, P5, P6, and P7 were weaker than responses to peptides P3 and P9, they were also significant when the highest peptide concentration was used. In some experiments, the response to peptide P5 was comparable to the responses to peptides P3 and P9 (data not shown). T-cell lines from cow C15 responded strongly to peptide P5 and also weakly, but significantly, to peptide P2 (P < 0.05). T-cell lines established from cow G3 responded significantly to peptides P5 and P7 (P < 0.05) and also responded to P6 (P = 0.06). These results indicate that the NT region of RAP-1 contains several different T-cell epitopes, the dominant ones of which are contained within peptides P3 and P9 (cow C97), P5 (all three cattle), and P7 (cow G3).
TABLE 2.
Synthetic peptides spanning RAP-1 NT2-1 and RAP-1 NT2-2
| Peptide | Amino acid positions | Peptide sequence |
|---|---|---|
| P1 | 134-163 | FRLFKESASNPAKNSVKREWLRFRNGANHG |
| P2 | 154-183 | LRFRNGANHGDYHYFVTGLLNNNVVHEEGT |
| P3 | 174-203 | NNNVVHEEGTTDVEYLVNKVLYMATMNYKT |
| P4 | 194-223 | LYMATMNYKTYLTVNSMNAKFFNRFSFTTK |
| P5 | 214-243 | FFNRFSFTTKIFSRRIRQTLSDIIRWNVPE |
| P6 | 234-263 | SDIIRWNVPEDFEERSIERITQLTSSYEDY |
| P7 | 254-283 | TQLTSSYEDYMLTQIPTLSKFARRYADMVK |
| P8 | 274-303 | FARRYADMVKKVLLGSLTSYVEAPWYKRWI |
| P9 | 294-316 | VEAPWYKRWIKKFRDFFSKNVTQ |
FIG. 3.
Proliferative responses of T-cell lines (CL) from the B. bovis-immune cattle C97, C15, and G3 against synthetic peptides (Table 2) spanning the RAP-1 NT region. Short-term T-cell lines from cows C97, C15, and G3 were stimulated with 1 and 10 μg of peptides P1 to P9/ml. As a negative control, a 30-mer peptide derived from A. marginale MSP-2 was used. Results are the mean counts per minute of triplicate cultures of T cells stimulated with antigen for 3 days ± 1 standard deviation. Responses significantly higher than those for A. marginale MSP-2 peptide are indicated with a † (P < 0.005) or ∗ (P < 0.05).
Epitope mapping with RAP-1 NT-specific T-cell clones.
T-cell clones derived from cow C97, which recognized the largest number of peptides, were used to more precisely define T-cell epitopes in the RAP-1 NT region. Clones specific for RAP-1 NT2-1 were obtained after the stimulation of T-cell lines with full-length RAP-1, followed by stimulation with RAP-1 NT2-1. Eight clones proliferated to B. bovis CM, RAP-1, and RAP-1 NT2-1 antigens, and three representative clones are shown in Table 3. When tested for IFN-γ production, clones 3B3 and 3E1, respectively, produced 40.9 ± 2.7 and 76.9 ± 0.4 U of IFN-γ per ml in response to B. bovis CM antigen, compared with 4.3 ± 0.1 and 2.4 ± 0.3 U of IFN-γ per ml in response to URBC antigen. Previously obtained RAP-1-specific CD4+-T-cell clones 1B9, 1E7, 1G12, and 4D10 (10; W. C. Brown, unpublished observations) were shown in the present study to respond to the NT2-2 region of RAP-1 (three clones are shown in Table 3). These clones also produced IFN-γ and little or no IL-4 (10). Importantly, these T cells respond to both recombinant protein and naturally processed epitopes, which suggests that as a vaccine component, peptides containing T-cell epitopes may be able to prime for a response that will be elicited upon exposure to the pathogen. Although the in vivo RAP-1-specific T-cell cytokine response has not been demonstrated, the type 1 cytokine profile exhibited by in vitro-cultured RAP-1-specific T-cell clones is consistent with a type 1 response elicited during acute infection (22).
TABLE 3.
Proliferative responses of B. bovis RAP-1 NT2-1- and RAP-1 NT2-2-specific CD4+-T-cell clones
| Antigena | Radioactivity incorporated byb:
|
|||||
|---|---|---|---|---|---|---|
| RAP-1 NT2-1 specific Th-cell clone:
|
RAP-1 NT2-2 specific Th-cell clone:
|
|||||
| 3B3 | 3C12 | 3E1 | 1E7 | 1B9 | 1G12 | |
| None (medium) | 4,958 ± 115 | 6,795 ± 512 | 2,537 ± 316 | 265 ± 5 | 4,525 ± 607 | 1,618 ± 36 |
| URBC | 4,817 ± 609 | 5,652 ± 94 | 2,631 ± 221 | 228 ± 73 | 5,706 ± 1,530 | 1,672 ± 616 |
| B. bovis CM | 27,157 ± 432d | 51,356 ± 1,093d | 37,254 ± 3,116d | 7,246 ± 977c | 16,163 ± 2,621d | 4,900 ± 333c |
| RAP-1 | 41,403 ± 1,601d | 48,123 ± 2,227d | 45,388 ± 303d | 16,774 ± 21d | 53,465 ± 1,075d | 5,560 ± 730c |
| RAP-1 NT2-1 | 29,028 ± 1,622d | 43,030 ± 1,063d | 23,183 ± 2,314c | 187 ± 3 | 4,547 ± 320 | 1,250 ± 16 |
| RAP-1 NT2-2 | 761 ± 473 | 3,807 ± 812 | 2,235 ± 2,175 | 5,826 ± 225d | 59,340 ± 4,174d | 4,447 ± 425c |
A final concentration of 25 μg of antigen per ml was used.
Results are the mean counts per minute of duplicate cultures of T cells stimulated with antigen for 3 days ± 1 standard deviation.
Significantly higher than the value for the medium control (P < 0.05).
Significantly higher than the value for the medium control (P < 0.005).
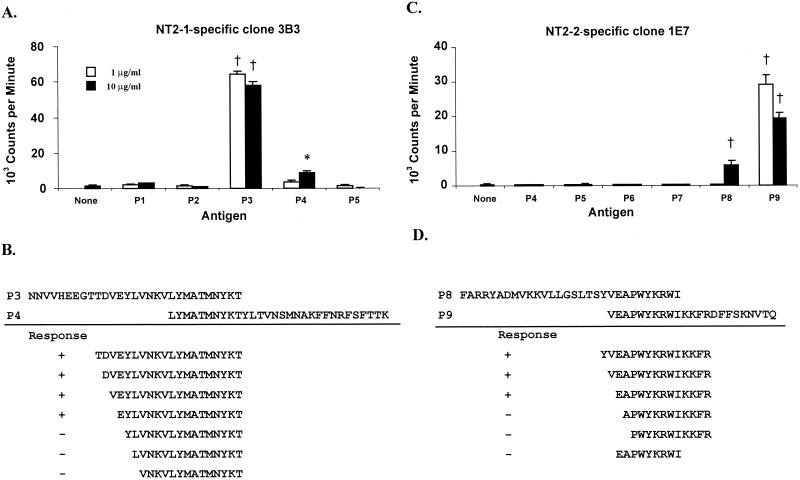
The epitopes recognized by RAP-1 NT-specific T-cell clones were mapped using synthetic peptides spanning the NT2-1 and NT2-2 fragments of RAP-1 (Table 2). All eight RAP-1 NT2-1-specific T-cell clones responded strongly to peptide P3 (aa 174 to 203) and weakly to peptide P4 (aa 194 to 223). Results from a representative clone (3B3) are shown in Fig. 4A. The T-cell epitope was defined to a 17-aa sequence, EYLVNKVLYMATMNYKT (aa 187 to 203), using a series of additional peptides (Fig. 4B and data not shown). E187 appeared to be essential for stimulation, and peptide P4 lacked the 7 NT aa (EYLVNKV) required for an optimal response. All four RAP-1 NT2-2-specific CD4+-T-cell clones responded to peptides P8 (aa 274 to 303) and P9 (aa 294 to 316), which overlap by 10 aa. Results from a representative clone (1E7) are shown in Fig. 4C. By using peptides containing the overlapping sequence, we identified a 13-aa sequence, EAPWYKRWIKKFR (aa 295 to 307), as a T-cell epitope for RAP-1 NT2-2-specific CD4+-T-cell clones (Fig. 4D and data not shown). Since the 9-aa sequence (EAPWYKRWI, aa 295 to 303) of peptide P8 overlaps the 13-aa epitope, this 9-mer peptide was also examined. Proliferation against this peptide was not observed, suggesting that although peptide EAPWYKRWI consists of the core T-cell epitope sequence, T-cell stimulation requires amino acid sequence extension from this core sequence towards either the N terminus or the C terminus.
FIG. 4.
T-cell-epitope mapping for RAP-1 NT2-1- and RAP-1 NT2-2-specific CD4+-T-cell clones. (A) Response of RAP-1 NT2-1-specific T-cell clone 3B3 to peptides P1 to P5; (B) summary of the response of clone 3B3 to smaller peptides spanning peptides P3 and P4; (C) response of RAP-1 NT2-2-specific T-cell clone 1E7 to peptides P4 to P9; (D) summary of the response of clone 1E7 to smaller peptides spanning peptides P8 and P9. For all assays, T cells were cultured in duplicate with medium or peptide and APC for 3 days. Responses significantly higher than those for T cells in the medium are indicated with a † (P < 0.005) or ∗ (P < 0.05).
T-cell epitopes within the RAP-1 CT region.
Although the response of C97 T cells to RAP-1 CT was weakly positive (Fig. 2A), this response was nevertheless significant and reproducible. To map T-cell epitope(s) in the CT region, C97 T-cell lines were stimulated with RAP-1 CT and cloned by limiting dilution. Five RAP-1 CT-specific CD4+-T-cell clones were obtained. All clones responded to recombinant full-length RAP-1, RAP-1 CT, and B. bovis CM antigens, demonstrating that recall responses to the CT region of RAP-1 were elicited by the native protein (Table 4, experiment 1).
TABLE 4.
Proliferative responses of B. bovis RAP-1 CT-specific CD4+-cell clones
| Expt | Antigena | Radioactivity incorporated by CT-region-specific Th-cell cloneb:
|
||||
|---|---|---|---|---|---|---|
| 2G8 | 2H1 | 2D11 | 3E5 | 2C3 | ||
| 1 | None (medium) | 1,879 ± 103 | 982 ± 78 | 127 ± 48 | 187 ± 32 | 208 ± 72 |
| URBC | 1,891 ± 84 | 1,027 ± 155 | 138 ± 21 | 202 ± 1 | 202 ± 43 | |
| B. bovis CM | 7,924 ± 254d | 8,746 ± 801c | 1,108 ± 87c | 2,393 ± 388c | 1,441 ± 359c | |
| RAP-1 | 13,914 ± 604d | 47,443 ± 836d | 14,913 ± 1,074d | 45,475 ± 2,011d | 22,889 ± 1,926d | |
| RAP-1 NT | 1,784 ± 40 | 1,753 ± 420 | 121 ± 2 | 153 ± 23 | 164 ± 46 | |
| RAP-1 CT | 11,214 ± 1,333d | 44,239 ± 1,071d | 17,882 ± 954d | 47,253 ± 54d | 34,001 ± 401d | |
| 2 | None (medium) | 480 ± 8 | 170 ± 28 | 680 ± 6 | 365 ± 35 | 370 ± 55 |
| CT-P1 | 489 ± 47 | 168 ± 23 | 830 ± 100 | 361 ± 41 | 391 ± 48 | |
| CT-P2 | 4,858 ± 383d | 9,337 ± 0d | 40,659 ± 3,083d | 29,111 ± 801d | 29,070 ± 1,793d | |
A final concentration of 25 μg of antigen per ml was used.
Results are the mean counts per minute of duplicate cultures of T cells stimulated with antigen for 3 days ± 1 standard deviation.
Significantly higher than the value for the medium control (P < 0.05).
Significantly higher than the value for the medium control (P < 0.005).
The CT region of RAP-1 is composed of seven degenerate 23-aa repeats within aa 317 to 534 (37). Peptides CT-P1 and CT-P2 (Table 5) were previously used for identification of B-cell epitopes (38) and were tested with RAP-1 CT-specific T-cell clones. All five CD4+-T-cell clones responded strongly to peptide CT-P2, representing aa 386 to 408 (Table 4, experiment 2). Two clones tested, 2G8 and 2H1, respectively produced 70.5 ± 0.2 and 61.1 ± 3.8 U of IFN-γ per ml when they were stimulated with B. bovis CM antigen, compared with 2.4 ± 0.1 and 0.7 ± 0.1 U of IFN-γ per ml when they were stimulated with URBC antigen. To further define the T-cell epitope(s) within peptide CT-P2, additional shorter peptides representing variations of the repeat were synthesized (Table 5) and tested for stimulation of the CT-P2-specific T-cell clones. A 12-aa sequence, FREAPQATKHFL (A397), present at positions 391 to 402, was identified as the epitope for the CT-P2-specific T-cell clones (data not shown). An exact repeat of the 12-aa sequence (A420) also exists at positions 414 to 425. We then determined if the other degenerate repetitive sequences could stimulate the T-cell clones (results are summarized in Table 5). None of the variant peptides present in the Mexico strain RAP-1 CT-region repeat sequences stimulated T-lymphocyte proliferation.
TABLE 5.
Summary of the responses of RAP-1 CT-specific CD4+-T-cell clones against variant peptides
| Amino acid positions | Peptide sequence or antigena | Response |
|---|---|---|
| 372-394 (CT-P1) | PQVTKHFFDENIGQPTKEFFREA | − |
| 386-408 (CT-P2) | PTKEFFREAPQATKHFLDENIGQ | + |
| 368-382 | FKEAPQVTKHFFDEN | − |
| 391-405 | FREAPQATKHFLDEN | + |
| 414-428 | FREAPQATKHFLGEN | + |
| 437-451 | FKDVPQVTKKVITEN | − |
| 391-402, 414-425 | FREAPQATKHFL | + |
| 414-425 (R1A strain) | FREAPQVTKHFL | − |
| 1-595 (R1A strain) | Recombinant full-length RAP-1 | + |
Bold letters mark amino acids different from those of the T-cell epitope FREAPQATKHFL.
Examination of the GenBank database showed that the rap-1 gene sequence for the Argentina R1A strain of B. bovis contains two versions of the CT-P2 T-cell epitope, FREAPQATKHFL (A397) and FREAPQVTKHFL (V420), so it was of interest to determine if this naturally occurring A397-to-V420 substitution would affect T-cell recognition (Table 5). The variant peptide did not elicit proliferation of the T-cell clones, whereas a full-length recombinant RAP-1 protein derived from strain R1A elicited strong T-cell proliferative responses by the clones (data not shown). The responses of the clones to the R1A strain RAP-1 protein are therefore directed against the stimulatory T-cell epitope FREAPQATKHFL (A397), and the presence of the variant epitope FREAPQVTKHFL (V420) in this strain does not appear to negatively influence this response. Together, these results suggest that nonstimulatory, degenerate repeat sequences in the CT region are not antagonistic for T-cell proliferation to the CT-region epitope defined in this study, at least not when in vitro recall responses are measured.
MHC class II restriction.
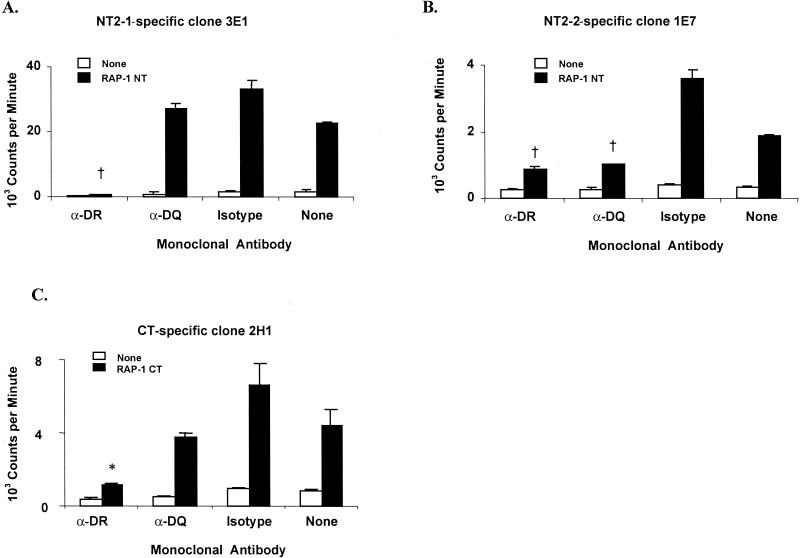
C97 T-cell clones specific for the three epitopes defined on RAP-1 were tested for responses to specific antigen in the presence of MAb directed against either the DRB3 or the DQ class II molecule. RAP-1 NT2-1-specific clones 3E1 and 3B3 and RAP-1 CT-specific clones 2H1 and 2G8 were significantly inhibited by MAb specific for bovine DQ (Fig. 5A and C and data not shown). In contrast, RAP-1 NT2-2-specific clones 1E7 and 4D10 were significantly inhibited by MAb against both DQ and DRB3 (Fig. 5B and data not shown). An isotype-matched IgG2a control MAb had no inhibitory effect on antigen-induced T-cell proliferation by these clones. These experiments were repeated at least three times with similar results.
FIG. 5.
Determination of MHC class II restriction patterns of C97 RAP-1-specific T-cell clones. (A) RAP-1 NT2-1-specific CD4+-T-cell clone 3E1; (B) RAP-1 NT2-2-specific CD4+-T-cell clone 1E7; (C) RAP-1 CT-specific CD4+-T-cell clone 2H1. Autologous APC (2 × 105) were incubated with a 4-μg/ml final concentration of MAb TH14B (anti-DR), MAb TH22A5 (anti-DQ), or isotype-matched control MAb Colis205D (isotype) in 96-well plates for 1 h before the addition of 3 × 104 T cells and 12.5 μg of antigen per ml. Results are the mean counts per minute of duplicate cultures ± 1 standard deviation. Antigen-specific responses that are significantly lower than those of T cells cultured without MAb are indicated with a † (P < 0.005) or ∗ (P < 0.05).
DISCUSSION
RAP-1 is a candidate for inclusion in a B. bovis vaccine based on its ability to induce partial protection in vaccinates, defined as a significant reduction in parasitemia upon challenge (44). RAP-1 is recognized by both antibody and T lymphocytes from cattle recovered from B. bovis infection that were immune to subsequent challenge (10, 38). Furthermore, the RAP-1 orthologue in B. bigemina also conferred protection against homologous challenge (8, 27). Examination of the cytokine response demonstrated that the RAP-1-specific CD4+ T cells of B. bovis and B. bigemina expressed predominantly IFN-γ, with little or no expression of IL-4 (9, 10, 30). IFN-γ is important for enhancing production of the opsonizing IgG2 isotype in cattle and for activating macrophages to produce toxic molecules such as nitric oxide, which inhibits the growth of B. bovis (9, 10, 12, 32, 35). However, it is unknown whether this in vitro RAP-1-specific cytokine response reflects the actual antigen-specific response in vivo. The present study has identified the nonrepetitive NT domain of B. bovis RAP-1 as the major domain to elicit memory T-cell responses in three immune cattle with different MHC class II haplotypes. Based on peptide-specific proliferative responses, at least four T-cell epitopes, which are found in peptides P3, P5, P7, and P9, are present in the NT region (aa 134 to 316) of RAP-1. The dominant response by C97 cell lines to peptides P3 and P9 was confirmed by the responses of two sets of T-cell clones which also were specific for epitopes present within these peptides. The rap-1 sequences that contain T-cell epitopes and are reported in the GenBank database are conserved among all B. bovis strains, including Texas, Mexico, Brazil, and Argentina (S2P and R1A strains), which is consistent with the previously reported recognition of different strains of B. bovis by the RAP-1 NT2-2-specific T-cell clones (9). These data directly support the inclusion of the NT region or defined epitopes in a multiepitope vaccine approach.
In this study, the NT and CT regions of B. bovis RAP-1 protein were considered two units because the molecular structures of these regions are distinct. The CT region is comprised of seven degenerate 23-aa repeats arranged in tandem that are not conserved among RAP-1 family members of different Babesia species. The NT region has a unique sequence, containing four cysteine residues and other sequence motifs that are highly conserved among RAP-1 orthologues from different babesial species (17, 33, 41). Several conserved oligopeptide motifs in the NT region were also found in the Plasmodium falciparum rhoptry protein, AMA-1/pf83, and members of the Pf60 multigene family (4, 41). The presence of such conserved motifs in apicomplexan parasites suggests that the NT region plays a critical role in the function of RAP-1, one that perhaps involves erythrocyte invasion (41). The findings that the predominant CD4+-T-cell response to RAP-1 is directed at epitopes in the NT region and that B-cell epitopes are located in both the NT and CT domains (38) suggest that it may be possible to eliminate the repetitive CT domain in a vaccine construct. Repetitive sequences are often serologically immunodominant and may skew the antibody response away from other domains (31). It has also been postulated that antigens containing repetitive sequences may induce T-cell-independent B-cell activation, resulting in the production of nonneutralizing antibody and immune evasion (31).
NT2-2-specific T-cell clones recognized epitope EAPWYKRWIKKFR (aa 295 to 307) as well as P8 (FARRYADMVKKVLLGSLTSYVEAPWYKRWI), which contained the 9-aa sequence EAPWYKRWI at the C terminus. This result suggests that EAPWYKRWI (aa 295 to 303) consists of a central core region for binding to the MHC class II groove. Amino acid E295 is likely anchored in the P1 pocket of the MHC class II groove, since deletion of E295 from EAPWYKRWIKKFR completely abolished the T-cell response. We also confirmed that the 9-mer peptide EAPWYKRWI did not stimulate the T-cell clones. Together, these results indicate that amino acid sequence extension from the core sequence towards either the N terminus or the C terminus is required for stimulation of RAP-1 NT2-2-specific T-cell clones. Flanking residues at the NT or CT ends of an antigenic core peptide can modulate its immunogenicity by changing the stability of the MHC class II molecule or T-cell receptor recognition (16, 28, 29, 43).
Although RAP-1 NT2-1- and RAP-1 CT-specific T-cell clones were apparently restricted by the MHC class II DRB3 molecule, both DRB3- and DQ-specific MAbs blocked responses of RAP-1 NT2-2-specific T-cell clones, suggesting that both molecules can present this peptide. In other studies using these MAbs and T-lymphocyte clones specific for A. marginale MSP1a and MSP2, either DQ- or DRB3-restricted responses showing that both antibodies are capable of blocking antigen presentation have been identified 11; W. C. Brown, unpublished observations). Further studies to define bovine MHC class II DRB3 and DQ alleles will be required to elucidate the promiscuous MHC class II restriction pattern observed with RAP-1 NT2-2-specific T cells.
In summary, we have demonstrated the presence of multiple, naturally processed T-helper-lymphocyte epitopes in the NT region of RAP-1, the region that is structurally conserved among different Babesia species. The presence of multiple T-helper-lymphocyte epitopes on RAP-1 NT antigen recognized by cattle with different MHC class II haplotypes may explain why B. bovis RAP-1 induces easily detectable memory CD4+-T-cell responses in immune cattle and supports the inclusion of RAP-1 NT epitopes in a B. bovis vaccine. Although the CT region was not immunodominant for T-cell responses from the cattle used in this study, its presence in the full-length antigen had no obvious negative or positive effects on RAP-1 NT-specific T-cell responses in vitro. Similarly, the presence of antigenically variant T-cell epitopes in the CT region did not adversely affect CT-specific T-cell responses. Future studies will compare the results of immunization of cattle with full-length RAP-1 or truncated RAP-1 NT protein missing the repetitive domain to determine whether the NT region mimics the ability of full-length RAP-1 to induce protection against clinical babesiosis upon challenge. RAP-1 T-cell epitopes may ultimately be useful in a multiple-epitope construct designed to elicit memory responses against several B. bovis antigens.
Acknowledgments
We thank Kimberly Kegerreis, Daming Zhu, and Deb Alperin for technical assistance; Harris Lewin and Colleen Olmstead for performing BoLA DRB3 typing; and Guy Palmer for helpful comments.
This research was supported by National Institutes of Health grant R01-AI30136.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Ababou, A., W. C. Davis, and D. Levy. 1993. The DA6-147 monoclonal antibody raised against the HLA-DR alpha chain identifies a cryptic epitope on the BoLA-DR alpha chain. Vet. Res. 24:402-407. [PubMed] [Google Scholar]
- 2.Ababou, A., J. Goyeneche, W. C. Davis, and D. Levy. 1994. Evidence for the expression of three different BoLA-class II molecules on the bovine BL-3 cell line: determination of a non-DR non-DQ gene product. J. Leukoc. Biol. 56:182-186. [DOI] [PubMed] [Google Scholar]
- 3.Beyer, J. C., R. W. Stich, W. C. Brown, and W. P. Cheevers. 1998. Cloning and expression of caprine interferon-gamma. Gene 210:103-108. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff, E., M. Guillotte, O. Mercereau-Puijalon, and S. Bonnefoy. 2000. A member of the Plasmodium falciparum P60 multigene family codes for a nuclear protein expressed by readthrough of an internal stop codon. Mol. Microbiol. 35:1005-1016. [DOI] [PubMed] [Google Scholar]
- 5.Brown, W. C., and D. J. Grab. 1985. Biological and biochemical characterization of bovine interleukin-2. Studies with cloned bovine T cells. J. Immunol. 133:3184-3190. [PubMed] [Google Scholar]
- 6.Brown, W. C., K. S. Logan, G. G. Wagner, and C. L. Tetzlaff. 1991. Cell-mediated immune responses to Babesia bovis antigens in cattle following infection with tick-derived or cultured parasites. Infect. Immun. 59:2418-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, W. C., K. S. Logan, S. Zhao, D. K. Bergman, and A. C. Rice-Ficht. 1995. Identification of Babesia bovis merozoite antigens separated by continuous-flow electrophoresis that stimulate proliferation of helper T-cell clones derived from B. bovis-immune cattle. Infect. Immun. 63:3106-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, W. C., T. F. McElwain, I. Hötzel, C. E. Suarez, and G. H. Palmer. 1998. Helper T-cell epitopes encoded by the Babesia bigemina rap-1 gene family in the constant and variant domains are conserved among parasite strains. Infect. Immun. 66:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, W. C., T. F. McElwain, G. H. Palmer, S. E. Chantler, and D. M. Estes. 1999. Bovine CD4+ T-lymphocyte clones specific for rhoptry-associated protein 1 of Babesia bigemina stimulate enhanced immunoglobulin G1 (IgG1) and IgG2 synthesis. Infect. Immun. 67:155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, W. C., T. F. McElwain, B. J. Ruef, C. E. Suarez, V. Shkap, C. G. Chitko-McKown, W. Tuo, A. C. Rice-Ficht, and G. H. Palmer. 1996. Babesia bovis rhoptry-associated protein 1 is immunodominant for T helper cells of immune cattle and contains T-cell epitopes conserved among geographically distant B. bovis strains. Infect. Immun. 64:3341-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, W. C., T. C. McGuire, D. Zhu, H. A. Lewin, J. Sosnow, and G. H. Palmer. 2001. Highly conserved regions of the immunodominant major surface protein 2 (MSP2) of the ehrlichial pathogen Anaplasma marginale are rich in naturally derived CD4+ T lymphocyte epitopes that elicit strong recall responses. J. Immunol. 166:1114-1124. [DOI] [PubMed] [Google Scholar]
- 12.Brown, W. C., and G. H. Palmer. 1999. Designing blood stage-vaccines against Babesia bovis and B. bigemina. Parasitol. Today 15:275-281. [DOI] [PubMed] [Google Scholar]
- 13.Brown, W. C., G. H. Palmer, T. F. McElwain, S. A. Hines, and D. A. E. Dobbelaere. 1993. Babesia bovis: characterization of the T helper cell response against the 42-kDa merozoite surface antigen (MSA-1) in cattle. Exp. Parasitol. 77:97-110. [DOI] [PubMed] [Google Scholar]
- 14.Brown, W. C., B. J. Ruef, J. Norimine, K. A. Kegerreis, C. E. Suarez, P. G. Conley, R. W. Stich, K. H. Carson, and A. C. Rice-Ficht. 2001. A novel 20-kilodalton protein conserved in Babesia bovis and B. bigemina stimulates memory CD4+ T lymphocyte responses in B. bovis-immune cattle. Mol. Biochem. Parasitol. 118:97-109. [DOI] [PubMed] [Google Scholar]
- 15.Brown, W. C., V. M. Woods, D. A. E. Dobbelaere, and K. S. Logan. 1993. Heterogeneity in cytokine profiles of Babesia bovis-specific bovine CD4+ T cell clones activated in vitro. Infect. Immun. 61:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carson, R. T., K. M. Vignali, D. L. Woodland, and D. A. A. Vignali. 1997. T cell receptor recognition of MHC class II-bound peptide flanking residues enhances immunogenicity and results in altered TCR V region usage. Immunity 7:387-399. [DOI] [PubMed] [Google Scholar]
- 17.Dalrymple, B. P., R. E. Casu, J. M. Peters, C. M. Dimmock, K. R. Gale, R. Boese, and I. G. Wright. 1993. Characterisation of a family of multi-copy genes encoding rhoptry protein homologues in Babesia bovis, Babesia ovis and Babesia canis. Mol. Biochem. Parasitol. 57:181-192. [DOI] [PubMed] [Google Scholar]
- 18.Dalrymple, B. P., J. M. Peters, R. Bose, and I. G. Wright. 1996. A polymerase chain reaction method for the identification of genes encoding members of the Bv60/p58 family of rhoptry protein homologues in the genus Babesia. Exp. Parasitol. 84:96-100. [DOI] [PubMed] [Google Scholar]
- 19.Dutia, B. M., L. MacCarthy-Morrough, E. J. Glass, R. L. Spooner, and J. Hopkins. 1995. Discrimination between major histocompatibility complex class II DQ and DR locus products in cattle. Anim. Genet. 26:111-114. [DOI] [PubMed] [Google Scholar]
- 20.Evavold, B. D., J. Sloan-Lancaster, and P. M. Allen. 1993. Tickling the TCR: selective T-cell functions stimulated by altered peptide ligands. Immunol. Today 14:602-609. [DOI] [PubMed] [Google Scholar]
- 21.Fell, A. H., and N. C. Smith. 1998. Immunity to asexual blood stages of Plasmodium: is resistance to acute malaria adaptive or innate? Parasitol. Today 14:364-369. [DOI] [PubMed] [Google Scholar]
- 22.Goff, W. L., W. C. Johnson, S. M. Parish, G. M. Barrington, W. Tuo, and R. A. Valdez. 2001. The age-related immunity in cattle to Babesia bovis infection involves the rapid induction of interleukin-12, interferon-γ, and inducible nitric oxide synthase mRNA expression in the spleen. Parasite Immunol. 23:463-471. [DOI] [PubMed] [Google Scholar]
- 23.Kuttler, K. L. 1988. World-wide impact of babesiosis, p. 1-15. In M. Ristic (ed.), Babesiosis of domestic animals and man. CRC Press, Inc., Boca Raton, Fla.
- 24.Mahoney, D. F. 1986. Studies on the protection of cattle against Babesia bovis infection, p. 539-541. In W. I. Morrison (ed.), The ruminant immune system in health and disease. Cambridge University Press, Cambridge, United Kingdom.
- 25.Mahoney, D. F., J. D. Kerr, B. V. Goodger, and I. G. Wright. 1979. The immune response of cattle to Babesia bovis (syn. B. argentina). Studies on the nature and specificity of protection. Int. J. Parasitol. 9:297-306. [DOI] [PubMed] [Google Scholar]
- 26.Mattern, T., A. Thanhäuser, N. Reiling, K.-M. Toellner, M. Duchrow, S. Kusumoto, E. T. Rietschel, M. Ernst, H. Brade, H.-D. Flad, and A. J. Ulmer. 1994. Endotoxin and lipid A stimulate proliferation of human T cells in the presence of autologous monocytes. J. Immunol. 153:2996-3004. [PubMed] [Google Scholar]
- 27.McElwain, T. F., L. E. Perryman, A. J. Musoke, and T. C. McGuire. 1991. Molecular characterization and immunogenicity of neutralization-sensitive Babesia bigemina merozoite surface proteins. Mol. Biochem. Parasitol. 47:213-222. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, C. A., S. J. Petzold, and, E. R. Unanue. 1993. Identification of two distinct properties of class II major histocompatibility complex-associated peptides. Proc. Natl. Acad. Sci. USA 90:1227-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, C. A., S. J. Petzold, and E. R. Unanue. 1994. Peptides determine the lifespan of MHC class II molecules in the antigen-presenting cell. Nature 371:250-252. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez, S. D., G. H. Palmer, T. F. McElwain, T. C. McGuire, B. J. Ruef, C. G. Chitko-McKown, and W. C. Brown. 1996. CD4+ T-helper lymphocyte responses against Babesia bigemina rhoptry-associated protein 1. Infect. Immun. 64:2079-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schofield, L. 1991. On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. Parasitol. Today 7:99-105. [DOI] [PubMed] [Google Scholar]
- 32.Shoda, L. K. M., G. H. Palmer, J. Florin-Christensen, M. Florin-Christensen, D. L. Godson, and W. C. Brown. 2000. Babesia bovis-stimulated macrophages express interleukin-1β, interleukin-12, tumor necrosis factor alpha, and nitric oxide and inhibit parasite replication in vitro. Infect. Immun. 68:5139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skuce, P. J., T. R. Mallon, and S. B. Taylor. 1996. Molecular cloning of a putative rhoptry protein homologue from Babesia divergens. Mol. Biochem. Parasitol. 77:99-102. [DOI] [PubMed] [Google Scholar]
- 34.Stich, R. W., A. C. Rice-Ficht, and W. C. Brown. 1999. Babesia bovis: common protein fractions recognized by oligoclonal B. bovis-specific CD4+ T cell lines from genetically diverse cattle. Exp. Parasitol. 91:40-51. [DOI] [PubMed] [Google Scholar]
- 35.Stich, R. W., L. K. M. Shoda, M. Dreewes, B. Adler, T. W. Jungi, and W. C. Brown. 1998. Stimulation of nitric oxide production by Babesia bovis. Infect. Immun. 66:4130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suarez, C. E., M. Florin-Christensen, S. A. Hines, G. H. Palmer, W. C. Brown, and T. F. McElwain. 2000. Characterization of allelic variation in the Babesia bovis merozoite surface antigen 1 (MSA-1) locus and identification of a cross-reactive inhibition-sensitive MSA-1 epitope. Infect. Immun. 68:6865-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suarez, C. E., T. F. McElwain, E. B. Stephens, V. S. Misha, and G. H. Palmer. 1991. Sequence conservation among merozoite apical complex proteins of Babesia bovis, Babesia bigemina and other apicomplexa. Mol. Biochem. Parasitol. 49:329-332. [DOI] [PubMed] [Google Scholar]
- 38.Suarez, C. E., G. H. Palmer, S. A. Hines, and T. F. McElwain. 1993. Immunogenic B-cell epitopes of Babesia bovis rhoptry-associated protein 1 are distinct from sequences conserved between species. Infect. Immun. 61:3511-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suarez, C. E., G. H. Palmer, I. Hötzel, and T. F. McElwain. 1998. Structure, sequence, and transcriptional analysis of the Babesia bovis rap-1 multigene locus. Mol. Biochem. Parasitol. 93:215-224. [DOI] [PubMed] [Google Scholar]
- 40.Suarez, C. E., G. H. Palmer, D. P. Jasmer, S. A. Hines, L. E. Perryman, and T. F. McElwain. 1991. Characterization of the gene encoding a 60-kilodalton Babesia bovis merozoite protein with conserved and surface exposed epitopes. Mol. Biochem. Parasitol. 46:45-52. [DOI] [PubMed] [Google Scholar]
- 41.Suarez, C. E., S. M. Thompson, T. F. McElwain, S. A. Hines, and G. H. Palmer. 1994. Conservation of oligopeptide motifs in rhoptry protein from different genera of erythroparasitic protozoa. Exp. Parasitol. 78:246-251. [DOI] [PubMed] [Google Scholar]
- 42.van Eijk, M. J. T., J. A. Stewart-Haynes, and H. A. Lewin. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23:483-496. [DOI] [PubMed] [Google Scholar]
- 43.Vignali, D. A. A., R. G. Urban, R. M. Chicz, and J. L. Strominger. 1993. Minute quantities of a single immunodominant foreign epitope are presented as large nested sets by major histocompatibility complex class II molecules. Eur. J. Immunol. 23:1602-1607. [DOI] [PubMed] [Google Scholar]
- 44.Wright, I. G., R. Casu, M. A. Commins, B. P. Dalrymple, K. R. Gale, B. V. Goodger, P. W. Riddles, D. J. Waltisbuhl, I. Abetz, D. A. Berrie, Y. Bowles, C. Dimmock, T. Hayes, H. Kalnins, G. Leatch, R. McCrae, P. E. Montague, I. T. Nisbet, F. Parrodi, J. M. Peters, P. C. Scheiwe, W. Smith, K. Rode-Bramanis, and M. A. White. 1992. The development of a recombinant Babesia vaccine. Vet. Parasitol. 44:3-13. [DOI] [PubMed] [Google Scholar]
- 45.Wright, I. G., B. V. Goodger, and I. A. Clark. 1988. Immunopathophysiology of Babesia bovis and Plasmodium falciparum infections. Parasitol. Today 4:214-218. [DOI] [PubMed] [Google Scholar]
- 46.Xu, A., M. J. T. van Eijk, C. Park, and H. A. Lewin. 1993. Polymorphism in BoLA-DRB3 exon 2 correlates with resistance to persistent lymphocytosis caused by bovine leukemia virus. J. Immunol. 151:6977-6985. [PubMed] [Google Scholar]