Abstract
By using the mutated gyrB gene from a spontaneous coumermycin A1-resistant Treponema denticola, an Escherichia coli-T. denticola shuttle vector that renders T. denticola resistant to coumermycin was constructed. The complete T. denticola flgE gene was cloned into the shuttle vector pKMCou, and the vector was transformed into the T. denticola ATCC 33520 flgE erythromycin-resistant knockout mutant HL210. The coumermycin-resistant transformants were motile and restored FlgE activity. This complementation system should prove useful in studying the virulence factors of T. denticola and uncultivable pathogenic spirochetes.
Treponema denticola has been shown to be associated with the severity of periodontal disease (18). Many potential virulence factors of T. denticola have been identified, and mutants of the corresponding virulence factors have been constructed and studied (5). Some of these putative virulence factors include the methyl-accepting chemotaxis DmcA (13) and DmcB (14) proteins; CheA (17), the central kinase in chemotactic signal transduction; Tap1, which might be involved in monitoring the flagellar hook length (16); the outer membrane-associated chymotrypsinlike protease Prtp (11); and Msp, which is an adhesin with pore-forming activity (6). However, studies using both the mutants and the complemented mutants would provide more accurate information on the role of the genes in the virulence of T. denticola. Previously, only an erythromycin resistance (Emr) marker could be used as a selective marker in T. denticola (14). Therefore, a second antibiotic marker is required for complementation of the Ermr mutants.
DNA gyrase is a type II topoisomerase which catalyzes the introduction of negative supercoils into DNA at the expense of ATP hydrolysis to ADP and phosphate (7, 20, 24). This enzyme plays an important role in DNA replication, transcription, and recombination. The enzyme is a tetramer composed of two A and two B subunits which are encoded by the gyrA and gyrB genes, respectively. The coumarin antibiotic coumermycin A1 inhibits DNA gyrase by competing with ATP for binding to the B subunits and blocks introduction of supercoils into relaxed DNA (1, 3, 19, 23).
The gyrB gene of the T. denticola ATCC 35405 strain has recently been cloned and sequenced (S. R. Greene and L. V. Stamm, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., p. 257, 1999). Five conserved type II topoisomerase motifs, an ATP binding site (Walker A), and amino acid residues that putatively interact with a nonhydrolyzable ATP analog are highly conserved in T. denticola GyrB. Spontaneous coumermycin A1-resistant T. denticola 35405 mutants were also isolated, and a single point mutation in the gyrB gene, changing Lys-136 to Glu or Thr, confers >20-fold higher resistance in the mutants relative to that in wild-type T. denticola (8). Therefore, we examined the possibility that the mutated gyrB gene which confers coumermycin resistance in T. denticola could serve as a second selectable marker in these spirochetes.
The periplasmic flagella, which contribute to the motility of spirochetes, are located between the outer and cytoplasmic membranes (9). In spite of its unique location, the general structure of the flagella is similar to that of other bacteria and contains filaments, a flagellar hook, a rod, and a basal body (21, 22). Previously in our laboratory, a mutant of the flagellar hook gene, flgE, of T. denticola 35405 was constructed (14). The mutant HL51 lost motility, and electron microscopy showed that the mutant lacks both the flagellar hook and filaments. In this study, we report the construction of a coumermycin A1-resistant E. coli-T. denticola shuttle vector based on our previously reported vector pKMR4PE (2) and the functional complementation of the T. denticola ATCC 33520 flgE mutant (14).
Construction of the coumermycin A1-resistant E. coli-T. denticola shuttle vector.
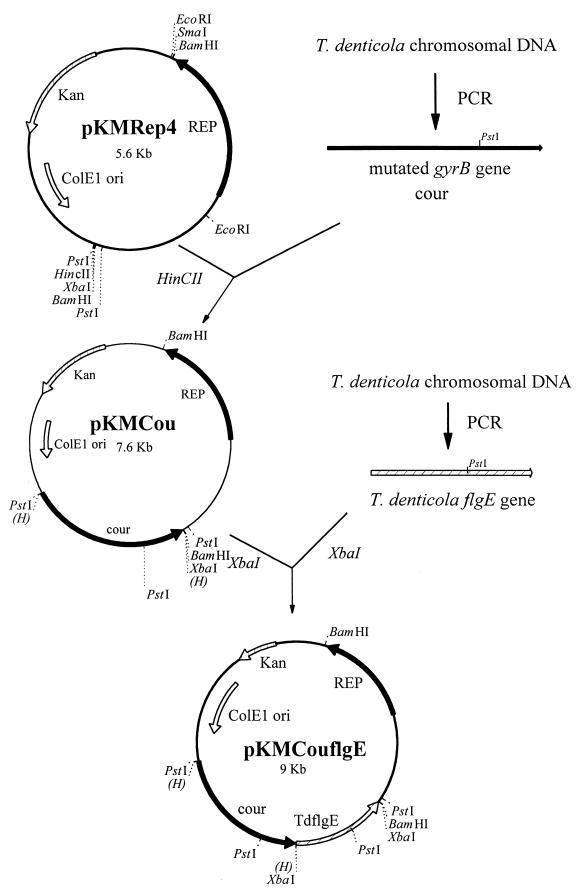
Spontaneous coumermycin A1-resistant T. denticola 33520 was induced according to Greene and Stamm (8), with some modifications. Stationary phase strain 33520 was inoculated in a 1-to-10 ratio in tryptone-yeast extract-gelatin-volatile fatty acids-serum (TYGVS) medium (20) containing 2 μg of coumermycin A1/ml (Sigma, St. Louis, Mo.). The culture was further passaged consecutively through a 10 μg/ml and then a 20 μg/ml concentration of coumermycin A1 in TYGVS medium. Single colonies of coumermycin-resistant T. denticola were isolated from TYGVS 0.8% Seaplaque agarose (Biowhittaker Molecular Applications, Rockland, Maine) plates containing 20 μg of coumermycin A1/ml. Chromosomal DNA was then isolated by using a Puregene DNA isolation kit (Gentra, Minneapolis, Minn.). The gyrB genes of both the wild-type and coumermycin-resistant strains were PCR amplified from the chromosomal DNA by using primers 5′-CCATTCACTCATAGCTCGCCA-3′ and 5′-AATTACGCATCACACATCCAG-3′. The approximately 2-kb fragments contained the 120-bp upstream sequence including the potential promoter region and ribosome binding site (8) and end immediately downstream of the stop codon. The fragments were then ligated into the HincII site (Fig. 1) of the plasmid pKMRep4 (2), which contains the open reading frame encoding the Rep protein of T. denticola cryptic plasmid pTS1, to generate pKMgyrB (not shown) and pKMCou (Fig. 1) plasmids, respectively. The wild-type and mutated gyrB genes were sequenced from the respective plasmids by using the above two primers and a third primer (5′-ATAAGATGGGCAGTGGATCC-3′) which is located 560 bp downstream of the start codon. The sequences revealed a single point mutation in the mutated gyrB gene which changed Lys-136 to Gln instead of to Glu or Thr as previously reported in coumermycin-resistant T. denticola 35405 (8). The gyrB genes of T. denticola 33520 and 35405 are about 99% identical, with a 17-nucleotide difference in the 1,917-bp gyrB open reading frame.
FIG. 1.
Construction of shuttle vectors pKMCou and pKMCouflgE. Relevant restriction sites are indicated.
The MICs of coumermycin A1 for the E. coli XL1-Blue (Stratagene, La Jolla, Calif.) pKMCou transformant and the original XL1-Blue host were determined. The respective bacteria from overnight cultures (106 CFU) were inoculated into 2 ml of Lennox L broth (Invitrogen, Carlsbad, Calif.) containing different concentrations of coumermycin A1, and the cultures were grown overnight. The MIC of coumermycin A1 is 6 μg/ml for the E. coli XL1-Blue pKMCou transformant versus 4 μg/ml for the original XL1-Blue cells. The difference in MIC between the wild-type XL1-Blue and the pKMCou transformant is too small for selection of the transformant in E. coli. Therefore, the plasmid is maintained in Lennox L broth containing 100 μg of fresh kanamycin/ml. The plasmid was isolated with the Wizard Plus Midiprep Kit (Promega, Madison, Wis.).
The plasmid pKMCou was then transformed into T. denticola 33520 to examine whether possession of the plasmid can render T. denticola coumermycin resistant. Strain 33520 (100 ml) was grown to mid- to late-logarithmic growth phase in TYGVS medium and heat treated at 50°C for 30 min before being chilled on ice for 15 min. This heat treatment greatly increases the efficiency of transformation (W. Shi, personal communication). The bacteria were then washed three times with 10% glycerol and resuspended in 10% glycerol to a 700-μl final volume. Eighty microliters of competent cells was next mixed with 0.5 to 1 μg of plasmid DNA and incubated on ice for 30 min. Electroporation was then carried out as described previously (14). Aliquots of competent cells can be frozen in a dry ice-alcohol bath and stored at −80°C for up to a year without a noticeable drop of efficiency. The transformants were selected on TYGVS 0.8% Seaplaque agarose plates supplemented with 10 μg of coumermycin A1/ml. Higher selection concentrations of coumermycin A1 greatly reduce the number of transformant colonies. The transformation efficiency was about 100 to 200 colonies per microgram of plasmid DNA. The individual colonies were isolated and inoculated into TYGVS medium containing 10 μg of coumermycin A1/ml and were analyzed for the possession of the shuttle plasmid by using the Wizard Miniprep Kit (Promega). Agarose gel electrophoresis of plasmid DNA extracted from the transformants showed that the transformants have an additional plasmid other than the cryptic plasmid pTD1 carried by strain 33520 (data not shown). The recombinant plasmid was then reintroduced into E. coli XL1-Blue cells. The restriction enzyme digestion patterns of the rescued plasmid were indistinguishable from those of the original plasmid (data not shown). These results suggest that the mutated gyrB gene can be used as a second antibiotic marker in T. denticola.
Complementation of T. denticola 33520 flgE mutant HL201.
Using both erythromycin and coumermycin A1 resistance markers, we next attempted to carry out complementation experiments with this novel shuttle vector. A T. denticola flgE mutant was chosen for complementation because the complemented bacteria are motile and the phenotype can be easily distinguished from that of the mutant. T. denticola 33520 flgE mutant HL201 was constructed with the same strategy as that used with the 35405 flgE mutant HL51 (14) and was confirmed to have an erythromycin cassette inserted in the flgE gene (data not shown). To complement HL201, the complete flgE gene, which begins 15 bp upstream of its ribosome binding site, was PCR amplified from T. denticola 33520 chromosomal DNA with flgE forward and reverse primers (5′-AAAAA TCTAGAGCAGATAACAATTAGGAGGCA-3′ and 5′-AAAAA TCTAGAC CATACTATCGTTTCAAGTTC-3′; XbaI sites in italics) and cloned into the XbaI site of the pKMCou plasmid downstream of the mutated gyrB gene in E. coli XL1-Blue. Since the flgE gene is in the flagellar operon downstream of the tap1 and flgD genes and has no promoter immediately upstream (22), the orientation of the cloned flgE gene was selected such that the flgE gene can be cotranscribed with the mutated gyrB gene from the gyrB promoter (Fig. 1).
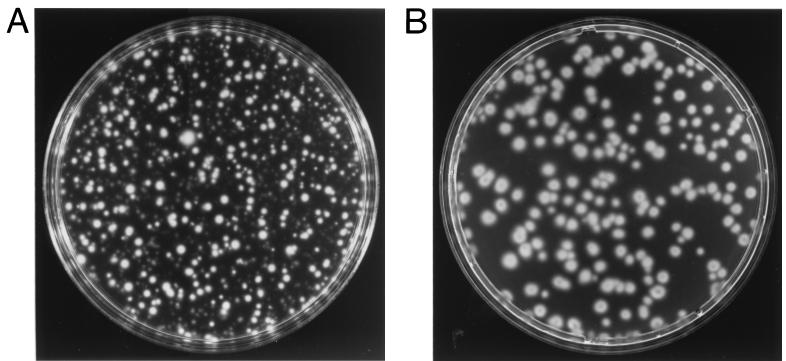
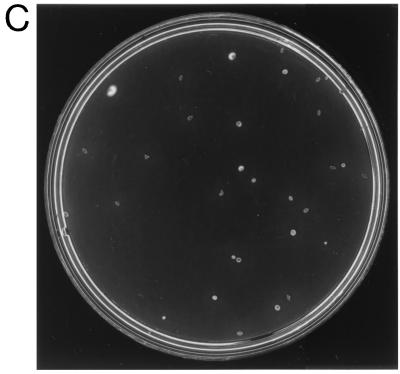
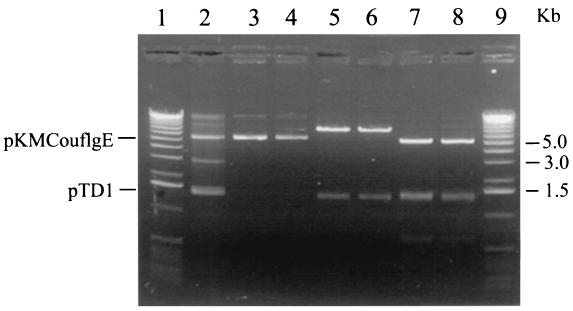
The resulting plasmid, pKMCouflgE, was transformed as described above into T. denticola HL201, and transformants were selected on 0.8% Seaplaque agarose plates containing 10 μg of coumermycin A1/ml and 40 μg of erythromycin/ml. The coumermycin- and erythromycin-resistant transformant colonies (Fig. 2B) grew larger over time and had diffuse edges like those of the wild-type strain (Fig. 2A). In contrast, the HL201 colonies were small with defined edges (Fig. 2C). The complemented transformants were also motile when viewed under a dark-field microscope (model E400; Nikon, Tokyo, Japan), whereas the HL201 colonies were nonmotile. The transformants had an irregular morphology like that of the wild-type strain, while the HL201 colonies had a helical morphology (21). The transformants were further analyzed for plasmids. As shown in Fig. 3, lane 2, the transformants had a plasmid the same size as the original pKMCouflgE (Fig. 3, lane 4) in addition to the cryptic plasmid pTD1. The plasmid from the transformants was then retransformed into E. coli XL1-Blue cells, and restriction endonuclease analysis confirmed that the plasmid was identical to the original pKMCouflgE (Fig. 3, lanes 5 to 8). To confirm that the transformants were complemented instead of the flgE mutation being reverted, ten of the above transformants were examined by PCR with the flgE forward primer (see above) and erythromycin cassette reverse primer (5′-GAAGCTGTCAGTAGTATACC-3′). The predicted size band was amplified, suggesting that the transformants still had a mutated flgE gene on the chromosome of the complemented strain (data not shown).
FIG. 2.
Colonies of wild-type T. denticola 33520 (A), T. denticola flgE mutant HL201 (C), and pKMCouflgE HL201 transformants (B) on 0.8% agarose plates.
FIG. 3.
Electrophoretic analysis of plasmids isolated from T. denticola HL201 pKMCouflgE transformants. Lanes 1 and 9, 1-kb DNA ladder; lane 2, plasmids from T. denticola HL201 pKMCouflgE transformants; lanes 3, 5, and 7, plasmids from E. coli XL1-Blue retransformed with the plasmids shown in lane 2; lanes 4, 6, and 8, original pKMCouflgE; lanes 3 and 4, undigested plasmids; lanes 5 and 6, XbaI digestion of the respective plasmids; lanes 7 and 8, PstI digestion of the respective plasmids.
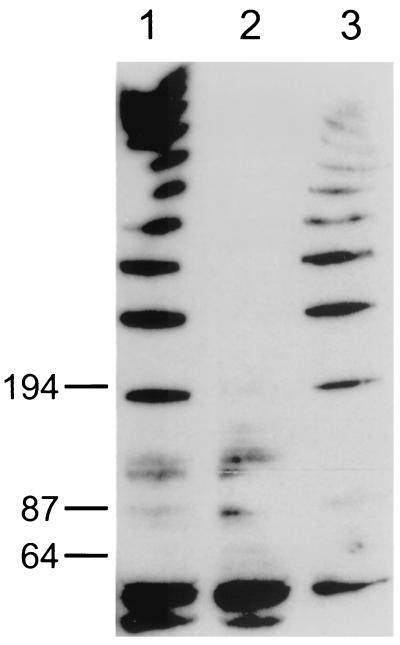
Western blot analysis was used to confirm that the complemented bacteria restored the expression of the FlgE protein. Antibody developed to the T. pallidum FlgE protein was used as the primary antibody (15). As shown in Fig. 4, lane 1, the wild-type T. denticola showed the characteristic polypeptide ladder of FlgE (15). In contrast, the cell extracts of mutant HL201 lacked the ladder pattern (lane 2). Presently, we do not know the nature of the bands around 64 kDa, which were also present in the cell extracts of mutant HL201. There could be some proteins that cross-react with the T. pallidum FlgE antibody. However, the complemented cell extracts showed the same ladder pattern as that of the wild type (lane 3). These results taken together suggest that the flgE gene mutation was functionally complemented from the shuttle vector in T. denticola.
FIG. 4.
Western blot analysis of T. denticola HL201 pKMCouflgE transformants. Cell extracts were separated on a sodium dodecyl sulfate-4 to 20% polyacrylamide gel electrophoresis gel and transferred to a Hybond C Extra membrane. Antibody developed to the T. pallidum FlgE protein was used as the primary antibody (1:3,000 dilution), and horseradish peroxidase-conjugated goat anti-mouse IgG (1:250,000 dilution) was used as the secondary antibody (Pierce, Rockford, Ill.). The blot was developed by using Supersignal West Dura Western blotting kits (Pierce). Lane 1, cell extract of wild-type T. denticola 33520; lane 2, cell extract of T. denticola flgE mutant HL201; lane 3, cell extract of T. denticola HL201 pKMCouflgE transformants. Numbers on the left indicate molecular masses in kilodaltons.
This is the first demonstration of functional complementation of a mutated gene in Treponema. The coumermycin A1-resistant shuttle vector should prove to be useful in identifying other virulence factors of T. denticola. Based on our recently reported data of heterologous gene expression from the shuttle vector in T. denticola (2), this system can now be used to study the virulence factors of uncultivable spirochetes, including T. pallidum, the causative agent for syphilis. A homologous gene in T. denticola can be inactivated to reduce the corresponding phenotype and the gene of interest from an uncultivable spirochete expressed from the coumermycin-resistant shuttle vector. The function of the potential virulence factors could then be studied in a T. denticola background.
E. coli overexpressing its own mutated gyrB gene from a plasmid is more resistant to coumermycin A1 (MIC, 40 μg/ml) (4) than the bacteria overexpressing the T. denticola mutated gyrB gene (MIC, 6 μg/ml). One reason for this may be that the T. denticola GyrB protein lacks sufficient homology for expressing increased resistance in E. coli. The GyrB proteins of T. denticola and E. coli are 66% homologous and 54% identical. The E. coli GyrB protein is also 166 amino acids longer than T. denticola GyrB (10). The function of the additional amino acids at the C-terminal region of E. coli GyrB is still unknown.
At present, although heterologous DNA can be transformed into T. denticola 35405 and mutants can be constructed, shuttle vector pKMCou does not replicate in this strain. One reason for this defect could be that strain 35405, lacking a cryptic plasmid, is incapable of maintaining plasmids. T. denticola 33520, which can be transformed with shuttle vectors, contains the cryptic plasmid pTD1 (12), which suggests that strain 33520 expresses a system for maintaining plasmids. Alternatively, the cryptic plasmid pTD1 may be important in maintaining the shuttle vector. Experiments are presently in progress to test these hypotheses.
Acknowledgments
This investigation was supported by National Institutes of Health grant DE09821.
The technical assistance of L. Wagner and S. Scanga is gratefully acknowledged.
Editor: E. I. Tuomanen
REFERENCES
- 1.Ali, J. A., A. P. Jackson, A. J. Howells, and A. Maxwell. 1993. The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32:2717-2724. [DOI] [PubMed] [Google Scholar]
- 2.Chi, B., S. Chauhan, and H. Kuramitsu. 1999. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect. Immun. 67:3653-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cozzarelli, N. R. 1980. DNA gyrase and the supercoiling of DNA. Science 207:953-960. [DOI] [PubMed] [Google Scholar]
- 4.del Castillo, I., J. L. Vizan, M. C. Rodriguez-Sainz, and F. Moreno. 1991. An unusual mechanism for resistance to the antibiotic coumermycin A1. Proc. Natl. Acad. Sci. USA 88:8860-8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenno, J. C., and B. C. McBride. 1998. Virulence factors of oral treponemes. Anaerobe 4:1-17. [DOI] [PubMed] [Google Scholar]
- 6.Fenno, J. C., G. W. Wong, P. M. Hannam, and B. C. McBride. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209-215. [DOI] [PubMed] [Google Scholar]
- 7.Gellert, M. 1981. DNA topoisomerases. Annu. Rev. Biochem. 50:879-910. [DOI] [PubMed] [Google Scholar]
- 8.Greene, S. R., and L. V. Stamm. 2000. Molecular characterization of the gyrB region of the oral spirochete, Treponema denticola. Gene 253:259-269. [DOI] [PubMed] [Google Scholar]
- 9.Holt, S. C. 1978. Anatomy and chemistry of spirochetes. Microbiol. Rev. 42:114-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, W. M. 1994. Type II DNA topoisomerase genes. Adv. Pharmacol. 29A:201-225. [DOI] [PubMed]
- 11.Ishihara, K., H. K. Kuramitsu, T. Miura, and K. Okuda. 1998. Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J. Bacteriol. 180:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivic, A., J. MacDougall, R. R. Russell, and C. W. Penn. 1991. Isolation and characterization of a plasmid from Treponema denticola. FEMS Microbiol. Lett. 62:189-193. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka, M., H. Li, S. Arakawa, and H. Kuramitsu. 1997. Characterization of a methyl-accepting chemotaxis protein gene; dmcA, from the oral spirochete Treponema denticola. Infect. Immun. 65:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, H., J. Ruby, N. Charon, and H. Kuramitsu. 1996. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J. Bacteriol. 178:3664-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Limberger, R. J., L. L. Slivienski, M. C. El-Afandi, and L. A. Dantuono. 1996. Organization, transcription, and expression of the 5′ region of the fla operon of Treponema phagedenis and Treponema pallidum. J. Bacteriol. 178:4628-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Limberger, R. J., L. L. Slivienski, J. Izard, and W. A. Samsonoff. 1999. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J. Bacteriol. 181:3743-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lux, R., J. N. Miller, N. H. Park, and W. Shi. 2001. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola. Infect. Immun. 69:6276-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh, P., and M. V. Martin. 1999. Oral microbiology. Wright, Woburn, Mass.
- 19.Maxwell, A. 1993. The interaction between coumarin drugs and DNA gyrase. Mol. Microbiol. 9:681-686. [DOI] [PubMed] [Google Scholar]
- 20.Ohta, K., K. K. Makinen, and W. J. Loesche. 1986. Purification and characterization of an enzyme produced by Treponema denticola capable of hydrolyzing synthetic trypsin substrates. Infect. Immun. 53:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 21.Ruby, J. D., H. Li, H. Kuramitsu, S. J. Norris, S. F. Goldstein, K. F. Buttle, and N. W. Charon. 1997. Relationship of Treponema denticola periplasmic flagella to irregular cell morphology. J. Bacteriol. 179:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamm, L. V., and H. L. Bergen. 1999. Molecular characterization of a flagellar (fla) operon in the oral spirochete Treponema denticola ATCC 35405. FEMS Microbiol. Lett. 179:31-36. [DOI] [PubMed] [Google Scholar]
- 23.Sugino, A., N. P. Higgins, P. O. Brown, C. L. Peebles, and N. R. Cozzarelli. 1978. Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc. Natl. Acad. Sci. USA 75:4838-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, J. C. 1985. DNA topoisomerases. Annu. Rev. Biochem. 54:665-697. [DOI] [PubMed] [Google Scholar]