Abstract
Many pathogens produce one or more superoxide dismutases (SODs), enzymes involved in the detoxification of endogenous and exogenous reactive oxygen species that are encountered during the infection process. One detectable cytoplasmic SOD was identified in the human mucosal pathogen Moraxella catarrhalis, and the gene responsible for the SOD activity, sodA, was isolated from a recent pediatric clinical isolate (strain 7169). Sequence analysis of the cloned M. catarrhalis 7169 DNA fragment revealed an open reading frame of 618 bp encoding a polypeptide of 205 amino acids with 48 to 67% identity to known bacterial manganese-cofactored SODs. An isogenic M. catarrhalis sodA mutant was constructed in strain 7169 by allelic exchange. In contrast to the wild-type 7169, the 7169::sodK20 mutant was severely attenuated for aerobic growth, even in rich medium containing supplemental amino acids, and exhibited extreme sensitivity to the redox-active agent methyl viologen. The ability of recombinant SodA to rescue the aerobic growth defects of E. coli QC774, a sodA sodB-deficient mutant, demonstrated the functional expression of SOD activity by cloned M. catarrhalis sodA. Indirect SOD detection assays were used to visualize both native and recombinant SodA activity in bacterial lysates. This study demonstrates that M. catarrhalis SodA plays a critical role in the detoxification of endogenous, metabolically produced oxygen radicals. In addition, the outer membrane protein (OMP) profile of 7169::sodK20 was consistent with iron starvation in spite of growth under iron-replete conditions. This novel observation indicates that M. catarrhalis strains lacking SodA constitutively express immunogenic OMPs previously described as iron repressible, and this potentially attenuated mutant strain may be an attractive vaccine candidate.
Moraxella catarrhalis is a gram-negative, aerobic diplococcus that is frequently identified as a commensal in nasopharyngeal flora, particularly in pediatric populations (18). However, the bacterium is now recognized as an important human mucosal pathogen of the upper and lower respiratory tracts. M. catarrhalis is the third leading cause of otitis media and sinusitis in American children and is associated with pulmonary exacerbations in adults with chronic lung disease (46). In addition, the bacterium can cause a variety of severe infections in immunocompromised hosts, and sporadic outbreaks of M. catarrhalis respiratory disease in hospital wards have established the organism as a nosocomial pathogen (13, 18, 47, 49, 53). The fact that M. catarrhalis was only recently recognized as an important human pathogen, combined with the lack of a relevant animal model, has delayed the elucidation of pathogenic factors involved in the establishment of M. catarrhalis infections (39, 50). Research progress over the past decade has focused on the identification and characterization of several M. catarrhalis surface antigens, including lipooligosaccharides, pili, and outer membrane proteins (OMPs) (for recent reviews, see references 34, 41, and 42). Currently, however, there is little information about the basic metabolic features of M. catarrhalis, such as the critically protective mechanisms involved in resistance to oxidative stress.
Natural by-products of aerobic metabolism include potentially toxic reactive oxygen intermediates, such as superoxide anion, hydrogen peroxide, and hydroxyl radicals. These highly reactive oxygen species (ROS) are capable of damaging a wide range of biomolecules, including DNA, proteins, and lipids (30, 33). Aerobic organisms have developed several defense mechanisms, involving both enzymatic and nonenzymatic strategies, to detoxify ROS (8, 15, 63). Several enzymes, including superoxide dismutases, catalases, peroxidases, glutathione synthase, and glutathione reductases, are believed to provide the primary protection of cellular components against oxidative stress (22, 57).
Superoxide dismutases (SODs), metalloenzymes that catalyze the dismutation of superoxide anions into molecular oxygen and hydrogen peroxide, represent a major cellular defense mechanism involved in the neutralization of ROS in both prokaryotic and eukaryotic organisms (23, 40). Three primary classes of SODs have been identified and are distinguished by the requirement for copper-zinc (Cu-Zn), iron (Fe), or manganese (Mn) as the metal cofactor necessary for enzymatic activity. Cu-ZnSOD (SodC) is primarily found in the cytosol of eukaryotic cells; however, it has recently been identified in the periplasm or associated with the periphery of several prokaryotes (1, 5, 24, 62). Mn- and Fe-cofactored SODs (SodA and SodB, respectively) have highly homologous amino acid sequences and are structurally and evolutionarily distinct from the Cu-ZnSODs (16, 44). Most bacteria possess one or both of the Mn- or Fe-cofactored SODs in the cytosol, although Mn-SOD is also present in eukaryotic mitochondria. These enzymes usually have a strict metal ion selectivity, and several amino acid residues have been identified as critical for discriminating between Fe- and Mn-binding proteins (6, 48). However, a small group of cambialistic SOD enzymes that can use either Mn or Fe in the active site have been identified; therefore, the definitive identity of the metal cofactor can be established only after analysis of the purified protein (14, 21, 56, 58). The cytosolic Mn- and Fe-SODs, encoded by the sodA and sodB genes, have been thoroughly studied, and the most detailed studies have been performed with Escherichia coli. Although E. coli mutants deficient in expression of either SodA or SodB were not affected in aerobic growth, sodA sodB double mutants exhibited depressed growth rates in rich medium, an auxotrophy for branched-chain and aromatic amino acids, extreme sensitivity to oxidative stress, and enhanced mutation rates (12, 20). Besides being critical for protection against oxidants and oxidative stress, SODs have also been implicated as bacterial virulence factors crucial to the survival of pathogens in the host environment (2, 3, 26, 51, 54).
Although M. catarrhalis requires oxygen for growth and survival, the antioxidant properties involved in the ability of this respiratory tract pathogen to defend against oxidative stress have not yet been described. In this study, we identified M. catarrhalis sodA, evaluated the ability of recombinant SodA to fully rescue the growth defects of an E. coli SOD-deficient mutant, and constructed and analyzed a sodA isogenic mutant. In addition to demonstrating the critically protective role of SodA in defense against intracellular superoxide flux, our study indicated that the loss of SodA activity in M. catarrhalis resulted in the constitutive expression of iron-repressible OMPs, a novel phenotypic observation that has not been reported for other bacterial Sod-deficient mutants.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The pediatric middle ear isolate M. catarrhalis 7169 (36, 37) was used to construct the sodA-deficient mutant 7169::sodK20. 7169 and 7169::sodK20 were routinely cultured on chocolate agar plates supplemented with 1% (vol/vol) IsoVitaleX at 35.5°C in 5% CO2 or in Haemophilus ducreyi broth (HD) (containing brain heart infusion [BHI], 25 μg of hemin per ml, 1% [vol/vol] IsoVitaleX, and 5% [vol/vol] heat-inactivated fetal bovine serum [11]) at 37°C with rotary shaking at 225 rpm. For broth-based growth analyses, chocolate agar plate-grown organisms were inoculated to an optical density at 600 nm (OD600) of 0.08 into GC broth (minimal salts medium lacking supplements), BHI, or HD. Each experimental culture was grown as described above, and bacterial growth was monitored spectrophotometrically (OD600) at 1-h intervals. M. catarrhalis strain KSA was obtained from Joel Bernstein (Buffalo General Hospital, Buffalo, N.Y.). E. coli XL1-Blue was used as the host strain for plasmid DNA manipulations. SOD-deficient E. coli QC774 (sodA sodB) and the parental strain GC4468 were kindly provided by Daniele Touati (Institut Jacques Monod, Paris, France). The E. coli strains were cultured on Luria-Bertani (LB) agar plates and broth or on minimal medium agar plates and broth (M63 supplemented with 0.4% glucose and 1 μg of thiamine per ml [12]). Antibiotics were added to growth media as required (ampicillin [100 μg/ml] and kanamycin [40 μg/ml] to E. coli cultures and kanamycin [25 μg/ml] to 7169::sod20K). All results shown for growth experiments are the averages of three independent assays.
General DNA manipulations.
Restriction endonucleases, T4 ligase, Pfu DNA polymerase, and standard molecular biology reagents were obtained from New England Biolabs, Inc. (Beverly, Mass.) or Stratagene (La Jolla, Calif.). Restriction enzyme digestions and analyses, ligations, and transformations with plasmid DNA were performed by standard methods. Plasmid isolation and gel purification of electrophoretically separated DNA fragments were performed with kits manufactured by Qiagen (Santa Clarita, Calif.). Isolation of chromosomal DNA, preparation of competent cells, and transformation by electroporation were performed as previously described (27, 28, 36, 55). PCR amplifications of chromosomal DNA were performed for 30 cycles with the GeneAMP PCR system 9700 (PE Applied Biosystems, Foster City, Calif.); annealing temperatures varied depending on the primer set used (described below). DNA nucleotide sequences of all constructs were obtained via automated DNA sequencing (RPCI Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, N.Y.) and analyzed with MacVector 6.0 and the Wisconsin sequence analysis package (Genetics Computer Group, Madison, Wis.).
Identification of sodA and construction of an isogenic sodA mutant.
The construction of the λTriplEx phagemid library of M. catarrhalis 7169 genomic DNA was described previously (37). The M. catarrhalis sodA homologue was identified by serendipity in the process of analyzing several recombinant pTriplEx plasmids that were plaque purified after the library was screened with a degenerate DNA probe designed to identify the gene encoding the M. catarrhalis phosphoglucomutase homologue. Sequence analysis of one of the recombinant clones identified the putative M. catarrhalis sodA within a 3.3-kb insert. A 1.48-kb DNA fragment containing the 618-bp sodA coding region was amplified from 7169 genomic DNA by PCR using primers 189 and 190 (5′-ATATATGAGCTCTCTACCAGAGTTGGGCTACAGC-3′ [sense] and 5′-CGCGCGGGTACCGCTCATCAAAATAATGCG-3′ [antisense], with engineered SacI and KpnI sites, respectively) and subcloned into pBluescript-KS, resulting in pBSKsod. Analysis of a second putative open reading frame (ORF), identified 422 nt downstream from the sodA ORF on pBSKsod, revealed no significant homology with nucleic acid or protein sequences currently deposited through GenBank at the National Center for Biotechnology Information.
The SmaI-digested aphA-3 nonpolar mutagenesis cassette from pUC18K (43) was inserted into the EcoRV site within the sodA coding region on pBSKsod such that the ATG codon 3′ of the kanamycin resistance determinant was placed in frame with the remainder of the sodA coding region, resulting in pBSsodKAN. pBSsodKAN was linearized by restriction endonuclease digestion and electroporated into competent M. catarrhalis 7169. 7169 sodA mutants were selected by kanamycin resistance and slow growth on chocolate agar plates. Insertional inactivation of sodA by the aphA-3 nonpolar mutagenesis cassette was verified by sequence analysis of PCR-amplified products obtained from chromosomal DNA preparations using primers designed to flank the predicted site of cassette insertion (primers 223 [5′-AATAACGCCAATGGTTGCTTG-3′] [sense] and 224 [5′-ACGCTTTTGTGCTTCGTCCC-3′] [antisense]) and specific to sequences within sodA. The sodA mutant was complemented to create a sodA wild-type revertant by electroporating a PCR-amplified product obtained from 7169 chromosomal DNA preparations with primers 223 and 224. Revertants were selected by 16 h of growth on minimal medium plates (GC agar base) and screened for loss of kanamycin resistance. Replacement of the entire aphA-3 nonpolar mutagenesis cassette by wild-type sodA was verified by sequence analysis. The sodA mutant and corresponding sodA-complemented revertant in strain KSA were constructed and analyzed by using the PCR-amplified products obtained with primers 223 and 224, as described above.
MV sensitivity assay.
The sensitivity of 7169::sodK20 to the redox-cycling agent methyl viologen (MV) was compared to that of 7169 with plate-based susceptibility assays essentially as described previously (17). Chocolate agar plate-grown 7169 and 7169::sodK20 were suspended in sterile phosphate-buffered saline to an OD600 of 0.2. Aliquots (100 μl) of serial dilutions were plated in triplicate on freshly prepared chocolate agar plates containing 0, 10, 20, or 30 μM MV. After 48 h of incubations, plates were examined for growth. A representative series of equivalent dilutions (10−4) was included in the results.
Analysis of OMPs.
Zwittergent-extracted OMP preparations, sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (PAGE), and Western blotting were performed according to standard procedures (9, 10). The M. catarrhalis TbpB-specific monoclonal antibody 11C6 was described previously (37).
Recombinant SodA (rSodA) complementation assays.
To examine the expression of M. catarrhalis SodA in the E. coli SOD-deficient mutant QC774, plasmid pMCSODA was constructed by inserting a PCR product containing the sodA coding region amplified from genomic DNA in frame with the lacZ of pUC18. Primers 246 and 247 (5′-GCGCGAATTCCGATGATTTTATGATTTGTCC-3′ [sense] and 5′-TATACTGCAGGGCTTATTTTATTGAGCG-3′ [antisense], with engineered EcoRI and BamHI sites, respectively) were designed to flank the predicted start and stop codons of M. catarrhalis sodA. As a negative control for the complementation assays, primers 246 and 247 were also used to clone insertionally inactivated sodA from 7169::sodK20 into pUC18, resulting in psodKAN. GC4468 and QC774 were each transformed with both pMCSODA and psodKAN and evaluated in broth-based growth assays essentially as described elsewhere (12), with the following modifications. Overnight LB-ampicillin cultures of each transformed strain were used to inoculate day cultures to an OD600 of approximately 0.04. Total culture volumes were less than 1/20 the total flask volume, and the cultures were vigorously shaken (250 rpm in a 37°C rotary shaker) to maintain high aeration throughout the assay. For the MV sensitivity assays, MV was added to a final concentration of 50 μM after 1 h of exponential growth in rich medium (LB). Growth was monitored spectrophotometrically at 1-h intervals.
Preparation of crude bacterial cellular extracts.
Crude cell extracts of M. catarrhalis 7169 and 7169::sodK20 grown on chocolate agar plates and of E. coli GC4468 and QC774 transformants grown in LB-ampicillin broth cultures were prepared as described previously (52), with the following modifications. Harvested cells were washed twice with 50 mM Tris-HCl (pH 7.4) and resuspended in 1.0 ml of lysis solution (50 mM Tris-HCl [pH 7.4] containing 0.1 mM EDTA and 0.2 mg of lysozyme per ml). After seven freeze-thaw cycles (each cycle consisted of alternating 1-min incubations in an ethanol-dry ice bath and a 42°C water bath), lysates were cleared by centrifugation at 21,000 × g for 15 min at 4°C, and total protein concentrations of each lysate were determined with the Lowry protein assay (Sigma Chemical Co., St. Louis, Mo.). Fifty micrograms of each soluble protein sample was resolved on a nondenaturing PAGE gel (native PAGE) as previously described (17).
SOD detection assays.
SOD activity was detected by the indirect assay method of Beauchamp and Fridovich, by which the presence of active SODs can be detected by the ability of the enzyme to scavenge superoxide from reaction mixtures and thereby inhibit superoxide-dependent reactions (4). In brief, SOD activity in M. catarrhalis and E. coli protein extracts was visualized by soaking native-PAGE gels in 0.2% nitroblue tetrazolium for 20 min in the dark and then in a solution containing 0.028 M N,N,N′,N′-tetramethylethylenediamine (TEMED; Sigma), 2.8 × 10−5 M riboflavin, and 0.036 M potassium phosphate (pH 7.8) for an additional 15 min in the dark. The gels were illuminated with fluorescent light, and the presence of SOD activity corresponded to achromatic zones in a uniformly deep blue background. Hydrogen peroxide (H2O2) was added to the second incubation step at a final concentration of 5 mM for the FeSOD inhibition assays (35). For N-terminal amino acid sequence analysis of SodA, proteins resolved by native PAGE were electrotransferred to polyvinylidene difluoride and localized by staining with Coomassie blue. The SodA and rSodA protein bands from the crude bacterial lysates that corresponded to the zones of SOD activity identified on the SOD detection gels were excised and subjected to protein microsequencing by Proseq, Inc. (Boxford, Mass.). A single, clean major signal with no ambiguities was identified, as described below.
Nucleotide sequence accession number.
The nucleotide sequence of M. catarrhalis 7169 sodA has been deposited in GenBank under accession no. AF 413524.
RESULTS AND DISCUSSION
Identification and analysis of M. catarrhalis sodA.
The gene encoding the M. catarrhalis SodA was identified in the process of probing a genomic phagemid library of M. catarrhalis 7169 with a degenerate probe to an unrelated gene. One positive clone selected for sequence analysis contained a 3.3-kb DNA insert. Analysis of the nucleotide sequence indicated that the cloned DNA fragment contained an ORF of 618 bp with a predicted gene product of 205 amino acids. Database searches with the deduced polypeptide sequence revealed high homology to known Mn- and Fe-cofactored SODs from other organisms, with the highest homology to prokaryotic Mn-SODs, termed SodA (48 to 69% identity). Amino acid sequence comparisons with both Mn- and Fe-SOD indicated that the M. catarrhalis homologue contained amino acid sequences typically conserved in known Mn-SODs and contained none of the key residues correlated with a Fe-cofactored enzyme. In particular, 18 of the 21 specific residues predicted to be discriminatory for an Mn-dependent enzyme were present in the M. catarrhalis SodA sequence (data not shown) (32, 48, 61). N-terminal amino acid sequence analysis of SodA (described in Materials and Methods) was consistent with the sequence deduced from the cloned DNA except for the absence of the initial formyl-methionine, as described for other bacterial SOD proteins (7, 31, 44, 48, 59).
Construction of an M. catarrhalis sodA mutant.
A sodA isogenic mutant was constructed in M. catarrhalis 7169 by integrating the nonpolar kanamycin resistance determinant from pUC18K into the sodA coding region in order to evaluate the physiologic effects of a SodA deficiency on this organism. In brief, a 1.48-kb fragment was subcloned from the 3.3-kb insert and the mutagenesis cassette was inserted into the sodA coding region such that the ATG codon situated 3′ of the resistance determinant was placed in frame with the stop codon of sodA. The linearized mutagenesis construct was electroporated into competent M. catarrhalis 7169, and subsequent PCR amplification, using primers designed to flank the predicted site of mutagenesis cassette insertion, of chromosomal DNA obtained from the kanamycin-resistant transformants was used to verify the disruption of sodA by the 850-bp aphA-3 determinant. DNA sequence analysis of these PCR products confirmed that the nonpolar cassette had been inserted into sodA at the predicted location, and one of the mutants, 7169::sodK20, was selected for further analysis.
An indirect SOD detection assay, which visualizes SOD activity in whole-cell lysates separated by native PAGE, was used to investigate the presence of SOD activity in M. catarrhalis 7169 and to confirm the absence of SodA activity in 7169::sodK20 (4). A single, achromatic band of SOD activity was visible in crude protein extracts from 7169 (Fig. 1, lane 1), and no activity bands were detected in cell lysates from 7169::sodK20 (lane 2), confirming the insertional inactivation of sodA in the mutant strain and suggesting that M. catarrhalis may contain only one SOD enzyme.
FIG. 1.

Native-PAGE gel stained to detect SOD activity from whole-cell extracts of M. catarrhalis 7169 (lane1) and the sodA isogenic mutant 7169::sodK20 (lane 2).
Growth effects of sodA disruption.
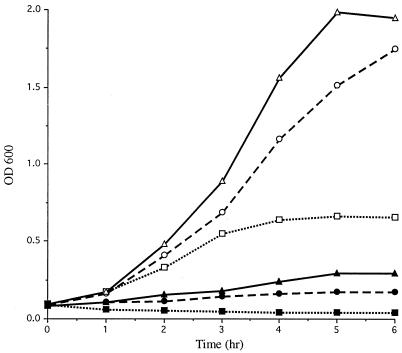
Comparative growth studies were performed to evaluate the effects of the SodA deficiency of 7169::sodK20 compared to the wild-type, 7169. The sodA isogenic mutant demonstrated an absolute requirement for rich, supplemented agar (chocolate agar plates). The mutants exhibited markedly slowed growth, requiring 48 h for visible single colonies on chocolate agar plates, whereas the wild-type 7169 formed single colonies on minimal medium plates within 16 h. To further evaluate the contribution of SodA to the oxidative-stress tolerance of aerobically grown M. catarrhalis, the wild-type and mutant strains were monitored spectrophotometrically for growth in vigorously shaking broth cultures. Figure 2 shows the aerobic growth of 7169 and 7169::sodK20 in three media with various degrees of nutritional richness. Growth of the SodA-deficient mutant 7169::sodK20 was severely impeded under all conditions compared to that of the parental strain, 7169. Although both strains exhibited the most vigorous growth in HD, the most nutritionally replete medium (containing BHI, IsoVitaleX supplements, and fetal bovine serum), the overall restricted ability of 7169::sodK20 to grow aerobically under standard broth culture conditions suggests the importance of SodA for the aerotolerance of M. catarrhalis.
FIG. 2.
Growth of 7169 (open symbols) and 7169::sodK20 (closed symbols) in a minimal salts medium (GC, dotted line), rich medium (BHI, dashed line), and rich, supplemented medium (HD, solid line) was monitored spectrophotometrically at 1-h intervals.
Effect of sodA mutation on MV susceptibility.
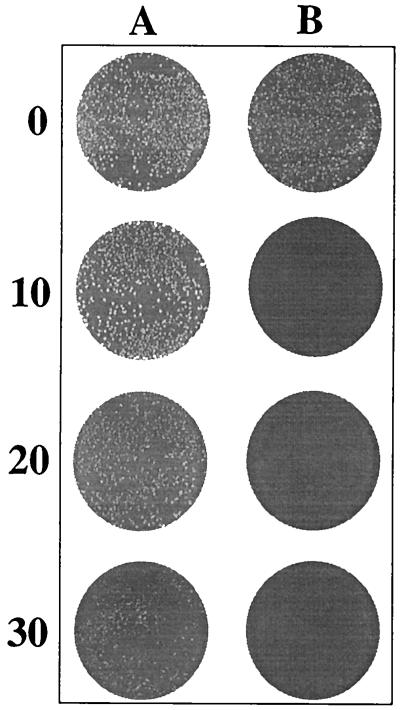
The effect of SodA deficiency on the oxidative stress response was further analyzed by investigating the susceptibility of 7169 and 7169::sodK20 to MV, a redox-cycling agent that generates an enhanced cytosolic superoxide flux. The sensitivities of the wild-type and SodA mutant strains to MV were assessed with a plate-based growth inhibition assay as described in Materials and Methods. Figure 3 shows that 7169 was capable of growth in the presence of MV, although the colonies were progressively smaller as the MV concentration increased. In contrast, 7169::sodK20 was completely inhibited for growth and did not survive exposure to MV under any of the experimental conditions. Together with the aerobic growth analyses, these studies demonstrated the importance of functional SodA activity for the ability of M. catarrhalis to resist cytosolic oxidative stress and indicated that the effects of sodA inactivation could not be rescued by an alternative cellular component.
FIG. 3.
Sensitivity of M. catarrhalis 7169 (A) and 7169::sodK20 (B) to the superoxide-generating agent MV. Final concentrations of MV (micromolar) are shown on the left.
Complementation of SOD-deficient E. coli by recombinant M. catarrhalis SodA.
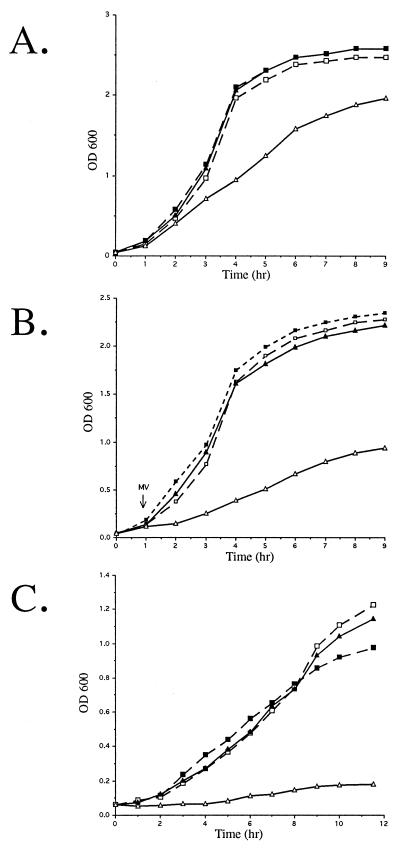
It is currently not possible to complement 7169::sodK20 in trans with a plasmid containing the sodA coding region, as no stable M. catarrhalis shuttle vectors have been identified to date. Therefore, E. coli QC774, a sodA sodB-deficient mutant, was used to demonstrate the functional expression of SOD activity by the cloned M. catarrhalis sodA in complementation studies. Previous studies demonstrated that although a single mutation in either the E. coli sodA or sodB gene does not affect growth of the organism, the double sodA sodB mutant grows slower aerobically than the wild type in rich media, is deficient for growth on glucose minimal media, and exhibits hypersensitivity to MV (12). To determine whether M. catarrhalis rSodA could rescue the QC774 growth defects, pMCSODA was constructed by PCR amplification of the predicted 7169 sodA ORF and cloned into pUC18. pMCSODA and the control psodKAN, containing the insertionally inactivated gene amplified from 7169::sodK20, were electroporated into both QC774 and the wild-type strain GC4468. QC774 transformed with pMCSODA, but not psodKAN, exhibited growth patterns comparable to those of the SOD-proficient GC4468 (Fig. 4). These data demonstrate that the M. catarrhalis sodA gene was functionally expressed in E. coli and was able to fully complement the growth defects of a sodA sodB-deficient strain.
FIG. 4.
Complementation of SOD-deficient E. coli (QC774) by the recombinant M. catarrhalis SodA. Growth in rich medium under aerobic conditions (A), in the presence of 50 μM MV (B), and in minimal medium (C) was measured at 1-h intervals. The sodA sodB mutant QC774 (solid lines) and the parental strain GC4468 (dashed lines) were transformed with pMCSODA (closed symbols) and the control vector psodKAN (open symbols).
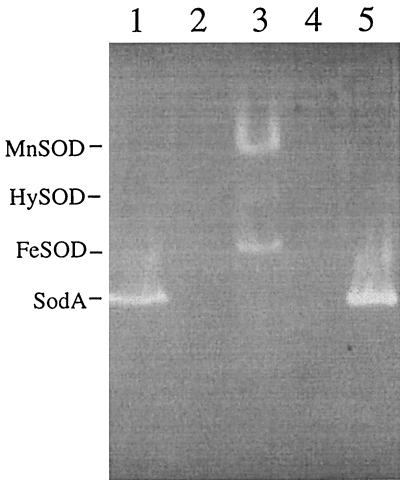
The SOD activity of rSodA was visualized by subjecting whole-cell protein extracts of E. coli QC774 transformed with pMCSODA to the native-PAGE SOD detection assay (Fig. 5). Unlike GC4468, which expresses both MnSOD and FeSOD, QC774 alone or transformed with psodKAN does not express any detectable SOD activity under these experimental conditions. pMCSODA was shown to encode a gene product with the same electrophoretic mobility as the native SodA from M. catarrhalis extracts, verifying that rSodA was expressed in E. coli QC774 and capable of restoring wild-type growth characteristics to the SOD-deficient strain. H2O2 treatment, which selectively inactivates Fe-cofactored SODs, reduced only the FeSOD activity of GC4468 (data not shown). The SOD activities of GC4468 MnSOD and M. catarrhalis 7169 SodA were relatively unaffected by H2O2 exposure, supporting the amino acid-based prediction that the M. catarrhalis enzyme is Mn cofactored.
FIG. 5.
Native PAGE of crude bacterial cell extracts from 7169 (lane 1), 7169::sodK20 (lane 2), GC4468/psodKAN (lane 3), QC774/psodKAN (lane 4), and QC774/pMCSODA (lane 5) stained to visualize SOD activity. Locations of the M. catarrhalis SodA and the E. coli MnSOD, FeSOD, and HySOD (Mn/Fe SOD hybrid) are indicated.
Effect of sodA disruption on OMP profiles.
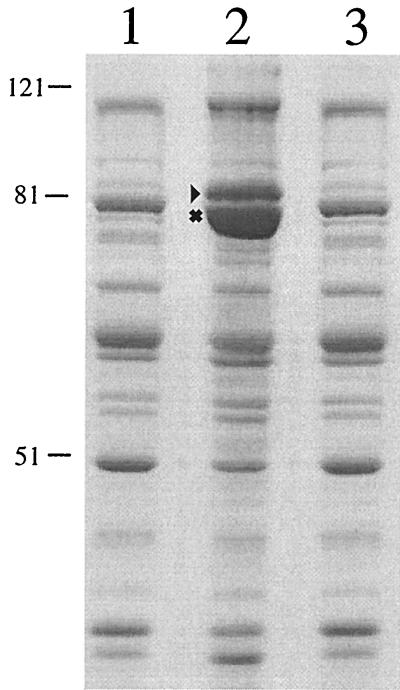
OMP preparations from the wild-type strain 7169 and the isogenic mutant 7169::sodK20 were made to assess whether the loss of SodA activity had an effect on the OMP profile. As shown in Fig. 6, the OMP profile of 7169::sodK20 (lane 2) was strikingly different from that of 7169 (lane 1) grown under identical conditions. Specifically, the OMP profile of 7169::sodK20 was characteristic of OMPs isolated from iron-starved M. catarrhalis cultures (34, 36, 37). Our laboratory demonstrated previously that there are several M. catarrhalis proteins with increased expression levels in response to iron starvation (9, 10, 36, 37). Two of the most prominent iron-regulated OMPs, TbpB (B1) and CopB (B2), have remarkably increased expression on the surface of 7169::sodK20 compared to 7169, even though both strains were cultured on nutritionally replete chocolate agar plates under identical growth conditions. Western blot analysis with the TbpB-specific monoclonal antibody 11C6 further confirmed the enhanced expression of this iron-regulated protein by the SodA-deficient mutant (data not shown).
FIG. 6.
A sodium dodecyl sulfate-PAGE gel, stained with Coomassie blue, comparing the OMP profiles of parental strain 7169 (lane 1), the 7169 sodA isogenic mutant 7169::sodK20 (lane 2), and a 7169 sodA-complemented revertant (lane 3). The iron-regulated OMPs TbpB (▸) and CopB (✖) and molecular sizes, in kilodaltons, are indicated.
Although it has been reported that expression of the M. catarrhalis iron-repressible proteins occurs solely as a result of iron starvation and not generalized environmental stress, the potential contribution of oxidative stress to this phenotype was not specifically evaluated (10). To address this, OMPs were prepared from 7169 exposed to 10, 20, and 30 μM MV. The OMP profiles of 7169 grown on chocolate agar plates under MV-induced oxidative stress did not correspond to an iron-stressed profile and were indistinguishable from the profiles of control cultures grown in the absence of MV (data not shown). Therefore, the phenotypic change observed in the OMPs isolated from 7169::sodK20 cannot be attributed to a general oxidative stress response resulting solely from an increased superoxide flux into the cytosol.
To confirm that the iron-stressed OMP phenotype of the mutant was attributed solely to the disruption of sodA, the wild-type sodA gene was used to complement 7169::sodK20 by replacing the aphA-3 nonpolar kanamycin resistance cassette with the intact sodA gene, thereby creating a wild-type revertant with restored SodA function. The OMP profiles of the complemented sodA revertant strains were indistinguishable from the wild-type 7169 OMP profile (a representative OMP preparation from one of the revertant strains is shown in Fig. 6, lane 3). Furthermore, an additional M. catarrhalis clinical isolate, strain KSA, was used to construct a second sodA mutant and a corresponding sodA wild-type revertant. The resulting OMP profiles of these strains demonstrated that the KSA-sodA mutant exhibited an iron-stressed OMP phenotype despite growth on iron-rich media, and this phenotypic alteration was restored to the wild-type OMP profile by reconstitution with the sodA gene alone (data not shown). These data suggest that the iron-stressed phenotype that is observed in M. catarrhalis sodA mutants is not a strain-specific event but appears to represent a general phenotypic alteration in strains lacking a functional SodA.
To our knowledge, this is the first study evaluating the OMP profiles of bacterial SOD-deficient mutants. Thus, we cannot speculate on the frequency or the significance of this novel observation, which correlates the loss of SodA activity with upregulation of iron-repressible protein expression at the bacterial surface. Multiple oxygen-dependent phenotypic alterations have been documented in the extensively analyzed E. coli SOD-deficient mutants, including defective amino acid biosynthesis, cell membrane structural instability, and high rates of spontaneous DNA mutagenesis, but increased expression of iron-regulated gene products has not been described (12, 19, 29, 38). Additional studies have identified six global transcriptional regulators that affect E. coli SodA expression, including the products of the fur, fnr, and arcA genes, the soxRS and soxQ loci, and integration host factor (12, 20, 29, 38). However, none of these regulatory elements have been identified in M. catarrhalis to date. Although iron-restricted aerobic growth conditions have been shown to induce SodA expression in other organisms (25, 45, 60), the relevance of SOD deficiency causing iron-regulated protein expression in M. catarrhalis and the mechanism by which this may occur remains unclear and warrants further investigation.
In summary, this study demonstrates that M. catarrhalis sodA encodes a functional SOD that is largely responsible for the aerotolerance and protection of this bacterial pathogen from internal oxidative stress, as assessed by a series of in vitro assays. The single band of SOD activity experimentally detected in M. catarrhalis extracts is the product of the sodA gene, and rSodA alone was capable of rescuing the growth defects of an E. coli sodA sodB-deficient mutant. The isogenic sodA mutant 7169::sodK20 exhibited severe in vitro growth deficiencies, demonstrating that SodA may be essential for bacterial survival in aerobic environments. Taken together, these data suggest that SodA may play a critical role in the pathogenesis of M. catarrhalis infections and support the hypothesis that SodA-deficient mutants may be attenuated for virulence in vivo. In addition, we report the intriguing finding that M. catarrhalis SodA-deficient mutants grown on nutritionally rich media exhibit OMP profiles consistent with those of organisms subjected to iron starvation. This novel observation is significant because several iron-regulated M. catarrhalis OMPs, including CopB, the lactoferrin binding proteins LbpA and LbpB, and the transferrin binding protein TbpA and TbpB, are currently under investigation as potential vaccine antigens (for recent reviews, see references 34 and 41). One of the most important attributes of the M. catarrhalis sodA mutant is that it appears to constitutively express many of these highly immunogenic, typically iron-repressed OMPs. Studies addressing the use of a potentially attenuated M. catarrhalis strain that stably exhibits an iron-stressed OMP phenotype as a putative vaccine candidate are under way.
Acknowledgments
We thank Daniele Touati for kindly providing the SOD-deficient E. coli strains and Katie J. Edwards for her assistance in constructing the KSA mutants.
This research was supported by grant AI46469 from the National Institutes of Health (NIAID) to A.A.C. N.R.L. is supported as a postdoctoral fellow by NIAID training grant AI07614.
Editor: D. L. Burns
REFERENCES
- 1.Battistoni, A., F. Pacello, S. Folcarelli, M. Ajello, G. Donnarumma, R. Greco, M. G. Ammendolia, D. Touati, G. Rotilio, and P. Valenti. 2000. Increased expression of periplasmic Cu,Zn superoxide dismutase enhances survival of Escherichia coli invasive strains within nonphagocytic cells. Infect. Immun. 68:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaman, B. L., C. M. Black, F. Doughty, and L. Beaman. 1985. Role of superoxide dismutase and catalase as determinants of pathogenicity of Nocardia asteroides: importance in resistance to microbicidal activities of human polymorphonuclear neutrophils. Infect. Immun. 47:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaman, L., and B. L. Beaman. 1990. Monoclonal antibodies demonstrate that superoxide dismutase contributes to protection of Nocardia asteroides within the intact host. Infect. Immun. 58:3122-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 5.Beck, B. L., L. B. Tabatabai, and J. E. Mayfield. 1990. A protein isolated from Brucella abortus is a Cu-Zn superoxide dismutase. Biochemistry 29:372-376. [DOI] [PubMed] [Google Scholar]
- 6.Beyer, W., J. Imlay, and I. Fridovich. 1991. Superoxide dismutases. Prog. Nucleic Acid Res. Mol. Biol. 40:221-253. [DOI] [PubMed] [Google Scholar]
- 7.Bowler, C., L. Van Kaer, W. Van Camp, M. Van Montagu, D. Inze, and P. Dhaese. 1990. Characterization of the Bacillus stearothermophilus manganese superoxide dismutase gene and its ability to complement copper/zinc superoxide dismutase deficiency in Saccharomyces cerevisiae. J. Bacteriol. 172:1539-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadenas, E. 1989. Biochemistry of oxygen toxicity. Annu. Rev. Biochem. 58:79-110. [DOI] [PubMed] [Google Scholar]
- 9.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campagnari, A. A., K. L. Shanks, and D. W. Dyer. 1994. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect. Immun. 62:4909-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campagnari, A. A., L. M. Wild, G. E. Griffiths, R. J. Karalus, M. A. Wirth, and S. M. Spinola. 1991. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect. Immun. 59:2601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Y., and E. M. Gregory. 1991. In vivo metal substitution in Bacteroides fragilis superoxide dismutase. Free Radic. Res. Commun. 12-13:313-318. [DOI] [PubMed] [Google Scholar]
- 15.Cortez, N., N. Carrillo, C. Pasternak, A. Balzer, and G. Klug. 1998. Molecular cloning and expression analysis of the Rhodobacter capsulatus sodB gene, encoding an iron superoxide dismutase. J. Bacteriol. 180:5413-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeShazer, D., J. D. Bannan, M. J. Moran, and R. L. Friedman. 1994. Characterization of the gene encoding superoxide dismutase of Bordetella pertussis and construction of a SOD-deficient mutant. Gene 142:85-89. [DOI] [PubMed] [Google Scholar]
- 17.D'Mello, R. A., P. R. Langford, and J. S. Kroll. 1997. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type b. Infect. Immun. 65:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright, M. C., and H. McKenzie. 1997. Moraxella (Branhamella) catarrhalis--clinical and molecular aspects of a rediscovered pathogen. J. Med. Microbiol. 46:360-371. [DOI] [PubMed] [Google Scholar]
- 19.Farr, S. B., R. D'Ari, and D. Touati. 1986. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc. Natl. Acad. Sci. USA 83:8268-8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabbianelli, R., A. Battistoni, F. Polizio, M. T. Carri, A. De Martino, B. Meier, A. Desideri, and G. Rotilio. 1995. Metal uptake of recombinant cambialistic superoxide dismutase from Propionibacterium shermanii is affected by growth conditions of host Escherichia coli cells. Biochem. Biophys. Res. Commun. 216:841-847. [DOI] [PubMed] [Google Scholar]
- 22.Graeff-Wohlleben, H., S. Killat, A. Banemann, N. Guiso, and R. Gross. 1997. Cloning and characterization of an Mn-containing superoxide dismutase (SodA) of Bordetella pertussis. J. Bacteriol. 179:2194-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassan, H. M. 1984. Determination of microbial damage caused by oxygen free radicals, and the protective role of superoxide dismutase. Methods Enzymol. 105:404-412. [DOI] [PubMed] [Google Scholar]
- 24.Hassan, H. M. 1989. Microbial superoxide dismutases. Adv. Genet. 26:65-97. [DOI] [PubMed] [Google Scholar]
- 25.Hassan, H. M., and I. Fridovich. 1977. Regulation of the synthesis of superoxide dismutase in Escherichia coli. Induction by methyl viologen. J. Biol. Chem. 252:7667-7672. [PubMed] [Google Scholar]
- 26.Heinzen, R. A., M. E. Frazier, and L. P. Mallavia. 1992. Coxiella burnetii superoxide dismutase gene: cloning, sequencing, and expression in Escherichia coli. Infect. Immun. 60:3814-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helminen, M. E., I. Maciver, J. L. Latimer, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect. Immun. 61:2003-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helminen, M. E., I. Maciver, M. Paris, J. L. Latimer, S. L. Lumbley, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J. Infect. Dis. 168:1194-1201. [DOI] [PubMed] [Google Scholar]
- 29.Imlay, J. A., and I. Fridovich. 1992. Suppression of oxidative envelope damage by pseudoreversion of a superoxide dismutase-deficient mutant of Escherichia coli. J. Bacteriol. 174:953-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 31.Inaoka, T., Y. Matsumura, and T. Tsuchido. 1998. Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis. J. Bacteriol. 180:3697-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson, S. M., and J. B. Cooper. 1998. An analysis of structural similarity in the iron and manganese superoxide dismutases based on known structures and sequences. Biometals 11:159-173. [DOI] [PubMed] [Google Scholar]
- 33.Janssen, Y. M., B. Van Houten, P. J. Borm, and B. T. Mossman. 1993. Cell and tissue responses to oxidative damage. Lab. Investig. 69:261-274. [PubMed] [Google Scholar]
- 34.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 35.Kroll, J. S., P. R. Langford, J. R. Saah, and B. M. Loynds. 1993. Molecular and genetic characterization of superoxide dismutase in Haemophilus influenzae type b. Mol. Microbiol. 10:839-848. [DOI] [PubMed] [Google Scholar]
- 36.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch, M., and H. Kuramitsu. 2000. Expression and role of superoxide dismutases (SOD) in pathogenic bacteria. Microbes Infect. 2:1245-1255. [DOI] [PubMed] [Google Scholar]
- 39.Marchant, C. D. 1990. Spectrum of disease due to Branhamella catarrhalis in children with particular reference to acute otitis media. Am. J. Med. 88:15S-19S. [DOI] [PubMed] [Google Scholar]
- 40.McCord, J. M., and I. Fridovich. 1969. The utility of superoxide dismutase in studying free radical reactions. I. Radicals generated by the interaction of sulfite, dimethyl sulfoxide, and oxygen. J. Biol. Chem. 244:6056-6063. [PubMed] [Google Scholar]
- 41.McMichael, J. C. 2000. Progress toward the development of a vaccine to prevent Moraxella (Branhamella) catarrhalis infections. Microbes Infect. 2:561-568. [DOI] [PubMed] [Google Scholar]
- 42.McMichael, J. C. 2000. Vaccines for Moraxella catarrhalis. Vaccine 19(Suppl. 1):S101-S107. [DOI] [PubMed]
- 43.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merkamm, M., and A. Guyonvarch. 2001. Cloning of the sodA gene from Corynebacterium melassecola and role of superoxide dismutase in cellular viability. J. Bacteriol. 183:1284-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moody, C. S., and H. M. Hassan. 1984. Anaerobic biosynthesis of the manganese-containing superoxide dismutase in Escherichia coli. J. Biol. Chem. 259:12821-12825. [PubMed] [Google Scholar]
- 46.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy, T. F. 1998. Lung infections. 2. Branhamella catarrhalis: epidemiological and clinical aspects of a human respiratory tract pathogen. Thorax 53:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker, M. W., and C. C. Blake. 1988. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 229:377-382. [DOI] [PubMed] [Google Scholar]
- 49.Patterson, T. F., J. E. Patterson, B. L. Masecar, G. E. Barden, W. J. Hierholzer, Jr., and M. J. Zervos. 1988. A nosocomial outbreak of Branhamella catarrhalis confirmed by restriction endonuclease analysis. J. Infect. Dis. 157:996-1001. [DOI] [PubMed] [Google Scholar]
- 50.Pelton, S. I., and J. O. Klein. 1999. The promise of immunoprophylaxis for prevention of acute otitis media. Pediatr. Infect. Dis. J. 18:926-935. [DOI] [PubMed] [Google Scholar]
- 51.Pesci, E. C., D. L. Cottle, and C. L. Pickett. 1994. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62:2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purdy, D., S. Cawthraw, J. H. Dickinson, D. G. Newell, and S. F. Park. 1999. Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in Campylobacter survival and colonization. Appl. Environ. Microbiol. 65:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richards, J. 1988. Evaluation of a rapid method for identifying Branhamella catarrhalis. J. Clin. Pathol. 41:462-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roggenkamp, A., T. Bittner, L. Leitritz, A. Sing, and J. Heesemann. 1997. Contribution of the Mn-cofactored superoxide dismutase (SodA) to the virulence of Yersinia enterocolitica serotype O8. Infect. Immun. 65:4705-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo, T. A., J. E. Guenther, S. Wenderoth, and M. M. Frank. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol. Microbiol. 9:357-364. [DOI] [PubMed] [Google Scholar]
- 56.Santos, R., S. Bocquet, A. Puppo, and D. Touati. 1999. Characterization of an atypical superoxide dismutase from Sinorhizobium meliloti. J. Bacteriol. 181:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 58.Takao, M., A. Yasui, and A. Oikawa. 1991. Unique characteristics of superoxide dismutase of a strictly anaerobic archaebacterium Methanobacterium thermoautotrophicum. J. Biol. Chem. 266:14151-14154. [PubMed] [Google Scholar]
- 59.Takeda, Y., and H. Avila. 1986. Structure and gene expression of the E. coli Mn-superoxide dismutase gene. Nucleic Acids Res. 14:4577-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsolis, R. M., A. J. Baumler, and F. Heffron. 1995. Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect. Immun. 63:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valderas, M. W., and M. E. Hart. 2001. Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J. Bacteriol. 183:3399-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, C. H., J. J. Tsai-Wu, Y. T. Huang, C. Y. Lin, G. G. Lioua, and F. J. Lee. 1998. Identification and subcellular localization of a novel Cu,Zn superoxide dismutase of Mycobacterium tuberculosis. FEBS Lett. 439:192-196. [DOI] [PubMed] [Google Scholar]
- 63.Yu, B. P. 1994. Cellular defenses against damage from reactive oxygen species. Physiol. Rev. 74:139-162. [DOI] [PubMed] [Google Scholar]