Abstract
Cell-mediated immunity plays a crucial role in host defenses against Cryptococcus (Filobasidiella) neoformans. Therefore, the identification of cryptococcal antigens capable of producing T-cell-mediated responses, such as delayed-type hypersensitivity (DTH) reactions, may be useful in the development of immune-based strategies to control cryptococcosis. In order to characterize DTH-producing antigens, culture supernatants from the unencapsulated Cap-67 strain were separated by anion-exchange chromatography. After further fractionation by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis, a purified protein with an apparent molecular mass of 25 kDa was found to produce DTH, as evidenced by increased footpad swelling in mice immunized with culture supernatants, relative to unimmunized mice. The 20-amino-acid N-terminal sequence of the 25-kDa protein was used to search data of the C. neoformans Genome Project. Based on the genomic DNA sequence, a DNA probe was used to screen a λ cDNA library prepared from strain B3501. Clones were isolated containing the full-length gene (d25), which showed homology with a number of polysaccharide deacetylases from fungi and bacteria. The recombinant d25 protein expressed in Escherichia coli was similar to the natural one in DTH-producing activity. Moreover, immunization with either the natural or the recombinant protein prolonged survival and decreased fungal burden in mice challenged with the highly virulent C. neoformans strain H99. In conclusion, we have described the first cryptococcal gene whose product, a 25-kDa extracellular polysaccharide deacetylase, has been shown to induce protective immunity responses.
Cryptococcus (Filobasidiella) neoformans is an encapsulated yeast that can infect both immunocompetent individuals and patients with a number of defects in antimicrobial defenses (21, 25). Cryptococcosis has emerged as one of the most frequent life-threatening infections in AIDS patients. Conventional antifungal therapy has major limitations, and patients may progress to fatal meningoencephalitis despite chronic treatment (21). Therefore, there is a need to develop alternative strategies to control cryptococcosis. Glucuronoxylomannan (GXM), the main capsular constituent, is a well-characterized virulence factor displaying antiphagocytic and tolerogenic activities. Accordingly, administration of anti-GXM antibodies can protect against infection (1, 23, 24).
Cell-mediated immunity plays a crucial role in controlling cryptococcal growth. In mice, as in humans, defects in cellular immunity lead to systemic and often fatal forms of the disease. Therefore, strategies based on cell-mediated immune responses could be useful to control cryptococcosis. Active immunization strategies for patients at risk of developing the disease must, of course, take into account the possibility of a progressive failure of important arms of the immune system, such as CD4+ T cells in AIDS patients. However, recent technical improvements in manipulation of CD4+ and CD8+ T cells, including their genetic modification, are opening new exciting possibilities in the field of adoptive immunization (33). A prerequisite for the successful application of these techniques is the identification of antigens capable of inducing protective cell-mediated responses.
In mouse models, delayed-type hypersensitivity (DTH) responses correlate well with restriction of C. neoformans systemic spreading (26) and have been extensively used to study cell-mediated immunity. DTH reactions can be elicited following intratracheal infection with viable yeasts (22) or immunization with either heat-killed yeasts (22) or concentrated culture supernatants known as CneF (3, 30). Immunization with CneF but not heat-killed C. neoformans produces significant protection in lethal experimental cryptococcosis (27). Little is known of CneF antigens capable of eliciting cell-mediated responses. CneF is known to contain, in addition to GXM, galactoxylomannan (GalXM) and mannoproteins (MPs) (29). MPs but not GXM or GalXM were found to induce DTH reactions (29). MPs, however, are heterogeneous proteins sharing the ability to bind to concanavalin A (ConA) columns. The structure and antigenic properties of individual MP components have been only partially characterized (4, 5). A 98-kDa MP, with homology to chitin deacetylases, was recently cloned and shown to stimulate a mouse CD4+-T-cell hybridoma to produce interleukin 2 (18).
Mandel et al. cloned a cryptococcal gene, termed DHA1, which encodes a secreted protein capable of inducing DTH (20). Since this protein only partially accounted for the DTH-inducing ability of whole CneF, it is likely that other cryptococcal antigens are also capable of producing cell-mediated responses. In the present study, we have purified a 25-kDa protein from cryptococcal culture supernatant and cloned the corresponding gene. This showed significant homology with chitin deacetylases from other fungi, as well as bacterial N-acetylglucosamine deacetylases and xylanases. After expression in Escherichia coli, the recombinant protein was able to produce not only DTH but also protective immunity responses that were similar in extent to those produced by the natural 25-kDa protein. To our knowledge, this is the first characterization of a cryptococcal protein capable of inducing protective immunity.
MATERIALS AND METHODS
Strains.
The encapsulated C. neoformans A9759 strain (serotype A), kindly donated by E. Reiss, Centers for Disease Control, Atlanta, Ga., was employed to produce culture supernatants used to immunize mice in DTH studies. The acapsular strain Cap-67, originally derived from serotype D strain B3501, was provided by E. Jacobson (Richmond, Va.) and was used for protein purification procedures. Cap-67 was used for two reasons. First, because it does not produce the high-molecular-weight GXM, thus simplifying supernatant concentration and protein purification. Second, a cDNA library prepared from the parent strain B3501 is commercially available (see below). Moreover, Cap-67 is closely related to strain JEC 21, which was chosen for sequencing by the Stanford University Technology Center C. neoformans Genome Project. The highly virulent serotype A C. neoformans strain H99 was used to establish a model of experimental cryptococcosis. Both C. neoformans H99 (ATCC 208821) and Blastomyces dermatitidis (ATCC 56123) were purchased from the American Type Culture Collection (Manassas, Va.).
Endotoxin-free conditions.
Endotoxin-free conditions were maintained by using glassware heated for 3 h at 180°C. Endotoxin-free water, produced with a Maxima Life Science USF Elga apparatus (distributed by Dasit S.p.A., Milan, Italy), was used for dialysis and preparation of culture media and buffers.
Cultures.
To obtain culture supernatants, cryptococci were grown at 30°C in a chemically defined, entirely dialyzable medium (3). Briefly, starter cultures were prepared by inoculating isolated colonies from Sabouraud agar plates (Difco; distributed by DID, Milan, Italy) to 200 ml of dialyzable medium.
The culture was incubated under agitation at 30°C for 2 days and used to inoculate 2-liter aliquots of fresh medium to a density of 104 cells per ml. Cultures were incubated under agitation with an orbital shaker at 30°C and harvested after 7 days. Yeast cells were removed by tangential filtration through 0.2-μm-pore-size cassettes (Pellicon System; Millipore S.p.A., Vimodrone, Italy). The supernatants were concentrated by tangential filtration with 10,000-Da-cutoff cassettes, dialyzed against water, and lyophilized (10, 11).
B. dermatitidis was reconstituted in sterile water, and the yeast phase was grown on Sabouraud agar plates. Five isolated colonies were inoculated in 200 ml of Sabouraud broth (Difco) and incubated at 35°C under agitation for 48 h. Yeast cells were collected by centrifugation, killed by heat treatment (60°C for 90 min), washed three times with phosphate-buffered saline (PBS) (0.01 M phosphate, 0.15 M NaCl, pH 7.4), and resuspended to a concentration of 108/ml.
DTH reactions.
Female BALB/c mice aged 8 to 10 weeks were obtained from Stefano Morini, S. Polo D'Enza, Italy. To sensitize mice with cryptococcal antigens, lyophilized supernatants from the A9759 strain were solubilized in PBS (1.4 mg/ml) and were emulsified with an equal volume of complete Freund’s adjuvant (CFA) (Sigma, Milan, Italy). Mice were immunized by subcutaneous injection at two different sites of 0.2 ml of the emulsion. Control nonsensitized mice received a PBS-CFA emulsion. Seven days later, mice were footpad tested by the inoculation of 25 μl of a PBS solution of the test antigen (right hind pad) and PBS only (left hind pad). In selected experiments, the endotoxin-inactivating agent polymyxin B (Sigma) was added to a final concentration of 20 μg/ml to the antigen solutions used for challenge. Twenty-four hours after challenge, mice were euthanatized. By using a surgical blade, both hind pads were cut off at the tarsus, which was recognized as a dent of bone above the articulatio tarsi transversa (17). Data were expressed as differences in weight between the right and left hind pads. DTH data presented here are means ± standard deviations of values observed in six mice during three separate experiments.
Anion-exchange chromatography.
Lyophilized supernatants from strain Cap-67 were dissolved in 0.01 M Tris-HCl buffer, pH 8.0, and loaded on a 30-ml DEAE column (Macro-prep DEAE-Support; Bio-Rad Laboratories, Milan, Italy). A flow rate of 1 ml/min was used for sample loading and throughout the subsequent chromatographic procedures. After absorbance (280 nm) values went back to the baseline, increasing NaCl concentrations (0.05, 0.5 and 1 M in 0.01 M Tris-HCl buffer, pH 8.0) were applied sequentially. Ten-milliliter fractions were collected, pooled according to the absorbance peaks, dialyzed against water, and lyophilized.
Analytical methods.
Protein concentration was determined by the Bradford protein microassay method (Bio-Rad) using bovine serum albumin (BSA) as a standard, according to manufacturer's instructions. Endotoxin levels in cryptococcal supernatants and fractions were measured by a Limulus amebocyte lysate assay kit (Associates of Cape Cod Inc.; distributed by International PBI, Milan, Italy). In all of the Cap-67 preparations endotoxin concentration was <5 pg/mg.
Electrophoresis and Western blots.
Analytical sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed with the PhastSystem using precast 10 to 15% gels, as per manufacturer's instructions (Amersham Pharmacia Biotech, Milan, Italy). Gels were stained with Coomassie according to the instructions of the gel manufacturer.
Preparative SDS-PAGE was performed to separate different bands from concentrated chromatographic fractions using a Protean II xi Slab Cell (Bio-Rad). After electrophoresis, protein recovery and removal of SDS from the acrylamide gel were performed using the Model 422 Electro-Eluter (Bio-Rad), according to the manufacturer's instructions. Purity of the obtained preparations was checked by analytical SDS-PAGE in overloaded gels (2 μg per lane) using the PhastSystem.
Amino acid sequencing.
The purified 25-kDa protein was subjected to SDS-PAGE electrophoresis in 20- by 20-cm gels, as described above, blotted onto polyvinylidene difluoride membranes (Immobilon P; Millipore), and stained according to the manufacturer's instructions. N-terminal amino acid sequencing was performed directly on excised bands at Midwest Analytical, Inc., St. Louis, Mo., using a Perkin-Elmer Biosystems (Foster City, Calif.) Model 477 Protein Sequencer. BLAST software (2) was used to conduct homology searches of the GenBank database and the C. neoformans Genome Project available at the web site of the Stanford Genome Technology Center (http://wwwsequence.stanford.edu/group/C.neoformans/index.html).
Sequence analysis was carried out at the BCM Search Launcher server (http://searchlauncher.bcm.tmc.edu/seq-util/seq-util.html) (35). Signal peptide cleavage site analysis was performed using SignalP (http://www.cbs.dtu.dk/services/SignalP/) (32). Domain analysis was done using the Pfam database (http://pfam.wustl.edu/).
cDNA library screening.
A sense primer (5′-CTATCAACAACTGTAATCAGC-3′), based on the genomic sequence coding for the N-terminal amino acids INNCNQ (http://wwwsequence.stanford.edu/group/C.neoformans/index.html), was used in conjunction with an antisense universal T7 primer to obtain a PCR product from a λ ZAP II phage C. neoformans cDNA library prepared from strain B3501 (Stratagene, La Jolla, Calif.). The PCR was perfomed with a Taq DNA polymerase (Roche, Monza, Italy) using 35 cycles and an annealing temperature of 54°C. The PCR product was then cloned using the TA cloning kit (Invitrogen, Carlsbad, Calif.) and transformed into Top10 cells (Invitrogen). Plasmid DNA was isolated from transformants by standard methods and was used for both nucleotide sequencing and preparing a labeled probe for cDNA library screening. Briefly, both DNA strands were sequenced by the Thermo Sequenase Fluorescent Labeled Primer Cycle Sequencing Kit (Amersham Pharmacia), using the ALFexpress DNA Sequencer (also from Amersham Pharmacia).
A 192-bp fragment was cut by restriction enzymes, [32P]dCTP labeled (High Prime kit; Boehringer GmbH, Mannheim, Germany), and used as a probe for cDNA library screening. One of the positive clones, termed pK-C, was selected for further analysis.
d25 gene expression in E. coli.
DNA from the pK-C clone was excised from the λ ZAP II vector using the manufacturer's recommendations and sequenced. A 684-bp sequence carrying the complete open reading frame was identified in pK-C and designated d25.
In order to obtain the product of the d25 gene, the latter was amplified from the pK-C clone using the gene-specific primers 5′- CGGGATCCGCGACCGTCGAAACTATCAACAACTG-3′ and 5′-GGAATTCATTGCCAAGAGCTATCCCTCTCTC-3′, where underlined nucleotides represent restriction sites for BamHI and EcoRI, respectively.
The gene was cloned in the expression vector pGEX-4T-1 (Amersham Pharmacia), and used to transform the E. coli BL21 strain. Protein expression and purification were performed as described earlier (34). Briefly, after induction with 1 mM isopropyl-β-d-thiogalactopyranoside, the d25-glutathione-S-transferase (GST) fusion protein was purified from lysed cells by absorption with glutathione-agarose beads (Sigma), followed by elution in the presence of 5 mM reduced glutathione (Sigma). The recombinant protein was subjected to SDS-PAGE and either stained with Coomassie or electroblotted onto nitrocellulose membrane for Western blot analysis using GST-specific polyclonal antibodies (Amersham Pharmacia), according to the manufacturer's instructions. The recombinant d25 protein was also analyzed by SDS-PAGE after cleavage of the GST moiety with the site-specific protease thrombin (Sigma), according to the protocols provided by the manufacturer.
RNA extraction and Northern blots.
For RNA extraction, Cap-67 cultures were grown as described above and the cells were harvested by centrifugation. The pellet was resuspended in extraction buffer (0.01 M Tris HCl, pH 7.5, 100 mM LiCl, 10 mM EDTA, and 0.2% SDS) and vortexed with glass beads in the presence of phenol-chloroform-isoamyl alcohol (25:24:1). After extracting the aqueous phase with chloroform-isoamyl-alcohol (24:1), RNA was precipitated with 1 volume of 5 M LiCl. For Northern blot analysis, the RNA preparation was size fractioned on a 1.3% agarose formaldehyde gel and transferred to a nylon membrane (Hybond N; Amersham Pharmacia). Hybridization was performed as described in the instructions provided with the membrane using the [32P]-labeled probe also employed for cDNA library screening.
Experimental cryptococcosis model.
To assess the ability of d25 to produce protective immunity, female BALB/c mice aged 8 to 10 weeks were immunized before challenge with a highly virulent C. neoformans strain (H99). Purified d25-GST fusion protein, GST, natural 25-kDa protein, or lyophilized supernatant from the Cap-67 strain, each at 0.2 mg/ml (protein concentration) in PBS, was mixed with an equal volume of CFA. Mice were immunized subcutaneously with 0.2 ml of emulsion and after 7 days were injected intravenously with 7.5 × 103 C. neoformans cells grown to the mid-log phase in synthetic medium. The lungs, spleens, and livers were removed at the indicated times after challenge, and the number of CFU in each organ was determined using standard methods. Three mice per group were used for CFU determinations, while 10 mice per group were observed daily for 30 days to measure survival time.
Statistical analysis.
All data are expressed as means ± standard deviations. Statistical significance of differences in footpad weight or number of organ CFU was assessed by one-way analysis of variance and the Student-Keuls-Newman test. When P values of less than 0.05 were obtained, the differences were considered statistically significant. Survival data were analyzed with Kaplan-Meier survival plots followed by the log rank test (JMP Software; SAS Institute, Cary, N.C.) on an Apple Macintosh computer.
Nucleotide sequence accession number.
The d25 gene sequence has been deposited with the EMBL Nucleotide Sequence Database (accession number AJ414580).
RESULTS
DTH induction by cryptococcal supernatants.
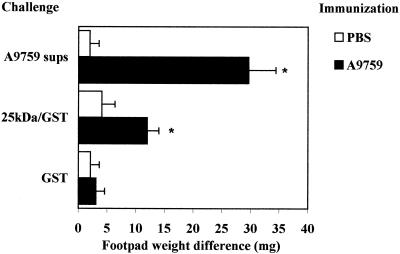
In initial experiments we ascertained whether supernatants obtained using Cap-67, the strain initially chosen for protein purification studies, could produce DTH in mice immunized with A9759 supernatants. The rationale for using unrelated strains for immunization and challenge (e.g., A9759 and Cap-67, respectively) in these and subsequent studies was to focus on antigenic epitopes shared by the strains that were, therefore, likelier to be widely expressed among cryptococcal isolates. In the experiments shown in Fig. 1A, mice were immunized with concentrated A9759 supernatants in CFA and challenged after 7 days in the right hind footpad with A9759 or Cap-67 supernatants. Control unimmunized (i.e., injected with CFA only) mice were challenged in the footpads exactly like the immunized ones.
FIG. 1.
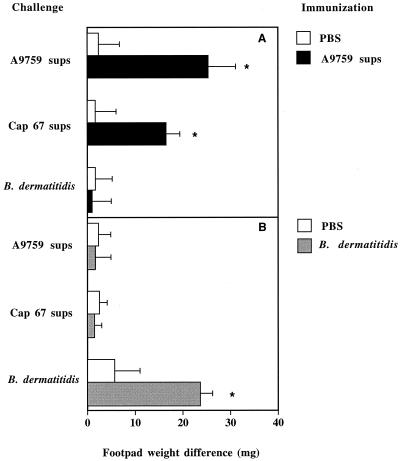
Induction of DTH by cryptococcal supernatants (sups). Mice were immunized with 280 μg of lyophilized supernatants from the A9759 C. neoformans strain (A9759 sups) (A) or with B. dermatitidis (107 cells) (B). Control nonimmunized mice received PBS only. Seven days later, mice were challenged in the right hind pad by the inoculation of A9759 or Cap-67 supernatants (35 μg) or B. dermatitidis (5 × 104 cells). Results indicate differences in weight between the right hind pad (challenged with the test preparation) and the left one (inoculated with vehicle only). Data represent means ± standard deviations of six observations conducted during three separate experiments. ∗, P < 0.05, relative to nonimmunized mice, by one-way analysis of variance and the Student-Keuls-Newman test.
A9759 supernatants produced significant footpad swelling in A9759-immunized animals but not in control, unsensitized mice (Fig. 1A). Significant responses in A9759-immunized animals were also produced by challenge with Cap-67 supernatants, indicating the presence of antigenic epitopes shared by the two strains. In contrast, significant reactions were not produced by B. dermatitidis in A9759-sensitized animals. Conversely, neither A9759 nor Cap-67 supernatants were able to produce significant DTH in mice sensitized with B. dermatitidis (Fig. 1B). As expected, B. dermatitidis-immunized mice developed a significant response upon challenge with the homologous antigen.
DTH induction by chromatographic fractions.
To identify DTH-producing components, Cap-67 supernatants were fractioned by anion-exchange chromatography into four protein-containing peaks (not shown). Peak 1 contained material that did not bind to the column, while peaks 2 to 4 were eluted after increasing NaCl concentrations (0.05, 0.5, and 1 M, respectively) were applied. Peak 4 material was not further studied because of its relatively low protein content.
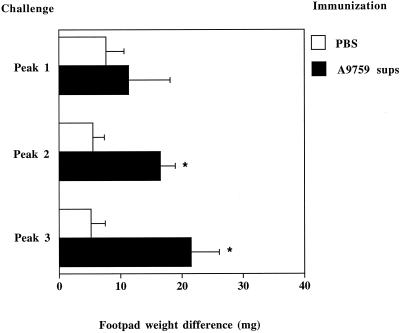
To test the Cap-67 fractions for DTH-inducing activity, mice immunized with concentrated A9759 supernatants in CFA or control, unimmunized (i.e., injected with PBS-CFA only) mice were challenged, in the right hind footpad with peak 1, peak 2, or peak 3 fractions (10 μg in 25 μl of PBS). The left footpad was inoculated with an equal volume of vehicle. Figure 2 shows that peak 3 had the highest potency in producing DTH reactions, as evidenced by footpad swelling in immunized but not in control mice. Peak 2 induced moderate DTH reactions, while peak 1 showed no activity (Fig. 2).
FIG. 2.
Induction of DTH by different anion-exchange chromatography fractions. Mice were immunized with 280 μg of concentrated supernatants from the A9759 strain (A9759). Control nonimmunized mice received PBS only. Seven days later, mice were challenged in the right hind pad by the inoculation of 10 μg (protein content) of peak 1, 2, or 3 preparations. Twenty-four hours after challenge, mice were sacrificed and hind pad weights were determined. Results indicate differences in weight between the right hind pad (challenged with the test preparation) and the left one (inoculated with vehicle only). Data represent means ± standard deviations of six observations conducted during three separate experiments. ∗, P < 0.05, relative to nonimmunized mice, by one-way analysis of variance and the Student-Keuls-Newman test.
DTH induction by preparative SDS-PAGE fractions.
When peak 2 or 3 fractions were subjected to analytical SDS-PAGE, different Coomassie-positive bands were found, the most prominent of which showed molecular masses of 96, 76, and 17 kDa (peak 2) and 200, 115, 83, 67, 50, 45, and 25 kDa (peak 3). Based on these findings, peak 2 and 3 proteins were subjected to preparative SDS-PAGE followed by electroelution from gel slices. Figure 3 shows the DTH-inducing activities of the various fractions (each at dose of 10 μg) in mice immunized with A9759 supernatants.
FIG. 3.
DTH induction by preparative SDS-PAGE fractions. Mice were immunized or injected with PBS as indicated in the legend to Fig. 2. Footpad testing was done with 10 μg of the designated antigen fractions. Data represent means ± standard deviations of six observations conducted during three separate experiments. ∗, P < 0.05, relative to nonimmunized mice, by one-way analysis of variance and the Student-Keuls-Newman test.
The highest activity was produced by the 20- to 30-kDa peak 3 fraction (Fig. 3). A single 25-kDa protein was present in this fraction, as shown by SDS-PAGE analysis using overloaded gels (Fig. 4A). Significant DTH was also produced by the 30- to 47-kDa peak 3 fraction (Fig. 3A). However, a 25-kDa band, in addition to 32- and 45-kDa ones, was detected in this fraction (not shown). Therefore, we could not exclude that the activity of the 30- to 47-kDa fraction was caused by contamination with the 25-kDa protein. Figure 3B shows that moderate DTH-producing activity was also present in the 70- to 100-kDa peak 2 fraction.
FIG. 4.
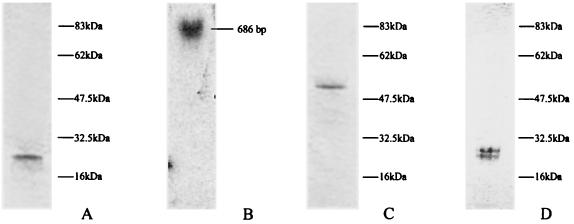
SDS-PAGE and Northern blot analysis of the 25-kDa deacetylase. (A) SDS-PAGE analysis of the purified 25-kDa natural protein. (B) d25 Northern blot analysis. The arrow indicates the size of the d25 mRNA band. (C) SDS-PAGE analysis of the purified d25-GST fusion protein. (D) SDS-PAGE analysis of the recombinant d25 protein after cleavage of the GST moiety.
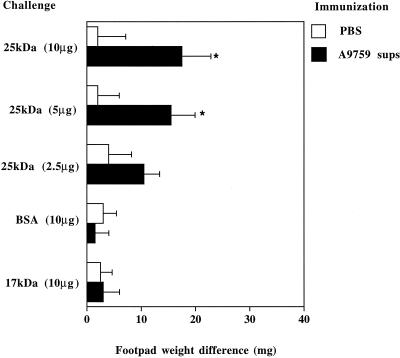
Since the 20- to 30-kDa peak 3 fraction showed the highest DTH-producing activity, further studies focused on this fraction. The 15- to 25-kDa peak 2 fraction, containing a single 17-kDa protein, as well as preparative SDS-PAGE-purified BSA, served as negative controls for further studies. Figure 5 shows that the DTH responses produced by the 25-kDa protein were dose dependent, while neither the 17-kDa protein nor BSA had any activity. The specificity of these results was further evidenced by the inability of the 25-kDa protein to produce DTH in mice sensitized with B. dermatitidis (not shown). Experiments were repeated after adding the endotoxin-inactivating agent polymyxin B (20 μg/ml) to the 25-kDa solution used for challenge. No differences were detected in footpad swelling induced by these preparations in the absence and in the presence of polymyxin B (not shown). These data indicated that contamination with endotoxin was unlikely to account for the DTH-inducing abilities of the 25-kDa protein.
FIG. 5.
Induction of DTH by the 25-kDa protein. Mice were immunized or injected with PBS as indicated in the legend to Fig. 2. Results indicate differences in weight between the right hind pad (challenged with the test preparation) and the left one (inoculated with vehicle only). Data represent means ± standard deviations of six observations conducted in three separate experiments. ∗, P < 0.05, relative to nonimmunized mice, by one-way analysis of variance and the Student-Keuls-Newman test.
Cloning and expression of the d25 gene.
The sequence of the 20 N-terminal amino acids of the 25-kDa protein was determined and used to search data of the C. neoformans Genome Project (http://www-sequence.stanford.edu/group/C.neoformans/index.html). The 20-amino-acid sequence obtained matched to the N terminus of the mature form of an extracellular protein after cleavage of the signal peptide. GenBank search and domain analysis identified the protein as a deacetylase. To obtain gene-specific DNA fragments, a combination of a forward sense primer and a T7 universal primer was used in PCR amplification from a C. neoformans λ ZAP II phage cDNA library prepared from strain B3501.
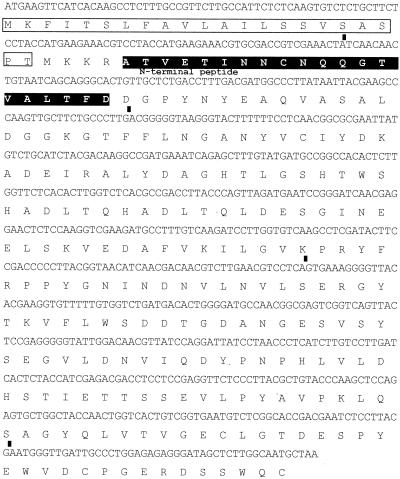
A fragment, approximately 600 bp in length, was obtained. Based on the DNA sequence of this fragment, a gene-specific probe was successfully used in screening of the cDNA library. A positive clone, termed pK-C, was selected, and the cDNA insert was sequenced. An open reading frame coding for a 226-amino-acid protein with a mass of about 25 kDa was identified and designated d25. The size of the d25 gene matched the expected value, as determined by Northern blot analysis of total Cap-67 RNA (Fig. 4B). Figure 6 shows the complete DNA sequence of the d25 gene and its corresponding deduced amino acid sequence.
FIG. 6.
Complete DNA and deduced amino acid sequence of the d25. The boxed sequence indicates a putative signal peptide, and the bars indicate introns of 55-, 53-, 56-, and 52-bp from the 5′ to 3′ end. White lettering on a black background indicates the N-terminal sequence obtained from the natural protein.
Additional analysis, conducted by comparing the d25 cDNA sequence with the genomic one found in the C. neoformans Genome Project database, indicated the presence of four introns of 55, 53, 56, and 52 bp from 5′ to 3′. Moreover, a putative signal peptide sequence was identified using SignalP. The putative cleavage site was predicted between amino acids 19 and 20 when setting parameters for yeasts. When setting parameters for gram-positive bacteria, for which longer leader peptides are considered, the cleavage site was predicted between amino acids 26 and 27. This cleavage site matched exactly the data obtained by amino acid sequencing of the mature natural protein, which indicated the Ala residue at position 27 as the most N-terminal amino acid.
Asparagine glycosylation sites were not detected, as evidenced by absence of the consensus sequence NASE. A BLAST search indicated that the protein has partial homology with a number of highly conserved polysaccharide deacetylases, including fungal chitin deacetylases, bacterial peptidoglycan N-acetylglucosamine deacetylases, and the rhizobial nodulation factor B proteins. The highest degrees of homology were found with a Streptococcus pyogenes deacetylase (37% identity), Blumeria graminis chitin deacetylase (36%), and Streptomyces coelicolor deacetylase (31%). A BLAST search of the C. neoformans Genome Project database indicated the presence of an additional gene with homology to chitin deacetylases, which was identical to the one described by Levitz et al. (18).
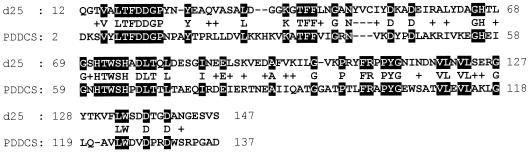
Figure 7 shows alignment of similar amino acid sequences from d25 and a polysaccharide deacetylase domain consensus sequence (pfam01522) from the Pfam database. The d25 region that displays homology to the polysaccharide deacetylase domain is located in the amino-terminal region (amino acids 14 to 140). The carboxyl-terminal region (amino acids 141 to 226) does not display any significant similarity to other proteins.
FIG. 7.
Alignment of similar amino acid sequences from d25 and polysaccharide deacetylase domain consensus sequence (PDDCS). White lettering on a black background indicates identity; + indicates conserved amino acid substitutions.
DTH activity of d25.
The d25 gene was subcloned into the bacterial expression vector pGEX-4T-1 to generate d25Z, and the expressed recombinant product was purified as indicated in Materials and Methods. The molecular weight of d25, after cleavage of the GST moiety with thrombin, exactly matched that of the natural purified protein (Fig. 4). The d25 protein was poorly or nonglycosylated, as determined using the Immun-Blot Kit for Glycoprotein Detection (Bio-Rad; data not shown).
To ascertain whether the recombinant d25 protein had the same DTH-producing activity as the natural one, mice immunized with A9759 supernatants were challenged in their footpad with 10 μg of the purified d25-GST fusion protein or GST alone. Figure 8 shows that the former but not the latter induced significantly higher increases in footpad weight of immunized mice than in unimmunized mice. These data indicated that the recombinant protein was similar to the natural one in DTH-producing activity.
FIG. 8.
Induction of DTH by the d25 recombinant protein. Mice were immunized or injected with PBS as indicated in the legend to Fig. 2. Results indicate differences in weight between the right hind pad (challenged with 10 μg of each test preparation) and the left one (inoculated with vehicle only). Data represent means ± standard deviations of six observations conducted in three separate experiments. ∗, P < 0.05, relative to nonimmunized mice, by one-way analysis of variance and the Student-Keuls-Newman test. sups, supernatants.
Protective effects of immunization with d25.
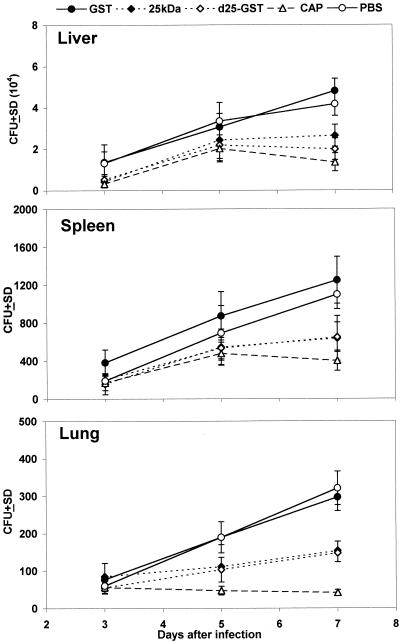
A murine cryptococcosis model was used to study the ability of the 25-kDa deacetylase to induce protective responses. To this end, mice were immunized with the d25-GST fusion protein, GST alone, or the natural 25-kDa protein, as detailed in Materials and Methods, and infected, 7 days later, with the highly virulent H99 strain. Additional groups of mice were immunized with whole Cap-67 supernatant, used as a positive control, or PBS, used as negative control. Groups of three mice were sacrificed at 3, 5, and 7 days after infection, and the numbers of CFU were measured in the lungs, spleens, and livers (Fig. 9). No significant differences between the experimental groups were detected in any of the organs at 3 days after infection. Similarly, no differences in liver and spleen colony counts were detected at 5 days after infection. At this time, however, significantly lower numbers of CFU were present in the lungs of mice immunized with Cap-67 supernatant, d25-GST, or the natural 25-kDa protein, relative to those treated with GST alone or PBS (P < 0.05). Similarly, at 7 days after infection, significantly lower numbers of CFU were detected in each organ of mice immunized with Cap-67 supernatant, d25-GST, or the natural 25-kDa protein than in those immunized with either GST alone or PBS (P < 0.05, Fig. 9). These data indicated that immunization with either the natural or the recombinant 25-kDa deacetylase resulted in enhanced clearance of cryptococci from the spleens, livers, and lungs of infected mice.
FIG. 9.
Mean numbers of cryptococcal CFU in livers, spleens, and lungs from immune and control mice at 3, 5, and 7 days after intravenous challenge with 7.5 × 103 viable C. neoformans cells. Three mice per group were used for CFU determinations. The data are from one representative experiment of three producing similar results. SD, standard deviations.
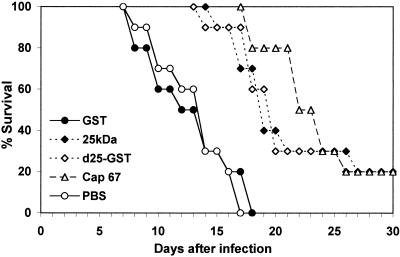
In further studies we looked at the effects of deacetylase immunization on survival. To this end, groups of 10 mice were immunized and infected as described above. Lethality was monitored daily for 30 days. Figure 10 shows that enhanced survival was observed in mice immunized with either Cap-67 supernatants, d25-GST, or the 25-kDa natural protein, relative to those immunized with GST alone or PBS. In fact, mean survival times were significantly higher in mice immunized with Cap-67 supernatants (23 ± 1.0 days), d25-GST (21 ± 1.3 days), or 25-kDa protein (21 ± 1.4 days) than in those treated with GST alone (13 ± 1.2 days) or PBS (13 ± 1.0 days; P < 0.05). The differences in survival time between Cap-67-immunized mice and those immunized with either d25-GST or the 25-kDa natural protein were not statistically significant.
FIG. 10.
Percent survival of GST-, 25-kDa-, d25-GST-, and Cap-67-immunized mice and control mice after challenge to the mice with 7.5 × 103 viable C. neoformans cells. Mice were treated 7 days before challenge with the designated preparations. Ten mice per group were observed over a 30-day period.
DISCUSSION
Cell-mediated immunity plays a crucial role in host defenses against cryptococcosis. However, little is known of C. neoformans antigens capable of inducing such responses. The characterization of protein antigens is useful not only in the development of control strategies based on cell-mediated immunity, including adoptive immunization (33), but also in the identification of potential carriers for the synthesis of GXM-protein conjugate vaccines.
DTH reactions have been classically used to detect cell-mediated responses to cryptococcal antigens in mice. These reactions have been produced mostly using heat-killed cryptococci or supernatants obtained from formalin-killed cultures and known as CneF (3, 26, 28, 30, 31). CneF but not killed yeasts produced protective responses in an experimental murine cryptococcosis model (27). The supernatants that we used as the starting material for purification are similar to previously described CneF, with the exception that formalin treatment was not used in our studies. Similarly to CneF (30), our supernatants consistently induced DTH reaction in mice immunized with the homologous antigen but not with B. dermatitidis (Fig. 1).
The major finding of the present study is the identification in cryptococcal culture supernatants of a 25-kDa protein capable of producing DTH and protective immunity responses. Both the natural protein and the corresponding recombinant product could produce significant footpad swelling in immunized animals but not in unimmunized controls or in those immunized with an heterologous antigen (B. dermatitidis). The specificity of these findings was further supported by the absence in immunized mice of DTH reactions to other cryptococcal products, including different biochemical fractions obtained from culture supernatants (Fig. 2, 3 and 5).
Therefore, the ability to induce DTH responses is apparently restricted to a relatively small number of cryptococcal proteins. This is in agreement with a previous study, also using biochemical separation of culture supernatants (20). Of course, our data do not exclude that other cryptococcal products are capable of producing DTH reactions. Rather, DTH production by whole supernatants may be accounted for by the combined activities of more than one extracellular protein. Mandel et al. have described a cryptococcal gene, termed DHA1, which encodes a DTH-producing peptide found in culture supernatants (20). Levitz et al. have recently characterized a 98-kDa MP that is capable of stimulating a mouse CD4+-T-cell hybridoma to produce interleukin 2 (18). Interestingly, both the latter protein and the one described here show homology with polysaccharide deacetylases. Studies are under way to ascertain whether the antigenic properties of these proteins reside in their deacetylase domain.
Since immunization with culture supernatants can not only sensitize mice for the development of DTH after challenge but can also protect them against experimental infection (27), we ascertained whether a deacetylase could produce protective immunity. We found here that both the natural and recombinant deacetylases prolonged survival and decreased fungal burden in mice challenged with the highly virulent C. neoformans strain H99. It is presently unclear whether the 25-kDa protein described here has any relationship to previously characterized antigens (6, 7, 15). None of the antigenic cryptococcal proteins described thus far was shown to elicit protective immunity responses.
Several cryptococcal products capable of binding human and/or rodent antibodies in Western blots have been previously described (6, 7, 13), although their ability to induce cell-mediated responses has not been tested. Sera from human immunodeficiency virus-positive patients with cryptococcosis frequently recognize protein antigens of 26, 52, 74, 110, and 114 kDa (13). Seventy-five- and 30-kDa bands were frequently detected in Western blots by sera of animals infected with C. neoformans strain 24067 (6). In addition, 34- to 38-kDa and 115-kDa proteins have been purified and characterized using specific monoclonal antibodies (14, 15). The 115-kDa protein was a mannose-rich protein with N-linked oligosaccharides (15).
The main constituents of cryptococcal supernatants have been previously characterized as GXM, GalXM, and MP (29). Of these, MPs but not GXM or GalXM were capable of inducing DTH (29). MPs are apparently composed of different proteins sharing the ability to bind ConA columns. Two MP fractions have been recently described as MP1 and MP2, with molecular masses of 35.6 and 8.2 kDa, respectively, although their DTH-inducing activities have not been tested (4, 5). It is unlikely that the 25-kDa deacetylase described in the present study is an MP. Firstly, the protein did not bind ConA in Western blots (unpublished observations). Secondly, reported molecular weights of MP1 and MP2 differ from that of the deacetylase described here. Moreover, the NASE N glycosylation consensus sequence, which is often found in mannose-rich proteins, was not found in d25. This is at variance with the deacetylase described by Levitz et al. (18).
The d25 deduced amino acid sequence showed significant homology with highly conserved polysaccharide deacetylases, including fungal chitin deacetylases, bacterial peptidoglycan N-acetylglucosamine deacetylases, and the rhizobial nodulation factor B proteins. Chitin deacetylase genes from Mucor rouxii, Saccharomyces cerevisiae, and Colletotrichum lindemuthianum have been cloned and characterized elsewhere (8, 16, 36, 37). Characterization of these and other fungal chitin deacetylases suggests that they can operate in tandem with chitin synthase and may have an important role in cell wall biosynthesis (8, 9, 12). Clearly, further studies are needed to test the hypothesis that the protein described here is involved in cell wall formation and is, therefore, a potential target for chemotherapy (19).
Acknowledgments
This work was supported by grants from the ISS (“Project AIDS” contract nos. 50a.O.34, 50 B.38, and 50 C.31) and, in part, from MURST (“Progetti di Rilevanza Nazionale”), CNR (“Progetto Finalizzato Biotecnologie”), and the European Commission (“HOSPATH” contract no. QLK2-CT-2000-00336). We acknowledge the contributions of the C. neoformans Genome Project, Stanford Genome Technology Center, funded by the NIAID/NIH under cooperative agreement U01 AI47087, and The Institute for Genomic Research, funded by the NIAID/NIH under cooperative agreement U01 AI48594.
We thank Bruno Maresca for his useful suggestions and assistance in performing the present work.
Editor: T. R. Kozel
REFERENCES
- 1.Abrahams, J., and T. G. Gilleran. 1960. Studies on actively acquired resistance to experimental cryptococcosis in mice. J. Immunol. 85:629-635. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan, K. L., and J. W. Murphy. 1993. Characterization of cellular infiltrates and cytokine production during the expression phase of the anticryptococcal delayed-type hypersensitivity response. Infect. Immun. 61:2854-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaka, W., A. F. M. Verheul, V. V. Vaishnav, R. Cherniak, J. Scharringa, J. Verhoef, H. Snippe, and I. M. Hoepelman. 1997. Cryptococcus neoformans and cryptococcal glucuronoxylomannan, galactoxylomannan, and mannoprotein induce different levels of tumor necrosis factor in human peripheral blood mononuclear cells. Infect. Immun. 65:272-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaka, W., A. F. M. Verheul, V. V. Vaishnav, R. Cherniak, J. Scharringa, J. Verhoef, H. Snippe, and A. I. M. Hoepelman. 1997. Induction of TNF-α in human peripheral blood mononuclear cells by the mannoprotein of Cryptococcus neoformans involves human mannose binding protein. J. Immunol. 159:2979-2985. [PubMed] [Google Scholar]
- 6.Chen, L. C., L. A. Pirofski, and A. Casadevall. 1997. Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect. Immun. 65:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L. C., L. D. Goldman, T. L. Doering, L. A. Pirofski, and A. Casadevall. 1999. Antibody response to Cryptococcus neoformans proteins in rodents and humans. Infect. Immun. 67:2218-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christodoulidou, A., V. Bouriotis, and G. Thireos. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 271:31420-31425. [DOI] [PubMed] [Google Scholar]
- 9.Davis, L. L., and S. Bartnicki-Garcia. 1984. Chitosan synthesis by the tandem action of chitin synthase and chitin deacetylase from Mucor rouxii. Biochemistry 23:1065-1073. [Google Scholar]
- 10.Delfino, D., L. Cianci, M. Migliardo, G. Mancuso, V. Cusumano, C. Corradini, and G. Teti. 1996. Tumor necrosis factor-inducing activities of Cryptococcus neoformans components. Infect. Immun. 64:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delfino, D., L. Cianci, E. Lupis, A. Celeste, M. L. Petrelli, F. Currò, V. Cusumano, and G. Teti. 1997. Interleukin-6 production by human monocytes stimulated with Cryptococcus neoformans components. Infect. Immun. 65:2454-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, X. D., T. Katsumoto, and K. Onodera. 1995. Purification and characterization of chitin deacetylase from Absidia coerulea. J. Biochem. 117:257-263. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton, A. J., I. Figueroa, L. Jeavons, and R. A. Seaton. 1997. Recognition of cytoplasmic yeast antigens of Cryptococcus neoformans and Cryptococcus neoformans var . gattii by immune human sera. FEMS Immunol. Med. Microbiol. 17:111-119. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton, A. J., L. Jeavons, P. Hobby, and R. J. Hay. 1992. A 34- to 38-kilodalton Cryptococcus neoformans glycoprotein produced as an exoantigen bearing a glycosylated species-specific epitope. Infect. Immun. 60:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton, A. J., and J. Goodley. 1993. Purification of the 115-kilodalton exoantigen of Cryptococcus neoformans and its recognition by immune sera. J. Clin. Microbiol. 31:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafetzopoulos, D., G. Thireos, J. N. Vournakis, and V. Bouriotis. 1993. The primary structure of a fungal chitin deacetylase reveals the function for two bacterial gene products. Proc. Natl. Acad. Sci. USA 90:8005-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamura, K. 1980. A footpad weight assay method to evaluate delayed-type hypersensitivity in the mouse. J. Immunol. Methods 39:277-283. [DOI] [PubMed] [Google Scholar]
- 18.Levitz, M. S., S. Nong, M. K. Mansour, C. Huang, and C. A. Specht. 2001. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc. Natl. Acad. Sci. USA 98:10422-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, R. K., and M. G. Rinaldi. 1999. In vitro antifungal activity of nikkomycin Z in combination with fluconazole or itraconazole. Antimicrob. Agent Chemother. 43:1401-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandel, M. A., G. G. Grace, K. I. Orsborn, F. Schafer, J. W. Murphy, M. J. Orbach, and J. N. Galgiani. 2000. The Cryptococcus neoformans gene DHA1 encodes an antigen that elicits a delayed-type hypersensitivity reaction in immune mice. Infect. Immun. 68:6196-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8:515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mody, C. H., R. Paine, C. Jackson, G. H. Chen, and G. B. Toews. 1994. CD8+ cells play a critical role in delayed type hypersensitivity to intact Cryptococcus neoformans. J. Immunol. 152:3970-3979. [PubMed] [Google Scholar]
- 23.Mukherjee, J., L. A. Pirofski, M. D. Scharff, and A. Casadevall. 1993. Antibody-mediated protection in mice with lethal intracerebral Cryptococcus neoformans infection. Proc. Natl. Acad. Sci. USA 90:3636-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee, J., M. D. Scharff, and A. Casadevall. 1992. Protective murine monoclonal antibodies to Cryptococcus neoformans. Infect. Immun. 60:4534-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, J. W. 1992. Cryptococcal immunity and immunostimulation. Adv. Exp. Med. Biol. 319:225-230. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, J. W. 1989. Cryptococcus, p. 93-138. In R. A. Cox (ed.), Immunology of fungal diseases. CRC Press, Inc., Boca Raton, Fla.
- 27.Murphy, J. W., F. Schafer, A. Casadevall, and A. Adesina. 1998. Antigen-induced protective and nonprotective cell-mediated immune components against Cryptococcus neoformans. Infect. Immun. 66:2632-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy, J. W. 1993. Cytokine profiles associated with induction of the anticryptococcal cell-mediated immune response. Infect. Immun. 61:4750-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, J. W., R. L. Mosley, R. Cherniak, G. H. Reyes, T. R. Kozel, and E. Reiss. 1998. Serological, electrophoretic, and biological properties of Cryptococcus neoformans antigens. Infect. Immun. 56:424-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, J. W., and N. Pahlavan. 1979. Cryptococcal culture filtrate antigen for detection of delayed-type hypersensitivity in cryptococcosis. Infect. Immun. 25:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muth, S. M., and J. W. Murphy. 1995. Effects of immunization with Cryptococcus neoformans cells or cryptococcal culture filtrate antigen on direct anticryptococcal activities of murine T lymphocytes. Infect. Immun. 63:1645-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen, H., S. Brunak, and G. von Heijne. 1999. Machine learning approaches to the prediction of signal peptides and other protein sorting signals. Protein Eng. 12:3-9. [DOI] [PubMed] [Google Scholar]
- 33.Sereti, I., and H. C. Lane. 2001. Immunopathogenesis of human immunodeficiency virus: implications for immune-based therapies. Clin. Infect. Dis. 32:1738-1755. [DOI] [PubMed] [Google Scholar]
- 34.Smith, D. B., and L. M. Corcoran. 2000. Expression and purification of glutathione-S-transferase fusion proteins, p. 16.7.1-16.7.7. In F. Ausubel et al. (ed.), Current protocols in molecular biology, vol. 3. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 35.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davison, and K. C. Worley. 1996. BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 36.Tokuyasu, K., et al. 1999. Cloning and expression of chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum. J. Biosci. Bioeng. 87:418-423. [DOI] [PubMed] [Google Scholar]
- 37.Tsigos, I., A. Martinou, D. Kafetzopoulos, and V. Bouriotis. 2000. Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotechnol. 18:305-312. [DOI] [PubMed] [Google Scholar]