Abstract
Owing to the degeneracy of the genetic code, protein-coding regions of mRNA sequences can harbour more than only amino acid information. We search the mRNA sequences of 11 human protein-coding genes for evolutionarily conserved secondary structure elements using RNA-Decoder, a comparative secondary structure prediction program that is capable of explicitly taking the known protein-coding context of the mRNA sequences into account. We detect well-defined, conserved RNA secondary structure elements in the coding regions of the mRNA sequences and show that base-paired codons strongly correlate with sparse codons. We also investigate the role of repetitive elements in the formation of secondary structure and explain the use of alternate start codons in the caveolin-1 gene by a conserved secondary structure element overlapping the nominal start codon. We discuss the functional roles of our novel findings in regulating the gene expression on mRNA level. We also investigate the role of secondary structure on the correct splicing of the human CFTR gene. We study the wild-type version of the pre-mRNA as well as 29 variants with synonymous mutations in exon 12. By comparing our predicted secondary structures to the experimentally determined splicing efficiencies, we find with weak statistical significance that pre-mRNAs with high-splicing efficiencies have different predicted secondary structures than pre-mRNAs with low-splicing efficiencies.
INTRODUCTION
Secondary structure can overlap protein-coding regions
Messenger RNAs (mRNAs) of eukaryotic genes are single-stranded RNA molecules whose main function is to provide information for protein synthesis. This is, most prominently, achieved by encoding the linear sequence of the protein's amino acids in a contiguous stretch of the mRNA which is flanked by 5′-untranslated region (5′-UTR) and 3′-UTR. In this so-called protein-coding or coding region of the mRNA, one nucleotide triplet or codon encodes one amino acid as determined by the genetic code. The genetic code is universal, i.e. the same for almost all organisms (but slightly different from that used in mitochondria), and degenerate, i.e. one amino acid is typically encoded by more than one type of codon which tend to differ in their third codon position. Owing to this degeneracy of the genetic code, the protein-coding region of an mRNA can thus comprise one or more overlapping layers of information that need not alter the underlying encoded amino acid sequence. The best known examples of extra information include overlapping protein-coding regions on the same or the antisense strand [not only in viral genomes (1), but also in bacteria in higher, eukaryotic organisms (2–4)] and overlapping RNA secondary structure. RNA secondary structure is formed through hydrogen bonds between complementary RNA nucleotides {G−C, A−U, G−U}. These interactions can bring distant, complementary sections of one RNA molecule into close spacial proximity. In the following, ‘secondary structure’ refers to RNA secondary structure.
Functional roles of secondary structure in gene expression
Gene expression on mRNA level is spatially and temporally regulated through a variety of mechanisms (5,6), some of which depend on RNA secondary structure. Secondary structure elements in 5′-UTRs can select translation initiation sites, can influence translation efficiency and inhibit translation (7–9), and they can also auto-regulate mRNA degradation (10). Endonucleolytic cleavage near one of the iron-responsive elements in the 3′-UTR of the human transferrin receptor mRNA is thought to be influenced by secondary structure (11). Artificial secondary structure elements can slow exonucleolytic digestion when inserted into the 3′-UTRs of yeast mRNAs (12). Proteins binding to secondary structure elements in the 5′-UTRs control the cellular response to biological signals from iron, oxygen, NO and growth factors in animals [for example, iron regulatory proteins binding iso-iron-responsive elements in animals (13)]. Small metabolites binding to riboswitches in UTRs provide a specific, fast and protein-independent way of adjusting gene expression according to the chemical environment in the cell by bringing about an allosteric rearrangement of the riboswitch's secondary structure [see (14) for an overview of riboswitches in eukaryotes and (15) for an example of a ribozyme in bacteria that cleaves its mRNA upon metabolite binding].
So far, functional secondary structure in the coding region of mRNAs has been found in Saccharomyces cerevisiae (16), where several bud-localized mRNAs contain the same secondary structure motif that is required by for protein-binding and correct mRNA localization. The use of short secondary structures as zip codes for mRNA localization has also been observed in Drosophila menalogaster (17), where these elements have been found in 3′-UTRs. As these elements typically show a low level of sequence conservation, but a high level of secondary structure conservation and as their presence in the coding regions of mRNAs has so far been thought to automatically prevent mRNA translation (which need not be the case, as shown in the example of S.cerevisiae above), many structural zip codes, some of them may be in coding regions, may have escaped the current analyses.
Functional secondary structure has also been found in the coding regions of some viral genomes. Hepatitis C has a linear, positive-sense RNA genome of ∼9600 bp length that contains one long open reading frame (ORF) encoding a poly-protein, which gives rise to nine proteins. One of the evolutionarily conserved secondary structure elements in the coding region (18) serves as a cis-acting replication element (19). There are also functional secondary structure elements in the UTRs: translation initiation depends on a highly structured internal ribosomal entry site (IRES) in the 5′-UTR (20) and a evolutionarily well-conserved stem–loop structure in the 3′-UTR is essential for virus replication (21,22). Furthermore, there is experimental evidence that long-range RNA–RNA interactions between parts of the 5′-UTR and parts of the coding region modulates IRES-dependent translation which may play a role in the regulation of viral gene expression (23). Experimental and theoretical studies of the avian influenza virus have shown that a secondary structure element in the coding region is related to the acquisition of virulence (24,25).
Existing bioinformatics studies of secondary structure in mRNAs
So far, there have been few bioinformatics studies that investigate secondary structure in the mRNAs of eukaryotic genes. Steffens and Digby (26) showed that the predicted minimum free energy (mfe) secondary structures of 51 mRNA sequences from plant, animal and bacteria are on average slightly, yet significantly more negative than those of randomized version of these mRNAs, for which codon choice is randomized while the mononucleotide composition is preserved [37 out of 51 native mRNAs (73%) have a lower mfe than their randomized versions, resulting in a, on average, 6.6% lower mfe, [−10.0, −3.2]% being the 95% confidence interval]. This result was shortly afterwards challenged by Workman and Krogh (27) who argue that dinucleotide rather than mononucleotide composition should be preserved in the randomization procedure when evaluating the significance of secondary structure. Investigating a similar set of 46 mRNAs, they find no statistical evidence that native mRNAs have on average lower predicted mfes than their randomized versions which have the same dinucleotide composition [but which do not preserve the encoded amino acid sequence as in Ref. (26)]. A later study of a large set of bacterial mRNA sequences (28) aimed to resolve this dispute by randomizing their sequences by preserving both, AI-nucleotide composition as well as the encoded amino acids (as well as codon usage). They define each mRNA's folding potential as the difference between the average mfe of native mRNA-subsequences and the average mfe of randomized versions of these mRNA-subsequences (subsequences of 50 bp length, step size of 10 bp) and also define a local folding potential based on the mfe difference of the real and the randomized subsequences. They observe a small, but statistically significant bias towards subsequences with lower than randomly expected mfe in the coding regions of many eubacteria.
Aims of our study
The aim of our study is to search for evolutionarily conserved secondary structure elements in eukaryotic mRNAs. Our method of choice is RNA-Decoder (29,30), a comparative secondary structure prediction method that is capable of detecting local and global secondary structure that has been evolutionarily conserved and that takes the known protein coding context of the coding mRNA regions explicitly into account when predicting secondary structure. The latter feature was shown to be essential for being able to reliably detect secondary structure in coding regions (30). As mRNAs do not have time to assume the mfe configuration within the living cell and as they may be bound by the ribosome, proteins or other RNA molecules, we do not expect the predicted mfe structure to be the structure that is realized in the cell and thus do not aim to predict it. In addition, the performance of programs such as Mfold (31,32) and RNAfold (33) that predict the mfe secondary structure is known to decrease with increasing sequence length (34), and we expect it to be fairly poor for sequences of typical mRNA length. By searching for evolutionarily conserved secondary structure, we aim to find secondary structure elements that are likely to play a functional role.
While analysing our set of mRNAs, Pagani et al. (35) published an experimental study of the effect of synonymous mutations on the splicing efficiency of exon 12 in the human CFTR pre-mRNA. As the CFTR gene is one of the genes in our dataset, we decided to extend our mRNA-based analysis by an analysis of the CFTR gene on pre-mRNA level in order to investigate whether the different, experimentally determined splicing efficiencies of Pagani et al. can be explained by differences in the predicted secondary structures.
MATERIALS AND METHODS
Dataset
Our dataset is derived from the multi-species dataset by Thomas et al. (36). The genomic sequences which cover 37 species of several vertebrae from the fugu fish to the human [Artibeus jamaicensis (Jamaican fruit-eating bat), Dasypus novemcinctus (nine-banded armadillo), Papio anubis (olive baboon), Felis catus (cat), Gallus gallus (chicken), Pan troglodytes (chimpanzee), Bos taurus (cow), Carollia perspicillata (Seba's short-tailed bat), Canis familiaris (dog), Sminthopsis macroura (stripe-faced dunnart), Callicebus moloch (Dusky titi), Takifugu rubripes (Fugu fish), Otolemur garnettii (small-eared galago), Gorilla gorilla gorilla (lowland gorilla), Atelerix albiventris (middle-African hedgehog), Equus caballus (horse), Homo sapiens (human), Lemur catta (ring-tailed lemur), Macaca mulatta (rhesus monkey), Callithrix jacchus (white-tufted-ear marmoset), Monodelphis domestica (grey short-tailed opossum), Microcebus murinus (grey mouse lemur), Mus musculus (house mouse), Muntiacus muntjak vaginalis (Indian Muntjac), Didelphis virginiana (North American opossum), Pongo pygmaeus (orangutan), Sus scrofa (pig), Ornithorhynchus anatinus (platypus), Oryctolagus cuniculus (rabbit), Rattus norvegicus (Norway rat), Ovis aries (sheep), Saimiri boliviensis boliviensis (Bolivian squirrel monkey), Tetraodon nigroviridis (Pufferfish), Gopherus agassizii (desert tortoise), Cercopithecus aethiops (African green monkey), Macropus eugenii (tammar wallaby), Danio rerio (zebrafish)]. These sequences are orthologous to a 1.8 Mb segment of the human chromosome 7q31.3, which contains the gene that is mutated in cystic fibrosis.
We cluster the protein-coding genes from all species into 11 groups according to their functional annotation: CAPZA2 gene (group 1), WNT2 gene (group 2), ST7a gene (group 3), MET gene (group 4), GASZ gene (group 5), CAV1 gene (group 6), ST7b gene (group 7), CAV2 gene (group 8), TES gene (group 9), CORTBP2 gene (group 10) and CFTR gene (group 11). Each group comprises between 14 and 28 different species and is called the Green dataset. We define an additional set of groups by retaining only the eutherian species. This dataset is called the Green-e dataset. The resulting 11 groups contain between 14 and 23 different species.
Sequence alignment
We generate an mRNA alignment for each of the above groups in a multi-step process: 5′-UTRs, encoded amino acid sequences and 3′-UTRs are first separately aligned with CLUSTALW1.83 (37). The protein-coding mRNA is then mapped onto the amino acid alignment. Finally, 5′-UTR alignment, protein-coding alignment and 3′-UTR alignment are merged into the mRNA alignment. As the alignment of the protein-coding mRNA regions is entirely based on the alignment of encoded amino acid sequences, gaps come in multiples of three and, more importantly, mis-alignments on RNA level should be avoided.
Alignment randomization
The mRNA alignments are randomized in order to test whether any of the predicted secondary structure is likely to be real or whether it can be attributed to a pattern that has arisen by chance.
Each mRNA alignment is randomized in a multi-step process: the 5′-UTR, 3′-UTR and protein-coding parts of the alignment are randomized separately. The 5′-UTRs and 3′-UTRs parts are randomized by permuting all columns and by permuting all letters and gaps within each column. The protein-coding part of the alignment is randomized only by randomizing each triplet column that corresponds to a codon. This is done by first deriving the set of encoded amino acids that are encoded in the triplet column and then by replacing each non-gap codon randomly by any of the codons that correspond to these amino acids. The number of gap codons in each triplet column is kept fixed, but their position in the triplet column is randomized. For example, if the original triplet colum contains the codons CTG (26 times, amino acid L), TAT (1 time, amino acid Y), CAT (1 time, amino acid H) and - - - (1 time);
codon position 1, CCCCCCCCCTCCCCCCCCCCCC-TCCC
codon position 2, TTTTTTTTTATTTTTTTTTTTT-TTTA
codon position 3, GGGGGGGGGTGGGGGGGGGGGG-GGGT
amino acid LLLLLLLLLYLLLLLLLLLLLL-LLLH
the codons for the randomized version of the triplet column will be randomly chosen from the union of codon sets {CAC, CAT} (amino acid H), {TTG, CTA, CTC, CTG, CTT, TTA} (amino acid L) and {TAC, TAT} (amino acid Y), resulting, for example, in
codon position 1 CCCCTCC-CTCCCCCCTTTTTCTCTCT
codon position 2 TATAATT-AAATTAATATTAATTATAA
codon position 3 CTTCCGG-CTTGCCCGTAGTCAACGCT
amino acid LHLHYLL-HYHLLHHLYLLYYLLHLHY
This randomization procedure keeps the set of encoded amino acids unchanged, but changes the underlying codons and their relative proportion. In particular, if there are base pairs between the codon positions of two triplet columns, they are likely to be destroyed in the randomization procedure. This randomization procedure may alter the dinucleotide distribution in each sequence. However, as RNA-Decoder's predictions do not depend on the dinucleotide distribution of each sequence (as it does not predict the mfe structure), but rather on the evolutionary pattern in the alignment of sequences, the dinucleotide distribution within each sequence does not have to be conserved in the randomization process.
Structure prediction
RNA-Decoder (29,30) is used to detect evolutionarily conserved secondary structure in the mRNA alignments. RNA-Decoder is the first and so far only prediction program that comparatively predicts secondary structure by explicitly taking the known protein-coding context of the input alignment into account. This is an important feature, as other comparative secondary structure prediction programs, such as RNAalifold (38) and Pfold (39), have difficulty in detecting conserved secondary structure in regions with two different evolutionary constraints (30).
For this study, we use a modified version of RNA-Decoder that requires a minimum stem length of 3 rather than only 2 consecutive base-pairs, thereby lowering the amount of noise.
Each mRNA alignment is partitioned into 600 bp long intervals using a step size of 200 bp. RNA-Decoder is used to predict conserved secondary structure for each sub-alignment. These individual predictions are then merged into one prediction along the entire alignment.
Evaluating the correlation between secondary structure and codon sparseness
We use the codon usage tables of http://www.kazusa.or.jp/codon/ (Source: GenBank Release 146.0, March 28, 2005). These tables provide information for 29 of the 37 species in our dataset. We define the sparseness of a codon c as
where aa(c) is the amino acid encoded by codon c, n(a) is the number of codons that encode amino acid a and fa(c) is the relative frequency that amino acid a is encoded by codon c [i.e. ∑{c|aa(c)=a} fa(c) = 1]. The sparseness value of a codon measures the deviation of the measured codon frequency from the frequency that would be expected if each codon would be used with equal probability to encode its amino acid. The sparseness value of a codon c, s(c), is negative, if its frequency fa(c) is s(c) × 100% lower than the expected frequency, 1/n(aa(c)), and positive, if is larger. It is contained in [−1, n − 1]. For an example, a sparseness value of 0.2 for codon TTG [amino acid L, n(L) = 6], means that this codon is 20% more frequent than the expected average frequency of 1/n(L) = 1/6.
We measure the average sparseness of codons whose third codon position is predicted to be base-paired (c3-paired), in two different ways by taking all mRNA alignments into account:
where is the number of codons whose third position is predicted to be base-paired, and
that is, giving each codon c a weight according to its number of base-paired positions, nbp(c), and using .
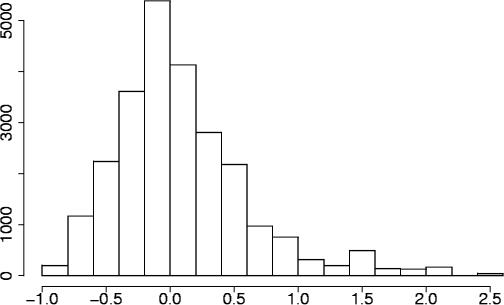
The distribution of sparseness values s(c3-paired) cannot be approximated by a normal distribution as is it not centred around zero and not symmetric (see Figure 4), and it can be shown that the expected value of this distribution is larger than zero. We, therefore, perform a randomization test to calculate P-values.
Figure 4.
Sparseness distribution. This figure shows the distribution of sparseness values for codons in the Green alignments whose third codon position is predicted to be base-paired.
We first compute the average sparseness for randomly drawn codons in two ways:
and
where and the codons ci are randomly drawn from the pool of all codons in all alignments.
We then calculate the average sparseness values for n = 10 000 sets of randomly drawn codons and compare each average sparseness value to the respective real sparseness value . If denotes the number of times that , the P-value pi = ni/n is the estimation of the probability that the average sparseness value for c3-paired codons is larger than the one for randomly chosen codons.
RESULTS
Conserved, local secondary structure can be found in the coding regions of eukaryotic mRNAs
We investigate the presence of evolutionarily conserved, local secondary structure in the mRNAs of 11 human protein-coding genes contained in a 1.8 Mb segment of the human chromosome 7q31.3, which contains the gene that is mutated in cystic fibrosis, see Materials and Methods for a detailed description of the dataset.
Each of the 11 genes is searched for conserved secondary structures by analysing two different mRNA alignments with RNA-Decoder, one alignment comprising only the orthologous mRNA sequences from eutherian species (Green-e alignment) and one alignment comprising all mRNA sequences (Green alignment). Every alignment is generated by separately aligning the 5′-UTRs, 3′-UTRs and protein-coding regions, the latter by basing the alignment of protein-coding mRNA sub-sequences on that of the encoded amino acid sequences, see Materials and Methods for a more detailed description. The mRNA alignments are typically too long to be analysed in one go by RNA-Decoder. We partition them into 600 bp wide segments and use a step size of 200 bp. RNA-Decoder takes each segment as fixed input alignment and predicts the most probable secondary structure elements that are supported by the evolutionary pattern and annotation of the alignment. The predictions for the individual segments are merged into one prediction along the entire mRNA alignment. Using this procedure, we can detect local, conserved secondary structure that is contained within at most 400 bp.
RNA-Decoder's prediction algorithm explicitly takes the known protein-coding regions of the input alignment into account, by detecting and disentangling potential dual evolutionarily constraints owing to overlapping amino acid and secondary structure information. Refer to Materials and Methods for a more detailed description of RNA-Decoder.
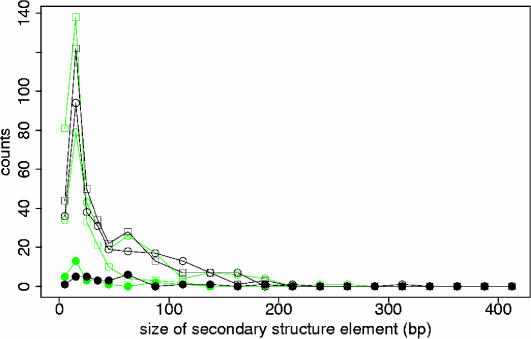
Local secondary structure is most abundant in 5′-UTRs and 3′-UTRs, but there also exist local secondary structure elements in the protein-coding regions of the mRNAs. Secondary structure elements cover on average 55% of the aligned 5′-UTRs ([13%, 96%]), 45% of the aligned coding regions ([18%, 68%]) and 68% of the aligned 3′-UTRs ([59%, 78%]). About one-third (33%) of the 5′-UTR columns are on average base-paired, as opposed to 27% of the coding and 40% of the 3′-UTR columns. Secondary structure elements in 5′-UTRs are on average longer (49 bp) than those in 3′-UTRs (40 bp) and coding regions of the alignment (33 bp). Even though the maximal length of secondary structure elements is limited to 400 bp owing to the fixed window size used to analyse each mRNA alignment, most secondary structure elements span <100 bp, confirming our initial expectation that evolutionarily conserved and potentially functional secondary structure should be local rather than global (see Figure 1).
Figure 1.
Length distributions of secondary structure elements in mRNA. This figure shows the length distributions of secondary structure elements in 3′-UTRs (open circles), 5′-UTRs (closed circles) and coding regions of the mRNA alignments (open squares), for the original alignments (in black) and for the randomized alignments (in green). The length of a secondary structure element is defined as the number of base pairs (bp) it bridges in the mRNA alignment. Note that the length of secondary structure elements is limited by 400 bp owing to the fixed window size used to analyse each mRNA alignment.
We investigate how the predicted secondary structure elements change if we randomize the mRNA alignments.
For the randomization, each mRNA alignment is partitioned into 5′-UTR, coding and 3′-UTR region, which are randomized separately. The 5′-UTR and 3′-UTR regions are each randomized by first permuting all columns and then by permuting all letters and gaps within each column, whereas the coding region is randomized by keeping the order of triplet columns fixed and by randomizing only within each triplet column. The latter is performed by replacing every codon in the triplet-column by a randomly picked codon from the set of all codons that are encoded by any of the amino acids encoded in that triplet-column, see Materials and Methods for a detailed description.
This procedure serves two purposes. First, it is meant to remove any codon bias, and, second, it is expected to destroy most of the base pairs between nucleotides of different codons.
When comparing the length distributions for the original and randomized mRNA alignments shown in Figure 1 we can see that the randomization introduces short, spurious secondary structure elements spanning <25 bp. In the coding regions, the excess of secondary structure elements spanning ∼50 bp essentially disappears, similarly for structures in the 5′-UTRs. For 3′-UTRs, the length distribution is also shifted towards smaller structures, decreasing the amount of structures spanning 90–140 bp and increasing those spanning 50–90 bp.
Secondary structure is suppressed in parts of the coding regions
As discussed above, the randomization procedure removes almost all of the secondary structure elements in the coding regions of the mRNA alignments, see for example the human ST7b mRNA sequence in Figure 2. However, few, mostly short novel secondary structure elements emerge in coding regions that were previously devoid of secondary structure. This can happen when the randomization procedure introduces so-far suppressed codons that can base pair with other randomly introduced and so far suppressed codons elsewhere (see Figure 3 for an example). In these cases, new base pairs typically emerge between codon positions 2–3 and 3–2. This is the most likely scenario as at least some of the newly introduced nucleotides in codon position 3 can pair with the nucleotides in codon position 2 that are more conserved and also less likely to be altered in the codon randomization procedure.
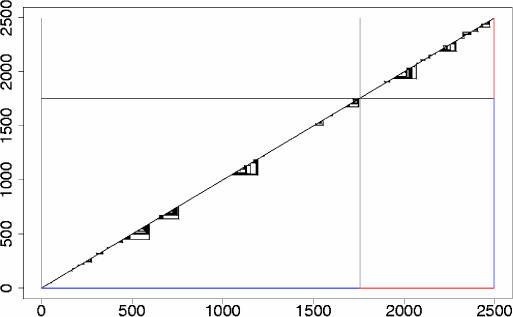
Figure 2.
Conserved secondary structure elements in the human ST7b mRNA, effects of alignment randomization. This figure shows the predicted evolutionarily conserved secondary structure elements for the human ST7b gene, once using the original mRNA alignment (lower right triangle) and once the randomized mRNA alignment (upper left triangle) as input to RNA-Decoder. The sequence positions of the mRNA are drawn along the x- and the y-axes, indicating the coding positions in blue and the 3′-UTR in red. Each predicted base pair corresponds to a triangle connecting the two base-paired positions. As can be seen, most of the predicted secondary structure elements disappear when the mRNA alignment is randomized. The horizontal and vertical lines indicate the boundary between the coding part of the mRNA and the 3′-UTR.
Figure 3.
Suppressed secondary structure in coding region. This figure shows part of the CAPZA2 alignment in the coding region, before (top) and after (bottom) randomization. The original alignment is devoid of secondary structure, whereas the randomized alignment contains a small, hairpin-like structure which mostly involves newly formed base pairs between codon positions 2 and 3. Newly formed base pairs are underlined in pink, altered base pairs in green and broken base pairs in yellow. Real and potential base-pairing partners are shown in upper case letters. The consensus base pairs for RNA molecules, i.e. {G−C, A−U, G−U}, correspond to {G−C, A−T, G−T} on DNA level, as depicted here. See Results for a detailed discussion.
Repeats often overlap secondary structure elements
The mRNA sequences of the human, house mouse, Norway rat, chicken, cow, pig, cat, dog, fugu fish and zebrafish were searched for repeats using the Repeatmasker Web server (40) (http://www.repeatmasker.org/) in a species-specific way. For most alignments, the repeats are almost exclusively found in the UTRs [CAPZA2, ST7a, MET, GASZ, CAV1, CAV2, CFTR (100% of repeat columns in UTRs) and ST7b (95.08%)]. However, for WNT2 (62.50% of repeat columns in UTRs), TES (78.85%) and especially CARTBP2 (35.87%), a sizeable fraction of the repeats overlaps the coding columns of the alignment. In UTRs, repeats often overlap secondary structure elements [from 34.15% (TES) to 100.00% (CARTBP2), average across all 11 genes is 58.69%]. In coding regions, this strongly depends on the individual gene [0% (WNT2, TES), 44.07% (CARTBP2) and 100.00% (ST7b)]. For about half of the columns that contain a repeat (54.41%), only one species contains the repeat. However, 11.29% contain repeats in two species and the remaining 34.30% contain repeats in three species or more, i.e. the position of repeats with respect to the gene structure is often evolutionarily conserved (note that some of the species could not be searched for repeats).
There is only a single Alu repeat in the 5′-UTR of the human CAV2 gene, whereas Alu repeats are abundant on pre-mRNA level, covering between 1 and 8% of the 11 human pre-mRNA sequences. It is known that pairs of inverted Alu repeats can be involved in A-to-I RNA editing on pre-mRNA level by forming hairpin-like secondary structures that are bound by the editing protein ADAR (41,42). Almost all Alu repeats are contained in introns in the coding regions or UTRs and are thus removed during splicing.
Secondary structure strongly correlates with sparse codons
The encoding of secondary structure in the protein-coding regions of an RNA is facilitated by the degeneracy of the genetic code. Codons that encode the same amino acid usually differ in their third codon position. If the third codon position of a codon is base-paired, it not only has to encode a certain amino acid, but also has to be able to form the desired base pair. These codons should thus show a biased distribution with respect to the codon frequencies observed in large datasets of protein-coding RNA.
To investigate this hypothesis, we calculate the sparseness for each codon whose third codon position is predicted to be base-paired and derive the average sparseness, see Materials and Methods. We compare this average sparseness to the average sparseness values that we obtain for 10 000 sets of randomly drawn codons, and derive the P-values for our observations. As shown in Table 1, the observed average sparseness for codons whose third position is predicted to be base-paired is significantly lower than that for randomly chosen codons, thus confirming our initial hypothesis. The measured sparseness values are ∼3 standard deviations away from the average sparseness values of the randomized sets.
Table 1.
Real and random sparseness averages
| i | pi | ||
|---|---|---|---|
| Green data set | |||
| 1 | 0.10100 | 0.11488 ± 0.002921 | <0.001 |
| 2 | 0.10650 | 0.11568 ± 0.003011 | 0.001 |
| Green-e data set | |||
| 1 | 0.05261 | 0.06602 ± 0.003076 | <0.001 |
| 2 | 0.05421 | 0.06345 ± 0.003257 | 0.003 |
This table shows the average values for the real sparseness, , as well as the mean values of the average random sparseness, , for both evaluation procedures i ∈ {1, 2}, see Materials and Methods for the detailed definitions. The pi-values estimate the probability that the average sparseness value is larger than the one for randomly chosen codons.
Local secondary structures can hinder access to the nominal start codon
For some genes, local secondary structure is bridging the 5′-UTR and the coding region that may affect access to the nominal start codon.
In the CAV1 gene, the nominal start codon is contained in the loop region of a hairpin-like secondary structure element (see Figure 5). The next ATG downstream of the nominal start codon is an in-frame codon at amino acid position 32, which is perfectly conserved in all species and is contained in a region that is devoid of secondary structure. The mouse CAV1 mRNA is known to yield two protein isoforms, alpha- and beta-caveolin 1, that derive from the use of two alternate start sites at amino acid position 1 and position 32, and that generate at least two distinct subpopulations of caveolae which may be differentially regulated by a specific caveolin-associated serine kinase (43). The short beta-caveolin 1 variant has also been observed in rat (GeneID: 25404; Refseq mRNA id: NM_133651; Refseq protein id: NP_598412) (44).
Figure 5.
Secondary structure in CAV1. This figure shows part of the CAV1 alignment around the nominal start codon (orange column on the left). Columns contained in secondary structure elements are shown in yellow, 5′-UTR columns are underlined in blue and columns in the coding region in green. The alternate start codon (orange columns on the right) that has been shown to give rise to the shorter protein variant (beta-caveolin 1) in mouse and rat (43,44) is perfectly conserved in all species and is contained in a region devoid of any conserved secondary structure. The last line in the alignment details the predicted secondary structure elements in bracket notation. The last but one line of the alignment contains the annotation for each column of the alignment (5, 5′-UTR; 1, first codon position; 2, second codon position; and 3, third codon position).
The WNT2 gene is another example where secondary structure overlaps the nominal start codon. The nominal start codon is located in a loop region of an extended secondary structure bridging 110 bp in the human mRNA, making an ATG 29 amino acids downstream the next in-frame start codon (the next out-of-frame start codon 75 bp downstream corresponds to an ORF of only 16 amino acids). The nominal start codons of the CAV2 and CORTBP2 genes are contained in the base-paired region of local secondary structure bridging 46 bp (human CAV2) and 65 bp (human CORTPB2). Similarly, the nominal start codons of the human CAPZA2, GASZ and CFTR genes are contained in smaller hairpin-like structures. It remains to tested in experiments whether or not these secondary structures play a functional role in gene expression.
Statistical evidence that secondary structure is required for the correct pre-mRNA splicing of the human CFTR gene
Nonsense, missense and even synonymous mutations within the exons of the human CFTR (cystic fibrosis transmembrane regulator) gene can induce skipping of the mutant exon during pre-mRNA splicing, which may lead to a non-functional protein product (45–47). The residual level of the normal splicing variant determines whether mutations result in severe cystic fibrosis or less severe phenotypes, such as male sterility. Pagani et al. (35) extensively evaluated the contribution of synonymous exonic mutations on the splicing efficiency of CFTR exon 12. They find that 25% of synonymous sites in that exon do not evolve freely because they are constrained by requirements which ensure correct splicing. However, Pagani et al. do not determine the nature of the constraints required for correct splicing or the mechanisms that lead to exon skipping.
Pre-mRNA splicing is a complex process that involves a variety of carefully orchestrated reactions. There exists experimental evidence that some pre-mRNA sequences of S.cerevisiae form secondary structure elements which are capable of promoting splicing (48), but there is no comparable evidence for human systems.
We analysed the wild-type and all mutated versions of the human CFTR exon 12 and its neighbouring introns with RNA-Decoder, in order to investigate whether secondary structure plays a role in determining if the exon can be correctly spliced.
For every wild-type and mutated version of the human CFTR exon 12, we prepared on input alignment for RNA-Decoder, resulting in 30 alignments. Each alignment has a length of 687 bp, contains the sequences of 19 species (including the human one) and comprises exon 12 as well as a stretch of 300 bp intron alignment on either side of the exon. The mutated alignments were generated by manually altering every sequence in the alignment in the same way as the human sequence. Each alignment was then analysed with RNA-Decoder in one go, i.e. without partitioning the alignment into shorter fragments.
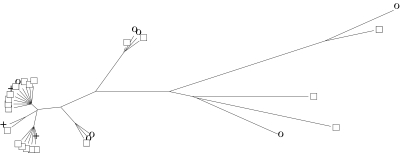
The 30 resulting predicted structures (one for each alignment) were projected onto the human sequence. These human structures were then clustered in a two-step process. We first determined the pairwise distances between the secondary structures using RNAforester (49) and then calculated a neighbour-joining tree (50), (see Figure 6).
Figure 6.
Clustering of secondary structures for the wild-type and mutated versions of CFTR exon 12. This neighbour-joining tree shows the clustering of predicted secondary structures for the wild-type and 29 mutated versions of CFTR exon 12. The distances between the secondary structures were calculated using RNAforester. Open squares denote secondary structures with a splicing efficiency over 80%, crosses those with an efficiency between 20 and 80% and circles those with an efficiency of <20%.
We ordered the predicted secondary structures according to their distance from the wild-type structure. The distribution of distances does not follow a normal distribution (data not shown). We, therefore, performed a one-tailed Mann–Whitney test in order to test whether secondary structures of sequences with low-splicing efficiencies are significantly different from the wild-type secondary structure. Since it is a priori not clear where the threshold between ‘high’ and ‘low’ splicing efficiencies should be placed, we performed two tests. In the first case, we use a threshold value of 20%. This results in two clusters, one containing 23 members with high efficiencies and the other containing 7 members with low efficiencies. We calculate an U-value of 117.5, which is marginally significant with a P-value of 0.035. In the second case, we cluster structures with an efficiency higher than 70% into one group having 20 members and those with an efficiency lower than 30% into another group having 8 members. This clustering yields an U-value of 103 which is not significant with a P-value of 0.129.
These results provide some explanation of the experimentally determined splicing efficiencies. Our main finding is that secondary structures having high-splicing efficiencies are marginally different from those having low-splicing efficiencies.
As the ‘mutated’ alignments were generated by replacing an entire column in the alignment with the nucleotide of the corresponding mutated human sequence, we have introduced some artificial conservation in the alignment which may decrease the predictive power of RNA-Decoder. The above results may thus have a lower than necessary statistical significance.
The predicted secondary structure for the wild-type sequence (see Figure 7) brings the two splice sites of the exon into close spatial proximity, which may aid the recognition of the splice sites by the spliceosome.
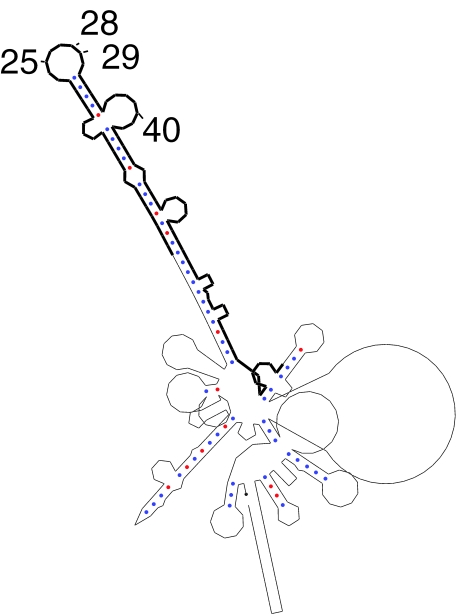
Figure 7.
Predicted, conserved structure for the wild-type version of CFTR exon 12. This is the predicted, conserved secondary structure for the wild-type version of CFTR exon 12. The sequence of exon 12 is highlighted with a thick black line. The exon positions indicated in the plot correspond to the same numbering scheme used in Pagani et al., starting with 1 at the most 5′ position of exon 12.
Two secondary structures with low-splicing efficiencies (25_A and 40_C) are the same as the wild-type structure. The two corresponding mutations both overlap a region of the pre-mRNA which is predicted to be single-stranded and which may be bound by a protein in a sequence-specific way. This suggests that some nucleotides in the exon may be as important for correct splicing as the formation of the correct secondary structure. The experimental finding that exon positions 28 and 29 play an important role in determining the splicing efficiencies (35) supports this hypothesis, as these positions are contained in the same predicted hairpin loop as position 25.
DISCUSSION
We predict and study, for the first time, evolutionarily conserved, local RNA secondary structure in the mRNA sequences of 11 protein-coding human genes. This is done by analysing mRNA alignments comprising overall 37 vertebra species in a comparative way using RNA-Decoder. This program predicts evolutionarily conserved global or local secondary structures by analysing the conservation pattern in the input alignment and by taking the known protein-coding regions of the input alignment explicitly into account. The latter feature has been shown to be essential (30) for reliably detecting secondary structure in coding regions, i.e. in regions of the alignment where two different evolutionarily constraints play a role.
We find well-defined, conserved RNA secondary structure elements in the coding regions of human mRNA sequences and investigate the significance of these structures by randomizing the mRNA alignments. This randomization destroys almost all of the secondary structures, confirming the significance of the predicted structures in the original alignments. The randomization also gives rise to few new, predominantly short secondary structure elements in part of the coding regions, where the choice of codon in the original alignments prevents the formation of secondary structures. This implies that parts of the coding regions are codon-biased in order to prevent secondary structure formation. We investigate the codons that are engaged in secondary structure formation and find that they significantly correlate with rare codons and that often overlap repetitive elements. For one of the eleven genes that we analyse, CAV1, the same mRNA is known to give rise to two protein variants in the mouse and rat (43,44), a long protein (alpha-caveolin 1) when the nominal start codon is used and short protein (beta-caveolin 1) when an alternative start codon at amino acid position 32 is used. These experimental findings are in line with our theoretical prediction that a conserved secondary structure element overlaps the nominal start codon, hindering access to the nominal start codon and making the next start codon downstream (which does not overlap secondary structure) the functional start codon. Thus, our predictions not only complement the experimental work, but also propose a potential mechanism for the regulation of gene expression.
Previous studies of mRNA sequences have so far only estimated the potential of mRNA sequences to form global or local structures, but have not studied the secondary structures themselves. These studies are entirely based on the predicted free energies of global (26,27) or local (28) mfe structures and rely on a number of assumptions. First and foremost, they assume that the mfe structure is the secondary structure that is realized in the cell. This is very unlikely to be true as the mRNA molecule (i) does not have time to reach the thermodynamic equilibrium and thus to assume the mfe structure (51); (ii) is likely to be bound by the ribosome, proteins or other RNA molecules (52) that may alter the mfe structure; (iii) does not assume one single structure with probability one (53); and (iv) can furthermore be influenced by the subtle effects of co-transcriptional secondary structure formation that not only affect the transcripts of RNA genes (54,55), but may also have an effect on the transcripts of eukaryotic, protein-coding genes. Second, the deviation between the mfe of the original sequence and a randomized version of that sequence is assumed to be a good indicator for the potential to form secondary structure. However, it is known (34) that the quality of the mfe structure prediction decreases with increasing sequence length. We, thus, have to expect a rather poor performance when investigating the global mfe structures of mRNA sequences. It is also known that the deviation between the mfe of the original sequence and a randomized version of that sequence is an unreliable indicator for local secondary structure in protein-coding regions (30). Third and last, the studies base their conclusions on properties that are directly derived from the predicted mfe structures (such as the free energy), but—on the other hand—do not study the structures themselves.
As we are currently not capable of simulating the numerous and complex structure defining and altering effects that an mRNA sequence encounters in the living cell, we only aim to identify secondary structure elements that have been conserved during evolution and that are therefore likely to play a functional role. We, therefore, employ a comparative method that analyses a given input alignment of related mRNA sequences. As we base this alignment on the known encoded amino acid sequences, we expect the alignment to correspond to the correct, structural alignment, at least in the coding regions. The method that we employ, RNA-Decoder, is implemented as a stochastic context free grammar that depends on probabilities rather than thermodynamic parameters such as free energies. It predicts secondary structures based on the pattern of conservation in the alignment and the known protein-coding context of each column in the alignment. It does not force each input alignment to assume a global secondary structure, but is also capable of modelling secondary structure elements that are separated by non-structural regions. One considerable disadvantage of our approach is that it requires a sufficiently large and evolutionarily diverse set of related sequences.
Repeat sequences are rarely observed in coding sequences as their insertion can cause frame shifts, give rise to in-frame stop codons or simply alter the encoded amino acids in a way which leads to a mal-functional protein. The strong correlation that we observe between secondary structure elements and repeat elements in coding regions may imply that repeat elements can be tolerated in coding regions if they serve a role in the formation of functional secondary structure.
Secondary structure elements in mRNAs can play a number of functional roles, in addition to the ones that we described in Introduction, for example, in regulating translation speed. It is already well known that the speed of translation is regulated by codon usage: frequent codons are used to encode alpha helices, which form quickly, whereas rare codons are frequently used to encode beta sheets and coils, whose proper formation needs more time (56–58). Artificial engineering of synonymous codons can significantly increase the expression level of protein products (59). Codon usage may also play a role in signal peptide recognition (60). The speed of translation depends not only on the concentration of tRNA molecules and amino acids, but also on the secondary structure of the mRNA sequences. Based on basic chemical reaction theory, the half-time of local secondary structures in coding region depends on the concentration of tRNA sequences, and the most efficient regulation can be achieved by the use of rare codons. This is one possible explanation for our observation that rare codons are preferred in local mRNA structures.
We complement our results on mRNA level by an investigation of the human CFTR gene on pre-mRNA level in order to determine whether secondary structure elements are required for correct pre-mRNA splicing. We study the wild-type version of the human CFTR gene as well as 29 variants with synonymous mutations in exon 12. We compare our predicted secondary structures to the experimentally determined splicing efficiencies and find that pre-mRNAs with high-splicing efficiencies have different predicted secondary structures than pre-mRNAs with low-splicing efficiencies. This result is marginally significant with a P-value of 0.035. We also identify several nucleotide positions in the exon that fall into a predicted hairpin loop and which may need to be bound in a sequence-specific way in order to ensure correct splicing.
Overall, this constitutes the first, albeit weak, statistical evidence that exon mutations can alter secondary structure elements around splice sites and that secondary structure elements are required for the correct splicing of the human CFTR gene.
Our aim was to show that theoretical investigations of evolutionarily conserved secondary structure elements can enhance our understanding of gene regulation on mRNA and pre-mRNA level and we hope to inspire future collaborations between experimentalists and theoreticians. All predicted secondary structures can be downloaded from www.ebi.ac.uk/~meyer/RNA_regulation.
Acknowledgments
We thank János Podani for discussing the randomization test, Jakob Pedersen for help with adjusting the minimum required stem length and Nick Goldman for supporting our research. I.M. is supported by a Békésy György postdoctoral fellowship. Funding to pay the Open Access publication charges for this article was provided by Nick Goldman, EBI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ernst H., Shatkin A.J. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc. Natl Acad. Sci. USA. 1985;82:48–52. doi: 10.1073/pnas.82.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merino E., Balbas P., Puente J.L., Bolivar F. Antisense overlapping open reading frames in genes from bacteria to humans. Nucleic Acids Res. 1994;22:1903–1908. doi: 10.1093/nar/22.10.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klemke M., Kehlenbach R.H., Huttner W.B. Two overlapping reading frames in a single exon encode interacting proteins—a novel way of gene usage. EMBO J. 2001;20:3849–3860. doi: 10.1093/emboj/20.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak M. Extensively overlapping reading frames in a second mammalian gene. EMBO Rep. 2001;2:768–769. doi: 10.1093/embo-reports/kve183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nature Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 6.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 7.Gray N.K., Wickens M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 8.Meijer H.A., Thomas A.A. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 2002;367:1–11. doi: 10.1042/BJ20011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkie G.S., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR binding factors. Trends Biochem. Sci. 2002;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 10.Diwa A., Bricker A.L., Jain C., Belasco J.G. An evolutionarily conserved RNA stem–loop functions as a sensor that directs feedback regulation of RNase E gene expression. Gene Dev. 2000;14:1249–1260. [PMC free article] [PubMed] [Google Scholar]
- 11.Binder R., Horowitz J.A., Basilion J.P., Koeller D.M., Klausner R.D., Harford J.B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decker C.J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Gene Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 13.Theil E.C., Eisenstein R.S. Combinatorial mRNA regulation: iron regulatory proteins and iso-iron-responsive elements (Iso-IREs) J. Biol. Chem. 2000;275:40659–40662. doi: 10.1074/jbc.R000019200. [DOI] [PubMed] [Google Scholar]
- 14.Sudarsan N., Barrick J.E., Breaker R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 16.Olivier C., Poirier G., Gendron P., Boisgontier A., Major F., Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol. Cell. Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snee M.J., Arn E.A., Bullock S.L., Macdonald P.M. Recognition of the bcd mRNA localization signal in Drosophila embryos and ovaries. Mol. Cell. Biol. 2005;25:1501–1510. doi: 10.1128/MCB.25.4.1501-1510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuplin A., Evans D.J., Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 19.You S., Stump D.D., Branch A.D., Rice C.M. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds J.E., Kaminski A., Carroll A.R., Clarke B.E., Rowlands D.J., Jackson R.J. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 21.Kolykhalov A.A., Feinstone S.M., Rice C.M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi M., Lemon S.M. 3′ Nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 2003;77:3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y.K., Lee S.H., Kim C.S., Seol S.K., Jang S.K. Long-range RNA–RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perdue M.L., Garcia M., Senne D., Fraire M. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997;49:173–186. doi: 10.1016/s0168-1702(97)01468-8. [DOI] [PubMed] [Google Scholar]
- 25.Perdue M.L., Suarez D.L. Structural features of the avian influenza virus hemagglutinin that influence virulence. Vet. Microbiol. 2000;74:77–86. doi: 10.1016/s0378-1135(00)00168-1. [DOI] [PubMed] [Google Scholar]
- 26.Steffens W., Digby D. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 1999;27:1578–1584. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Workman C., Krogh A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res. 1999;27:4816–4822. doi: 10.1093/nar/27.24.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz L., Burge C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen J.S., Forsberg R., Meyer I.M., Hein J. An evolutionary model for protein-coding regions with conserved RNA structure. Mol. Biol. Evol. 2004;21:1913–1922. doi: 10.1093/molbev/msh199. [DOI] [PubMed] [Google Scholar]
- 30.Pedersen J.S., Meyer I.M., Forsberg R., Simmonds P., Hein J. A comparative method for finding and folding RNA secondary structures in protein-coding regions. Nucleic Acids Res. 2004;32:4925–4936. doi: 10.1093/nar/gkh839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M. Mfold webserver for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer S., Tacker M., Schuster P. Fast folding and comparison of RNA secondary strutures. Monatsh. Chem. (Chemical Monthly) 1994;125:167–188. [Google Scholar]
- 34.Morgan S.R., Higgs P.G. Evidence for kinetic effects in the folding of large RNA molecules. J. Chem. Phys. 1996;105:7152–7157. [Google Scholar]
- 35.Pagani F., Raponi M., Baralle F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofacker I.L., Fekete M., Stadler P.F. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen B., Hein J. Pfold: RNA secondary structure prediction using stochastic context-free grammars. Nucleic Acids Res. 2003;31:3423–3428. doi: 10.1093/nar/gkg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smit A.F.A., Hubley R., Green P. 2005. Repeatmasker open-3.1.0—Webserver.
- 41.Blow M., Futreal P.A., Wooster R., Stratton M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athanasiadis A., Rich A., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLOS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scherer P.E., Tang Z., Chun M., Sargiacomo M., Lodish H.F., Lisanti M.P. Caveolin isoforms differ in their N-terminal protein sequence and subcellular distribution. Identification and epitope mapping of an isoform-specific monoclonal antibody probe. J. Biol. Chem. 1995;270:16395–16401. doi: 10.1074/jbc.270.27.16395. [DOI] [PubMed] [Google Scholar]
- 44.Ratajczak P., Oliviero P., Marotte F., Kolar F., Ostadal B., Samuel J.L. Expression and localisation of caveolins during postnatal development in rat heart. Implication of thyroid hormone. J. Appl. Physiol. 2005;99:244–251. doi: 10.1152/japplphysiol.01292.2004. [DOI] [PubMed] [Google Scholar]
- 45.Pagani F., Buratti E., Stuani C., Baralle F.E. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580–26588. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 46.Pagani F., Stuani C., Tzetis M., Kanavakis E., Efthymiadou A., Doudounakis S., Casals T., Baralle F.E. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 47.Aznarez I., Chan E.M., Zielenski J., Blencowe B.J., Tsui L.C. Characterization of disease-associated mutations affecting an exonic splicing enhancer and two cryptic splice sites in exon 13 of the cystic fibrosis transmembrane conductance regulator gene. Hum. Mol. Genet. 2003;12:2031–2040. doi: 10.1093/hmg/ddg215. [DOI] [PubMed] [Google Scholar]
- 48.Buratti E., Baralle F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Höchsmann M., Töller T., Giegerich R., Kurtz S. Proc. IEEE Comput. Syst. Bioinform. Conf. 2003. Local similarity in RNA secondary structures; pp. 159–168. [PubMed] [Google Scholar]
- 50.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Wickiser J.K., Winkler W.C., Breaker R.R., Crothers D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 52.Neugebauer K.M. On the importance of being co-transcriptional. J. Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 53.Miklós I., Meyer I.M., Nagy B. Moments of the Boltzmann distribution for RNA secondary structures. B. Math. Biol. 2005;67:1031–1047. doi: 10.1016/j.bulm.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Heilman-Miller S.L., Woodson S.A. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–733. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyer I.M., Miklós I. Co-transcriptional folding is encoded within RNA genes. BMC Mol. Biol. 2004;5:e10. doi: 10.1186/1471-2199-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thanaraj T.A., Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thanaraj T.A., Argos P. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 1996;5:1973–1983. doi: 10.1002/pro.5560051003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makhoul C.H., Trifonov E.N. Distribution of rare triplets along mRNA and their relation to protein folding. J. Biomol. Struct. Dyn. 2002;20:413–420. doi: 10.1080/07391102.2002.10506859. [DOI] [PubMed] [Google Scholar]
- 59.Kang T.J., Loc N.H., Jang M.O., Yang M.S. Modification of the cholera toxin B subunit coding sequence to enhance expression in plants. Mol. Breeding. 2004;13:143–153. [Google Scholar]
- 60.Power P.M., Jones R.A., Beacham I.R., Bucholtz C., Jennings M.P. Whole genome analysis reveals a high incidence of non-optimal codons in secretory signal sequences of Escherichia coli. Biochem. Biophys. Res. Commun. 2004;322:1038–1044. doi: 10.1016/j.bbrc.2004.08.022. [DOI] [PubMed] [Google Scholar]