Abstract
The Shigella bacteria cause bacillary dysentery, which remains a significant threat to public health. The genus status and species classification appear no longer valid, as compelling evidence indicates that Shigella, as well as enteroinvasive Escherichia coli, are derived from multiple origins of E.coli and form a single pathovar. Nevertheless, Shigella dysenteriae serotype 1 causes deadly epidemics but Shigella boydii is restricted to the Indian subcontinent, while Shigella flexneri and Shigella sonnei are prevalent in developing and developed countries respectively. To begin to explain these distinctive epidemiological and pathological features at the genome level, we have carried out comparative genomics on four representative strains. Each of the Shigella genomes includes a virulence plasmid that encodes conserved primary virulence determinants. The Shigella chromosomes share most of their genes with that of E.coli K12 strain MG1655, but each has over 200 pseudogenes, 300∼700 copies of insertion sequence (IS) elements, and numerous deletions, insertions, translocations and inversions. There is extensive diversity of putative virulence genes, mostly acquired via bacteriophage-mediated lateral gene transfer. Hence, via convergent evolution involving gain and loss of functions, through bacteriophage-mediated gene acquisition, IS-mediated DNA rearrangements and formation of pseudogenes, the Shigella spp. became highly specific human pathogens with variable epidemiological and pathological features.
INTRODUCTION
Shigella is a group of Gram-negative, facultative intracellular pathogens. Recognized as the etiologic agents of bacillary dysentery or shigellosis in the 1890s, Shigella was adopted as a genus in the 1950s and subgrouped into four species: Shigella dysenteriae, Shigella flexneri, Shigella boydii and Shigella sonnei (also designated as serogroups A to D) (1). The bacteria are primarily transmitted through the faecal-oral route and therefore continue to threaten public health mainly in developing countries where sanitation is poor. The estimated annual number of episodes of shigellosis is 160 million, with 1.1 million deaths, mostly children under 5 years old in developing countries (2). Owing to the emerging multiple resistance strains that have compromised antibiotic treatment, development of effective novel vaccination strategies is urgently required (3).
There are very few biochemical properties that can distinguish Shigella from enteroinvasive Escherichia coli (EIEC), which are also a major cause of dysentery. Indeed, some O-antigens associated with EIEC are identical to those found in Shigella spp. (4), and many plasmid-associated virulence determinants are common to both EIEC and Shigella (5).
The virulence plasmid encodes the ∼30 kb Mxi-Spa type III secretion system (TTSS) and invasion plasmid antigens (Ipa proteins) required for invasion of the colonic and rectal epithelial cells, and the release of the bacteria into the host cell cytosol. IpaB in particular induces apoptosis in host macrophages and dentritic cells, leading to inflammatory infection (6). The virulence plasmid also encodes a surface protein, IcsA, responsible for actin-based motility required for intra- and inter-cellular spread of the bacteria (7). In addition to the plasmid, many chromosomal genes, such as those encoded by the Shigella pathogenicity island (SHI)-1 and SHI-2, also contribute to virulence (8–10). For a better understanding of the genetic basis of Shigella pathogenicity, we and others previously sequenced the genome of Shigella flexneri 2a, the most prevalent of the Shigella species (11,12). The genome of S.flexneri includes a virulence plasmid and shares a large proportion of chromosomal genes with closely related non-pathogenic and enterohemorrhagic E.coli (EHEC) strains. This is in good agreement with a study based on multilocus sequencing (MLS) that places Shigella within E.coli (13). However, in the S.flexneri chromosome, there are hundreds of pseudogenes, numerous lateral acquired S.flexneri-specific sequences, as well as insertion sequence (IS)-mediated deletions, translocations and inversions, which extensively reshaped the genome presumably for the benefit of a fuller expression of virulence.
Despite the availability of the S.flexneri genome sequence, questions remain about the distinctive epidemiological and pathological features that Shigella species/strains exhibit. While S.flexneri (six serotypes) is primarily endemic in developing countries, S.sonnei (1 serotype) is largely associated with episodes in industrialized nations. S.boydii (18 serotypes) is mainly endemic to the Indian subcontinent. Shigella dysenteriae serotype 1, which possesses the cytotoxic Shiga toxin (Stx), causes deadly epidemics in many of the poorer countries (6). For understanding of these viable features, we have determined the complete nucleotide sequences for the genomes of S.dysenteriae serotype 1 (strain 197), S.boydii serotype 4 (strain 227) and S.sonnei (strain 046) for comparison with the previously reported S.flexneri serotype 2a (strain 301) genome. Our study has revealed extensive diversity among the Shigella genomes, forming the genetic basis to explain the species/strain specific epidemiological and pathological features. Furthermore, many of the putative novel virulence genes identified may offer possible targets for the development of new treatment and prevention strategies.
MATERIALS AND METHODS
Shigella strains
S.dysenteriae serotype 1 (strain 197), S.boydii serotype 4 (strain 227) and S.sonnei (strain 046) were subjected to complete genome sequencing and are abbreviated to Sd197, Sb227 and Ss046, respectively. All strains were isolated from epidemics in China during the 1950s and were kindly provided by The Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine.
Sequencing and analysis
The whole genome sequence shotgun libraries for all strains were established as described previously (11), and ABI3730 automated sequencers were used for sequence collection. For each genome, we generated over 48 000 paired-end shotgun reads with estimated 8- to 9-fold coverage. The initial genome assembly was processed by phred/phrap program with the Q20 criteria (14). As there were large numbers of IS-elements present in each of the genomes, to avoid mis-assembly contigs obtained by phrap were split at each dubious IS locus and their relationships were rebuilt manually based on paired-end reads location information using Consed (15). Approximately 4500–6000 sequencing reads were generated for primer-walking of large clones or for PCR amplicons during the finishing phase for each of the genomes. To verify the final assembly, we designed overlapping primer pairs covering the whole genome sequence using genomic DNA as template for PCR amplifications. The genome annotations were performed as described previously (11), and GenomeComp was used for genomic comparison with default parameters (16). Each pairwise comparison figure used in Figure 2 was exported from GenomeComp with a 1000 bp filter setting along with the scale setting of 3000 and 300 for chromosomes and virulence plasmids, respectively. The KEGG database was used for the metabolic pathways analysis (17).
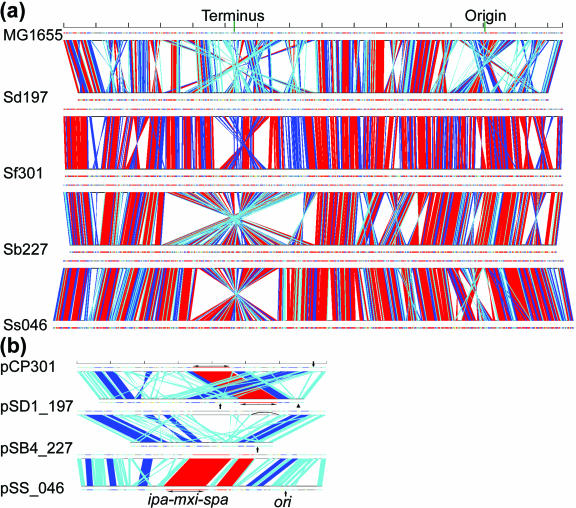
Figure 2.
Comparison of the Shigella chromosomes (a) and the virulence plasmids (b) (to scale). The chromosomes are compared to that from the E.coli K12 strain MG1655 (top). The virulence plasmids comparisons are made with pCP301 from Sf301 (always on the top). Each marker length denotes 300 and 30 kb for chromosome and plasmid comparisons, respectively. Colour code donates maximal length of the paired segments: red, >10 kb; blue, 5–10 kb; cyan, 1–5 kb. The replication origin, ori, is indicated by an arrow for each plasmid, and the cell-entry regions are marked with horizontal double-arrowhead lines. The arrowhead indicates the locus of the truncated ori sequence in Sd197, and the arched line indicates the corresponding region in pCP301 that is deleted from pSB4_227 nearby the cell-entry region (see main text).
Data accessibility
Complete genome sequences have been deposited in the GenBank. The accession numbers for chromosomes and virulence plasmids are Sd197, CP000034 and CP000035; Sb227, CP000036 and CP000037; Ss046, CP000038 and CP000039. In addition, genome annotation and comparative analysis can be obtained at ShiBASE (http://www.mgc.ac.cn/ShiBASE/) (18).
RESULTS
General features of the Shigella genomes
In common with the reported S.flexneri strain Sf301 and 2457T, the genomes of the newly sequenced Shigella strains all contain a virulence plasmid and a single chromosome (Table 1 and Figure 1). Note that, we included data of both Sf301 and 2457T genomes in Tables 1–5 for a complete comparison. However, since variations between the 2457T and the Sf301genomes are minute (12), we use Sf301 genome only for comparison with newly sequenced genomes to avoid redundant descriptions. All the virulence plasmids have nearly identical (R100-like) replication origins and maintenance genes, including repA, copA and copB for replication, and the parA and parB genes for partitioning. The plasmid from S.flexneri is also known to have post-segregation killing systems ccdA/ccdB and mvpA/mvpT (19,20). While the ccdA/ccdB is absent from pSS_046 in Ss046 only, the mvpA/mvpT is intact in all the virulence plasmids. The ∼30 kb cell-entry region, encoding the Mxi-Spa TTSS and Ipa proteins, is generally conserved in virulence plasmids pCP301, pSD1_197 and pSS_046. But, there is some polymorphism, with ipaD showing the most with 41 polymorphic sites (4.1% of the coding sequence). However, a pairwise analysis on the ipaD coding sequences showed that only in the case of pCP301 and pSS_046 is the synonymous to non-synonymous substitution ratio (Ks/Ka) smaller than 1 (0.71). The Ks/Ka rations are 1.85 and 1.73 for pCP301 and pSD1_197, and pSS_046 and pSD1_197, respectively. In fact, the Ks/Ka ratios for the majority of genes in the cell-entry regions are greater than 1 (data not shown), suggesting that the selection pressure from the host has maintained the conservation of these coding sequences. This is in contrast to the notion that many plasmid genes that encode secreted effector proteins (e.g. ospB, ospC2, ospD2, ospF and mxiL) outside of the cell-entry region are under pressure for non-synonymous substitution (21). The cell-entry region is bracketed by IS100 and IS600 in all the virulence plasmids, suggesting the transmission of a common ancestral form of the virulence plasmid to all Shigella, or alternatively, the cell-entry region was transmitted to all the virulence plasmids from related sources. The cell-entry region and the icsA gene are, however, deleted from pSB4_227 in strain Sb227. Additionally, pSB4_227 lacks a segment of ∼30 kb corresponding to the region between icsA and ccdA in pCP301 (Figure 2b). Since Sb227 was positive in the Sereny test when isolated and ipaB was detected previously by PCR (China CDC record), loss of the cell-entry region and icsA has probably occurred during long-term storage. Thus, caution needs being taken for interpreting gene decay via deletions in the Shigella genomes as the presence of the huge numbers of IS-elements can riddle the genomes considerably during storage.
Table 1.
General features of the Shigella genomes compared with the genome of E.coli K12 MG1655
| Chromosome | MG1655a | Sd197 | Sf301b | 2457Tc | Sb227 | Ss046 |
|---|---|---|---|---|---|---|
| Total length (bp) | 4 639 675 | 4 369 232 | 4 607 203 | 4 599 354 | 4 519 823 | 4 825 265 |
| No. of total ORFs | 4254 | 4557 | 4434 | 4456 | 4353 | 4434 |
| No. of pseudogenes | 12 | 285 | 254 | 372 | 217 | 210 |
| Percentage of CDS (%) | 87.3 | 77.2 | 80.4 | 77.2 | 80.5 | 80.5 |
| G+C content (%) | 50.79 | 51.25 | 50.89 | 50.91 | 51.21 | 51.01 |
| No. of ribosomal RNA (16S/23S/5S) | 7/7/8 | 7/7/8 | 7/7/8 | 7/7/8 | 7/7/8 | 7/7/8 |
| No. of transfer RNA | 86 | 85 | 97 | 98 | 91 | 97 |
| Deletions (kb)d | – | 955 | 639 | 709 | 746 | 518 |
| Insertions (kb)d | – | 411 | 444 | 479 | 441 | 490 |
| Translocations and inversionsd | – | 43 | 13 | 15 | 23 | 11 |
| IS-elements (percentage) | 44 (1%) | 623 (12%) | 314 (7%) | 280 (7%) | 403 (9%) | 394 (8%) |
| Virulence plasmid | pSD1_197 | pCP301b | pINV-2457Tc | pSB4_227 | pSS_046 | |
| Total length (bp) | 182 726 | 221 618 | ∼218 000 | 126 697 | 214 396 | |
| No. of total ORFs | 224 | 267 | ND | 149 | 241 | |
| Percentage of CDS (%) | 76.03 | 76.24 | ND | 74.18 | 79.06 | |
| G+C content (%) | 44.80 | 45.77 | ND | 47.41 | 45.27 | |
| IS-elements (percentage) | 78 (27%) | 88 (32%) | ND | 72 (38%) | 96 (33%) |
aData are obtained from a recently updated version of U00096.
bData are obtained from Jin et al. (11).
cData are obtained from Wei et al. (12); the virulence plasmid pINV-2457T was reported in the communication but the sequence is not yet publicly available.
dOnly those with DNA segments > 5 kb are listed.
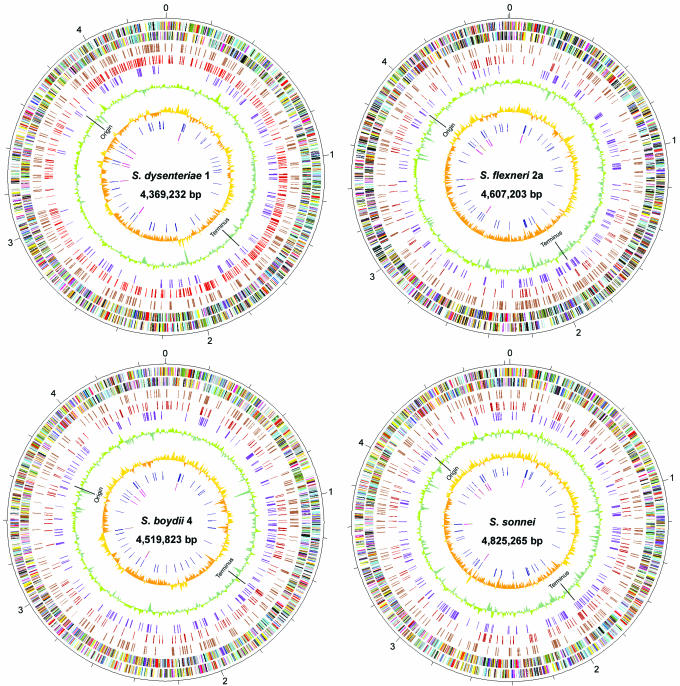
Figure 1.
Circular representations of the Shigella genomes. The outer scale is marked every 200 kb. Circles range from 1 (outer circle) to 9 (inner circle). Circles 1 and 2, ORFs encoded by leading and lagging strands, respectively, with colour code for functions: salmon, translation, ribosomal structure and biogenesis; light blue, transcription; cyan, DNA replication, recombination and repair; turquoise, cell division; deep pink, posttranslational modification, protein turnover and chaperones; olive drab, cell envelope biogenesis; purple, cell motility and secretion; forest green, inorganic ion transport and metabolism; magenta, signal transduction; red, energy production; sienna, carbohydrate transport and metabolism; yellow, amino acid transport; orange, nucleotide transport and metabolism; gold, co-enzyme transport and metabolism; dark blue, lipid metabolism; blue, secondary metabolites, transport and catabolism; grey, general function prediction only; black, function unclassified or unknown. Circle 3, distribution of pseudogenes. Circles 4 and 5, distribution of IS1/IS1N and other IS-species, respectively. Circles 6 and 7, G+C content and GC skew (G-C/G+C), respectively, with a window size of 10 kb. Circles 8 and 9, distribution of tRNA genes and rrn operons, respectively. The replication origin and terminus are indicated for each. (The circular map for Sf301 was created based on the updated annotation.)
Table 5.
ORFs in each Shigella genome related to main clinical biochemical reactions
| Reaction | Gene | Product | Sd197 | Sf301 | 2457T | Sb227 | Ss046 |
|---|---|---|---|---|---|---|---|
| Indol | tnaA | Tryptophanase | – | SF3754 | S4017 | SBO3667a | – |
| Ornithine | speC | Ornithine decarboxylase | SDY3107 | SF2962 | S3165 | SBO3024a | SSO3230 |
| Lactose | lacY | Galactoside permease | SDY0376a | – | – | – | SSO0300a |
| lacZ | Beta-D-galactosidase | SDY0378 | – | – | – | SSO0299 | |
| Lysine | cadA | Lysine decarboxylase | SDY4466a | – | – | – | SSO4308a |
| cadB | Lysine/cadaverine transport protein | SDY4465a | – | – | – | SSO4315a | |
| Hydrogen sulfide | phsA | Hydrogen sulfide production: membrane anchoring protein | – | – | – | – | – |
| phsB | Hydrogen sulfide production: iron–sulfur subunit; electron transfer | – | – | – | – | – | |
| phsC | Hydrogen sulfide production: membrane anchoring protein | – | – | – | – | – | |
| Citric acid | citT | Citrate:succinate antiporter | – | SF0530a | S0536a | SBO0477 | SSO0564 |
| citC | Citrate lyase synthetase | – | SF0535 | S0542 | SBO0483 | SSO0571a | |
| citD | Citrate lyase acyl carrier protein (gamma chain) | – | SF0534 | S0541 | SBO0482 | SSO0569 | |
| citE | Citrate lyase beta chain (acyl lyase subunit) | – | SF0533 | S0540 | SBO0481 | SSO0568 | |
| citF | Citrate lyase alpha chain | – | SF0532 | S0539 | SBO0480 | SSO0567 | |
| citA | Sensory histidine kinase, regulation of citrate fermentation, senses citrate | – | – | – | SBO0484a | SSO0572 | |
| citB | Response regulator, regulation of citrate fermentation | – | SF0660 | S0683 | SBO0485 | SSO0573 | |
| Acetate | aceA | Isocitrate lyase | SDY4328 | SF4081 | S3649 | SBO4035a | SSO4187 |
| aceB | Malate synthase A | SDY4329a | SF4080a | S3650a | SBO4034 | SSO4186 | |
| aceK | Isocitrate dehydrogenase kinase/phosphatase | SDY327 | SF4082a | S3648a | SBO4036 | – | |
| d-mannitol | cmtA | PTS system, mannitol permease II, BC component | SDY3144 | – | SBO3056 | SSO3087a | |
| cmtB | PTS system, mannitol permease II, A component | SDY3143 | – | SBO3055 | SSO3086 | ||
| mtlA | PTS system, mannitol permease II, ABC components | – | SF3633 | S4135 | SBO3597 | SSO3809 | |
| mtlD | Mannitol-1-phosphate dehydrogenase | – | SF3634 | S4134 | SBO3598 | SSO3808 | |
| d-sorbitol | srlA | PTS system, glucitol/sorbitol-specific II, C component | SDY2898 | SF2725a | S2916a | SBO2816 | SSO2846 |
| srlE | PTS system, glucitol/sorbitol-specific II, B component | SDY2899a | SF2726 | S2917 | SBO2815 | SSO2847 | |
| srlB | PTS system, glucitol/sorbitol-specific enzyme II, A component | SDY2901 | SF2727 | S2918 | SBO2814 | SSO2848 | |
| srlD | Glucitol (sorbitol)-6-phosphate dehydrogenase | SDY2902 | SF2728 | S2919 | SBO2813 | SSO2849a | |
| d-xylose | xylA | d-xylose isomerase | – | SF3609a | S4160a | SBO3573 | SSO3820 |
| xylB | Xylulokinase | – | SF3608 | S4161 | SBO3572 | SSO3821 | |
| xylF | d-xylose transport system substrate-binding protein | SDY4336a | SF3610 | S4159 | SBO3574 | – | |
| xylG | d-xylose transport system ATP-binding protein | – | SF3611 | S4158 | SBO3575 | – | |
| xylH | d-xylose transport system permease protein | – | SF3612 | S4157 | SBO3576 | – |
aPseudogenes.
The Shigella chromosomes have the same replication origin and terminus as those of MG1655 (22), indicating that they probably have the same replication mechanism as E.coli. In all the Shigella genomes, the rRNA operons map to approximately the same relative positions as in MG1655, indicating that there is no DNA recombination between rRNA operons as observed previously in some Shigella strains (23). All currently sequenced Shigella and E.coli genomes available from GenBank have in total ∼3 Mb in common (or backbone). This potentially encodes 2790 proteins accounting for 65% of coding capacity of the MG1655 genome, which likely includes genes essential for bacterial survival and growth. Within the backbone there are 2393 orthologous genes shared by all four genomes. Details about orthologous genes between each pair of the genomes can be found in ShiBASE at http://www.mgc.ac.cn/ShiBASE/Orth_order.htm#table. Within the backbone there are 313 genes which are pseudogenes in at least one of the Shigella genomes (Supplementary Table S1), indicating that probably none of these is essential for survival and growth. The chromosomes of Sd197, Sf301 and Sb227 are smaller than MG1655, while that of Ss046 is larger (Table 1). This variation in genome size is due mainly to deletions and insertions of >5 kb.
IS-elements and dynamics of the Shigella genomes
The most striking feature of the Shigella genomes is their highly dynamic nature due to the presence of hundreds of IS-elements in each of the genomes (Table 2). IS-elements are capable of causing many kinds of DNA rearrangements (24) and the presence of the many rearrangements (deletions as well as translocations and inversions) are a likely the result of the copious numbers of IS-elements. The Sd197 genome shows the most rearrangements and is considerably smaller than the MG1655 genome due to a large number of deletions (Table 1). The genome of this Shigella strain also possesses the greatest number of IS-elements, mainly in the form of IS1N (Table 2), which may be responsible for many of these rearrangements.
Table 2.
IS-elements identified in Shigella genomes and E.coli K12 MG1655 chromosome
| Length (bp) | No. of ORFs | No. of intact elements | No. of partial elementsa | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MG1655 | Sd197 | Sf301 | 2457T | Sb227 | Ss046 | pCP301 | pSD1_197 | pSB4_227 | pSS_046 | MG1655 | Sd197 | Sf301 | 2457T | Sb227 | Ss046 | pCP301 | pSD1_197 | pSB4_227 | pSS_046 | |||
| IS1 | 768 | 2 | 7 | 151 | 108 | 105 | 160 | 167 | 2 | 3 | 6 | 3 | 0 | 10 | 9 | 3 | 14 | 8 | 1 | 0 | 0 | 1 |
| iso-IS1(IS1N) | 803 (766) | 2 | 0 | 273 | 0 | 1 | 1 | 1 | 0 | 8 | 0 | 0 | 0 | 27 | 1 | 0 | 0 | 0 | 5 | 5 | 4 | 5 |
| IS2 | 1331 | 2 | 6 | 25 | 30 | 29 | 33 | 27 | 1 | 2 | 3 | 2 | 1 | 7 | 5 | 4 | 10 | 16 | 2 | 4 | 4 | 4 |
| IS3 | 1258 | 2 | 5 | 0 | 5 | 5 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 3 | 1 | 0 | 1 | 7 | 6 | 4 | 6 |
| IS4 | 1428 | 1 | 1 | 10 | 18 | 19 | 16 | 28 | 1 | 1 | 3 | 1 | 0 | 2 | 3 | 3 | 10 | 5 | 1 | 1 | 0 | 1 |
| IS5 | 1198 | 1 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| iso-IS10R | 1329 | 1 | 0 | 0 | 13 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IS21 | 2131 | 2 | 0 | 0 | 0 | 0 | 0 | 17 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 4 | 3 | 3 | 1 | 5 |
| IS30 | 1221 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| IS91 | 1830 | 1 | 0 | 0 | 3 | 5 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 2 | 6 | 4 | 2 | 6 |
| IS100 | 1963 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 3 | 3 | 5 |
| IS150 | 1443 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 2 | 0 | 1 | 1 |
| IS186 | 1372 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| IS600 | 1264 | 2 | 0 | 54 | 35 | 35 | 20 | 51 | 3 | 2 | 1 | 6 | 1 | 28 | 17 | 12 | 17 | 23 | 10 | 8 | 4 | 4 |
| IS629 | 1310 | 2 | 0 | 0 | 10 | 12 | 41 | 3 | 8 | 4 | 5 | 3 | 0 | 2 | 11 | 2 | 0 | 1 | 3 | 5 | 4 | 6 |
| IS630 | 1164 | 1 | 0 | 0 | 0 | 0 | 0 | 16 | 1 | 0 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 | 4 |
| IS911 | 1250 | 2 | 0 | 12 | 16 | 16 | 26 | 7 | 1 | 0 | 2 | 0 | 4 | 9 | 0 | 4 | 22 | 0 | 0 | 1 | 4 | 1 |
| IS1294 | 1689 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 2 | 0 | 2 | 3 | 3 | 0 | 0 | 7 | 2 | 7 | 6 |
| ISSfl1 | 923 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 5 |
| ISSfl2 | 1374 | 1 | 0 | 4 | 6 | 5 | 8 | 9 | 2 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 1 |
| ISSfl3 | 1302 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| ISSfl4 | 2754 | 3 | 0 | 0 | 3 | 6 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 7 | 5 | 3 | 0 | 2 | 5 | 4 | 4 |
| ISEc8 | 2506 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 2 | 3 | 0 | 1 | 1 | 1 |
| ISSbo6 | 2506 | 3 | 0 | 0 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 1 | 0 | 1 | 1 | 1 |
| Total | 37 | 529 | 247 | 238 | 314 | 327 | 26 | 26 | 24 | 28 | 7 | 94 | 67 | 42 | 89 | 67 | 62 | 52 | 48 | 68 | ||
aOnly those with IS fragments ≥ 100 bp are listed.
The deletions are often associated with translocations and inversions, which interrupt the collinearity of the Shigella and the MG1655 chromosomes (Figure 2a). Sd197 and Sb227 have more translocations and inversions involving DNA segments >5 kb than Sf301 and Ss046 (Table 1). Therefore, the collinearity with MG1655 chromosome is more severely interrupted in Sd197 and Sb227. All Shigella chromosomes have inversions at the replication origins and termini, which have been suggested to be recombination hotspots (25). The rearrangements at the termini can be very complex. Although a single inversion, probably mediated by IS1, is present in Ss046, several inversions and translocations of different sizes, and probably mediated by different IS-elements, are present in the other genomes (Figure 2a). This suggests that the rearrangements at the termini were formed through independent recombination events among the Shigella genomes.
GC skew is the measurement of mononucleotide frequencies ([fG − fC]/[fG + fC]). The GC compositional strand bias observed in the E.coli genome, which gives rise to two distinctive replichores, has been hypothesized to reflect the biased mutational traits in codon positions in the leading and lagging strands under natural selection (22). Owing to the many inversions, the GC skew has been distorted in the Shigella genomes particularly in Sd197 and Sb227 (Figure 1).
Inversions are often accompanied by deletions. The ompT gene is removed by inversion-associated deletions from all four genomes. This is the basis of so called kcp locus necessary for Shigella to cause keratoconjunctivitis in guinea pigs because OmpT reduces IcsA expression (26). The cadA gene responsible for converting lysine to cadaverine that in turn attenuates virulence (27) is missing from Sf301 and Sb227 by inversion-associated deletions. In Sd197 and Ss046 cadA is inactivated via a frameshift and an IS insertion, respectively.
Compared with MG1655, Shigella strains not only have many more copies of IS-elements but also have additional IS-species, such as IS1N, IS600 and IS629 (Table 2). Within the Shigella genomes, IS1 is predominant in the Sf301, Sb227 and Ss046 chromosomes whereas IS1N is copiously present in the Sd197 chromosome. Intact IS21 and IS630 are present only in Ss046, while the newly identified ISSbo6 is found mainly in Sb227 chromosome. ISSbo6 is similar to ISEc8 found adjacent to the locus of enterocyte effacement (LEE) pathogenicity island in EHEC (28). Furthermore, most copies of the ISSbo6 are located within SHI-1, SHI-2 and ipaH islands (see below) in the Sb227 genome. The virulence plasmids and chromosomes share most of the IS-species, suggesting that inter- and intra-replicon translocation and replication has occurred, leading to large numbers of IS-elements in the genomes.
The virulence plasmids also display a dynamic nature with many IS-mediated deletions, translocations and inversions. Plasmid pSS_046 from Ss046 shows the closest collinearity to pCP301 (Figure 2b). Apart from IS-mediated inversions and translocations, the collinearity is interrupted downstream to the replication origin, ori, due to a 13 kb insertion in pSS_046 that carries genes for O-antigen synthesis as described previously (29). pSB4_227 from Sb227 also shows the collinearity with pCP301 except for the ∼80 kb deletion including the cell-entry region (see above). Plasmid pSD1_197 from Sd197 has a notable translocation involving a DNA segment of ∼50 kb associated with duplication of the ori sequence and nearby repA and copAB genes. As a result, the cell-entry region is sandwiched by two sets of ori, repA and copAB (Figure 2b). The original ori sequence is truncated, so that plasmid replication is probably performed by the functional duplicated ori sequence.
A number of notable loci present in pCP301 are absent from pSS_046. These include sepA, (which codes for an extracellular serine protease involved in tissue invasion) (30), phoN (encoding a non-specific phosphatase), stbAB (encoding one of the two partition systems) and ipgH (encoding a sugar phosphate). The sepA gene has been shown by DNA hybridization to be absent from a number of other S.sonnei virulence plasmids (21).
Diversity of the virulence genes: gain of functions
The distribution of putative virulence genes shows diversity among the Shigella genomes (Tables 3 and 4). The so-called Shigella pathogenicity island (SHI)-1 in Sf301 encodes three characterized proteins: the autotransporter proteases Pic and SigA, and the enterotoxin ShET1 which is encoded by the setAB genes and which are entirely within, and on the complementary strand of, the pic coding sequence. Pic is implicated in mucinase activity, serum resistance, and hemagglutination (31), while SigA is capable of casein degradation, is cytopathic for HEp-2 cells and along with ShET1 contributes to fluid accumulation in rabbit ileal loops (32,33). SHI-1 is wholly absent from Sd197, and in Sb227 and Ss046, although sigA is present, the pic/setAB coding sequence is missing. A second copy of sigA is also present in Sb227.
Table 3.
Known and putative virulence genes in the Shigella chromosomes
| Product | Gene | Location | Function | Sd197 | Sf301 | 2457T | Sb227 | Ss046 |
|---|---|---|---|---|---|---|---|---|
| Toxins | ||||||||
| Shiga toxin | stxAB | stx-phage P27 | N-glycosidase, block protein synthesis | SDY1398,1390 | – | – | – | – |
| ShET1 | set1A, set1B | SHI-1 | Ion secretion | – | SF2973a, 2973b | NDa | – | – |
| ShET2 | senB | Ion secretion | – | – | – | – | SSO2665 | |
| Protease | ||||||||
| Serine protease | pic | SHI-1 | Mucinase | – | SF2973 | S3178 | – | SSO3595a |
| Serine protease | sigA | SHI-1 | Ion secretion | – | SF2968 | S4824 | SBO0233, 4150 | SSO3223 |
| Others | ||||||||
| Aerobactin | iutA, iucABCD | SHI-2/SHI-3 | Iron acquisition | – | SF3719, 3715–3718 | S4052, 4053–4056 | SBO4314, 4337–4340 | SSO3605, 3601–3604 |
| Siderophore receptor | iroN, iroBCDE | Iron acquisition | SDY1022, 1023–1026 | – | – | – | – | |
| ABC transporter | sitABCD | Iron acquisition | SDY1454–1457 | SF1362–1365 | S1964–1967 | SBO1691–1694 | SSO1750–1753 | |
| Hemin receptor | shuA, shuS, shuTWXYUV | Iron acquisition | SDY3547–3555 | – | – | – | – | |
| ABC transporter | Iron acquisition | SDY1240–1242 | SF1192–1194 | S1278–1280 | – | – | ||
| Invasion plasmid antigen | ipaH | ipaH island | Unknown | SDY0834, 1062, 2001, 2003, 2753 | SF0722, 1383, 1880, 2022b, 2610 | S0761b, 0934, 1268, 1947, 2119, 2330b, 2782 | SBO0653, 0953, 1026, 1256, 1619, 2084 | SSO0751, 1272, 1317, 2179, 2646 |
| Putative adhesin | yadA-like | OI#144-like island | Unknown | – | SF3641 | S4127b | SBO3605 | SSO3803 |
| Putative chaperone | clp-like | OI#7-like island | Unknown | – | – | – | – | SSO0242 |
| Inner memberane protein | IcmF-like | OI#7-like island | Unknown | – | – | – | – | SSO0236 |
| Exoprotein | RTX-like | OI#28-like island | Unknown | SDY0420–0424 | – | – | – | – |
| Transport system | OI#28-like island | Unknown | SDY0416, 0417 | – | – | – | – | |
| T2SS | gspC-M | Unknown | SDY3092–3102 | – | – | SBO3011b, 3012b, 3013–3021 | – | |
aSequences exist in the genome but are not recognized as coding genes by the current annotation.
bPseudogenes.
Table 4.
Known and putative virulence genes in the Shigella virulence plasmids
| Product | Gene | Function | pSD1_197 | pCP301 | pSB4_227 | pSS_046 |
|---|---|---|---|---|---|---|
| TTSS | mxi-spa region | Invasion and internalization | SDYP174–193 | CP0136–0156 | – | SSOP098–117 |
| TTSS secreted protein | ipaA | Actin depolymerization | SDYP163 | CP0125 | – | SSOP087 |
| ipaB | Inducing apoptosis | SDYP166 | CP0128 | – | SSOP090 | |
| ipaC | Actin polymerization, activation of Cdc42 and Rac | SDYP165 | CP0127 | – | SSOP089 | |
| ipaD | Forming a complex with IpaB, control the flux of proteins through the type III secretion | SDYP164 | CP0126 | – | SSOP088 | |
| ipgD | Inositol 4-phosphatase, membrane blebbing | SDYP171 | CP0133 | – | SSOP095 | |
| icsB | Camouflaging IcsA from autophagic host defense system | SDYP170 | CP0132 | – | SSOP094 | |
| virA | Microtubule destabilization, membrane ruffling | SDYP211 | CP0181 | – | SSOP142 | |
| ospF/mkaD | Unknown | SDYP013 | CP0010 | SBOP017 | SSOP009 | |
| ipaH7.8 | Facilitating the escape of the bacteria from phagocytic vacuole of macrophages | SDYP038 | CP0078 | SBOP067 | SSOP058 | |
| ipaH9.8 | Transported to the nucleus, function unknown | SDYP099 | CP0226 | SBOP113 | SSOP167 | |
| ipaH4.5 | Unknown | SDYP037 | CP0079 | SBOP066 | SSOP059 | |
| ipgB | Unknown | SDYP168 | CP0130 | – | SSOP092 | |
| ospG | Unknown | SDYP101 | CP0227 | – | SSOP170 | |
| Toxins | ||||||
| ShET2 | senA | Ion secretion | SDYP056 | CP0093 | SBOP076 | SSOP050 |
| senB | Homologues of ShET2 | SDYP010 | CP0009 | SBOP016 | SSOP008 | |
| Enzymes | icsP/sopA | Cleavaging of IcsA | SDYP224 | CP0271 | SBOP149 | SSOP241 |
| sepA | Tissue invasion | – | CP0070 | – | – | |
| msbB | Fatty acyl modification of O-antigen | SDYP110 | CP0238 | SBOP119 | SSOP182 | |
| apy | ATP-diphosphohydrolase | SDYP004 | CP0004 | SBOP006 | SSOP004 | |
| phoN-Sf | Non-specific acid phoshatase | SDYP067 | CP0190 | – | – | |
| rfbU | O-antigen biosynthesis | SDYP108 | CP0236 | – | SSOP180 | |
| ushA | UDP-sugar hydrolase (5′-nucleotidase) | SDYP064 | CP0185 | – | SSOP147 | |
| Regulators | virF | Activating transcription of virB and icsA | – | CP0046 | SBOP052 | SSOP041 |
| virK | Post-transcriptional regulation of icsA expression | SDYP109 | CP0237 | SBOP118 | SSOP181 | |
| virB | Activating ipa, spa, and mxi operons | SDYP161 | CP0123 | – | SSOP085 | |
| Others | icsA/virG | Nucleation of actin filaments | SDYP214 | CP0182 | – | SSOP143 |
SHI-2 was originally identified at the selC tRNA locus in S.flexneri. It carries the iut/iuc operon encoding an aerobactin system for iron acquisition (10). SHI-2 is present in Ss046 but unlinked with the selC gene, which appears to be caused by an inversion near the replication origin in Ss046, as evidenced by the fact that selC tRNA gene is located in the leading strand in MG1655 and Sf301 but situated in the lagging strand in Ss046. The previously reported S.boydii SHI-3 is present in Sb227. SHI-3 carries the same iut/iuc operon as SHI-2 but is linked with the pheU tRNA locus (34). Sd197 has neither SHI-2 nor SHI-3 but solely possesses the shu and the iro operons (Table 3). The shu operon encodes a TonB-dependent heme transport system (35). The iro genes were originally identified in Salmonella enterica as a ferric iron transport system (36).
One of the interesting features of the Sf301 genome is the presence of 12 copies of the ipaH genes: 5 on the plasmid and 7 on the chromosome, of which 5 are located in ipaH-islands which are apparently acquired via phage-mediated lateral gene transfer (11). All the ipaH products have a conserved C-terminal half of 260 amino acid residues but variable N-terminal halves, within which are leucine-rich repeat regions implicated in protein–protein interaction. The plasmid encoded IpaH7.8 is involved in the escape of Shigella from phagocytic vacuoles in macrophages (37), and the bacteria express more IpaH9.8 when inside host cells (38). Hence, the multiple ipaH are an interesting phenomenon. It is now confirmed that there are multiple ipaH genes in all Shigella genomes (Tables 3 and 4), except that ipaH1.4 and ipaH2.5 are absent in pSB4_227 and pSS_046, respectively, and that the coding region of ipaH1.4 in pSD1_197 is truncated due to an IS629 insertion. Thus, the necessity for these two ipaH genes in virulence is in doubt. The chromosomal ipaH genes are mostly within ipaH islands and are always next to a gene that encodes a protein homologous to a bacteriophage P27 protein (accession no. NP_543109) of unknown function. However, the plasmid ipaH genes from all strains are unlinked with the phage gene. Hence, the virulence plasmids and the chromosomes may have acquired the ipaH genes from different sources or by different mechanisms.
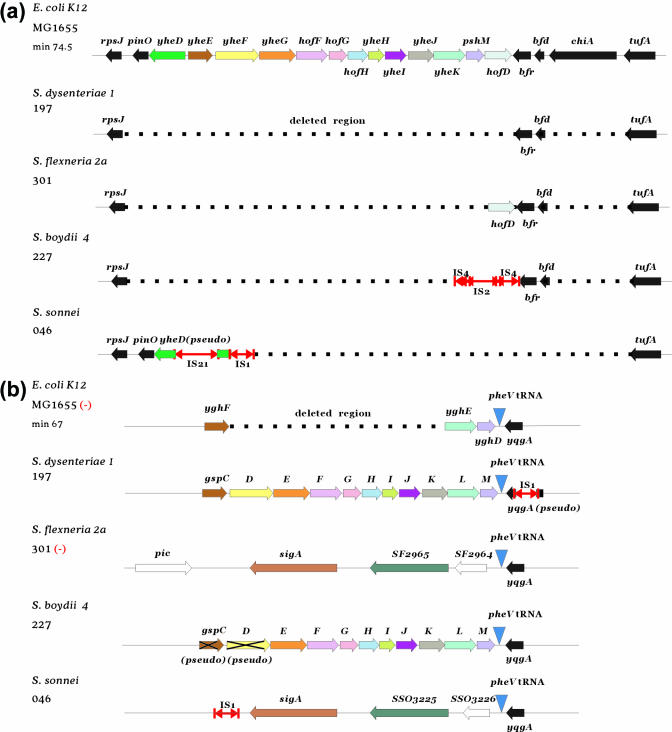
The type II secretion system (T2SS) encoded by genes of the general secretion pathway (gsp) is widely distributed in Gram-negative bacteria (Supplementary Figure S1). The known E.coli T2SS encoded by the yhe genes at 74.5 min of the MG1655 chromosome is absent in all four sequenced Shigella genomes (Figure 3a). However, there is a novel set of gsp genes in the Sd197 and Sb227 chromosomes, which is absent in MG1655 and the other Shigella genomes (Figure 3b). The gsp products from Sd197 and Sb227 show some similarity to those of the E.coli yhe genes, but significantly greater similarity to those of the gsp genes from enterotoxigenic E.coli (ETEC) and Vibrio cholerae responsible for secreting the E.coli heat labile toxin (Ltx) and cholera toxin (Ctx), respectively (39,40) (Supplementary Figure S1). Shiga toxin, Stx, is encoded by a prophage at a distance locus from gsp as described previously (41). The Sb227 T2SS is likely to be inactive due to a frameshift in gspC and a nonsense mutation in gspD. Interestingly, the gsp genes are present as an island at the pheV tRNA locus in both Sd197 and Sb227. However, in Sf301 and Ss046, this locus is occupied by SHI-1 (Figure 3b). So, the pheV tRNA locus in Shigella strains may be a hotspot for insertion of laterally transferred genes.
Figure 3.
Graphic representation of the different T2SS loci in E.coli K-12 MG1655 and Shigella genomes (to scale). (a) The yhe locus at 74.5 min of the MG1655 chromosome and the corresponding regions in the Shigella genomes. (b) The pheV tRNA locus at 67 min of MG1655 and the corresponding loci in Shigella genomes where the gsp genes are located. A strain name followed by a minus sign (−) means the reverse complement strands of the genome sequences were used for the diagram.
In the Shigella genomes, there are regions similar to the ‘O-islands’ (OI) from EHEC EDL933 (42). Some of these O-island-like sequences may be of significance to virulence. In Sd197, the open reading frames (ORFs) SDY0416-SDY0425 may encode a RTX-toxin-like exoprotein and a transporter similar to those encoded by OI #28. In Sd197 and Sf301, there are ORFs (SDY1240-SDY1242 and SF1192-SF1194, respectively) which encode a putative iron compound ABC transporter similar to those from EDL933 (Z1964-Z1966). In Sf301, Sb227 and Ss046, there are genes encoding a putative adhesin similar to that encoded by the EDL933 OI #144 (Z5029), which belong to the Yersinia YadA family that mediates bacterial adherence and invasion through binding to fibronectin and β1 integrin (43) and induces the production of interleukin-8 (44). However, only Ss046 encodes a protein of 1616 amino acids similar in length to that of EDL933 of 1588 amino acids. Sf301 encodes a protein with only 990 amino acids at the C-terminus, whereas Sb227 encodes a protein with a truncation of more than 200 amino acids at the C-terminus, both of which may not be functional.
Deletions and pseudogenes: loss of functions
Deletions and pseudogenes are effective mechanisms for loss of functions, and the inactivation of the ompT and cadA genes provide examples of how loss of some functions may increase the pathogenicity of Shigella (see above). Additionally, all the sequenced strains of Shigella have lost flagellar function due to mutations in many different genes (Supplementary Table S2). Fimbriae are also absent from these Shigella. In the EHEC genome (NC_002655), there are 14 loci involved in fimbrial biogenesis. None of the counterpart loci in the Shigella genomes is intact (Supplementary Table S2).
The central intermediary metabolism is conserved in all four Shigella species and MG1655. However, considerable variations have been found in carbohydrate and amino acid metabolism (Supplementary Table S2). Shigella bacteria do not synthesize lysine decarboxylase due to inactivation of cadA gene (see above), do not produce hydrogen sulfide from thiosulfate, do not produce gas from carbohydrate, do not use citric acid as a sole carbon source and do not grow on sodium acetate. Table 5 is a summary of the genetic basis for these negative Shigella properties in main clinical biochemical reactions (see Supplementary Table S2 for a complete list). Note that only Sb227 carries all genes necessary for utilization of d-mannitol, d-sorbitol and d-xylose (Table 5).
Lactose fermentation is a biochemical property commonly used for distinguishing Shigella from E.coli. However, some S.dysenteriae 1 and S.sonnei isolates ferment lactose slowly, which now can be explained genetically. In the genomes of Sd197 and Ss046 the key gene, lacZ (encoding β-d-galactosidase), is intact though lacY (encoding galactose permease) is a pseudogene (both of them are deleted from Sf301 and Sb227). Additionally, Sd197 and Ss046 have ORFs SDY2556 and SSO2450, respectively, which encode proteins similar to the sucrose permease from EHEC (NP_288931) sharing a conserved LacY domain and overall 34% identity with the lactose permease from Klebsiella pneumoniae (JT0487). This unspecialised galactoside transport function may compensate partially for the loss of LacY in Sd197 and Ss046 leading to slow lactose fermentation.
DISCUSSION
An MLS study (13) and the previous reported S.flexneri genomes (11,12) have suggested strongly that Shigella is within the species of E.coli. The complete genomes of S.dysenteriae, S.boydii and S.sonnei have provided additional supporting evidence; they all have ∼3 Mb of genomic DNA in common with all published E.coli and Shigella genomes. The extensive diversity of the Shigella genomes revealed by the whole genome sequences supports the hypothesis that Shigella have emerged from diverse origins of E.coli. Recently, Lan et al. (45) have presented evidence based on MLS that EIEC strains are also derived from different origins of E.coli. One of the EIEC strains (serotype O112ac) is grouped into Shigella Cluster 2, and outliers of Shigella strains, S.dysenteriae type 1 and S.sonnei, are more closely related to EIEC strains. Based on the comparative genomic hybridization microarray, Fukiya et al. (46) have also shown that three out of four EIEC strains are closely related to three Shigella strains (S.flexneri 2a, S.sonnei and S.boydii) but more distance to EPEC, ETEC, EHEC and UPEC strains. Thus, there is little doubt now that Shigella and EIEC form a single pathovar of E.coli.
The extensive diversity of the Shigella genomes appears to be multi-factorial. First, Shigella has evolved from diverse genomic backgrounds of E.coli. Particularly, we must remember that Sd197 and Ss046 are outside of three main Shigella phylogenetic groups (13). Therefore, the diversity of the four sequenced genomes does not reflect the entire genome diversity within the Shigella. Second, putative virulence genes have been transferred by bacteriophages to selected genomes. The distribution of SHI-1 and SHI-2 provide an example of this. Third, convergent evolution has been facilitated by IS-mediated rearrangements. For example, ompT and cadA were inactivated by deletions involving different DNA segments in different genomes, and 14 orthologous fimbrial systems identified in the EHEC genome have all been inactivated in different ways in the Shigella genomes. Fourth, creation of independent pseudogenes, e.g. different genes for utilization of d-sorbitol are inactivated in different genomes (Table 5).
The diversity of the genomes provides a basis for further investigations into pathogenesis, epidemiology and microbial evolution. For example, it is known that S.dysenteriae produces Shiga toxin (47). However, it is unknown until now that there is a T2SS. Given that Stx has an overall similar structure to Ctx and Ltx and that T2SS from Sd197 shows extensive homology to those from V.cholerae and ETEC, it is highly likely that Stx is actively secreted from Sd197. Therefore, the S.dysenteriae T2SS ought to contribute significantly to pathogenicity as it enables toxin molecules to reach the target host cells from proliferating bacteria. Otherwise, the accumulated toxin molecules can be released only upon bacterial lysis. Of course, it is also of interest to investigate whether the Shigella T2SS secretes other putative virulence factors in addition to Shiga toxin.
A comparative genomic hybridization test indicates a wide distribution of the gsp genes among strains from all phylogenetic groups (J. Peng, X. Zhang, J. Yang, J. Wang, E. Yang, W. Bin, C. Wei, M. Sun and Q. Jin, unpublished data), which suggests that many strains possessed Stx before their subsequent loss, and thereafter in the case of Sb227 the T2SS was inactivated. It has been reported that an Stx-expressing prophage from an S.sonnei strain is able to form plaques on a number of different Shigella species and serotypes (48). We found in this study that Ss046 possesses remnants of the previously reported Stx-phage Φ P27 (49) which has a different gene content and organization to the Sd197 Stx-phage, and Sf301 and Sb227 possess remnants of the Stx-phage of Sd197. Taken together, many Shigella strains have probably gained and then lost the Stx genes in the evolutionally past. Perhaps, loss of Stx genes has provided advantages to the bacteria for better adaptation to the human hosts, as causing severer disease offers little benefit to the organisms for long term survival. Alternatively, according to the hypothesis by Escobar-Paramo et al. (50) the integration, retention and expression of certain virulence factors may be the result of the interaction between the newly introduced genes and the bacterial genomic background. Hence, perhaps only S.dysenteriae 1 and a few S.sonnei strains have the right genomic background to retain and express Stx stably.
In addition to Shiga toxin and the T2SS, S.dysenteriae type 1 alone possesses two iron acquisition systems, shu and iro. Though the shu system, responsible for heme uptake, is not essential for invasion and proliferation in cultured Henle cells, it still can be very important in vivo (51). Recently, Skaar et al. (52) showed that Staphylococcus aureus preferably imports heme iron over transferrin in vivo and that mutant strains defective in heme transport are severely attenuated in a Caenorhabditis elegans infection model. Thus, there is a need to establish whether S.dysenteriae also prefers heme over other iron sources during infection. In addition, it is important to identify the genetic basis of the previously observed heme transport activity in S.flexneri (53). A comparison of that yet unidentified heme transport system with the Shu system is necessary for a better understanding of the iron acquisition strategies that Shigella employs.
The iro genes were originally identified in S.enterica as a ferric iron transport system (36). As the receptor, the iroN product, has affinity to some iron-containing substrates produced by soil microbes, the iro system has been speculated to facilitate the growth of S.enterica in soil. Whether this system offers an advantage to S.dysenteriae for environmental survival over other strains or plays a role during infection requires further investigations. We must emphasize that the observed differences in gene content between the different species here are not necessarily characteristic for the different species. For example, the iro genes are only present in serotype 1 but not in other S.dysenteriae strains (J. Peng, X. Zhang, J. Yang, J. Wang, E. Yang, W. Bin, C. Wei, M. Sun and Q. Jin, unpublished data).
In general, the virulence of Shigella is in the order, S.dysenteriae > S.flexneri > S.sonnei (54). The lack of ShET1 and Pic, and SepA in Ss046 may collectively make S.sonnei lesser virulent than S.flexneri, as these are probably the major determinants involved in the diarrhoeal phase of the infection. On the other hand, S.dysenteriae 1 infection generally has only a very limited diarrhoeal phase, but abrupt onset of acute dysentery (54). This may be due to its possession of factors, such as the very potent Shiga toxin, and thus lack of SHI-1 is insignificant to S.dysenteriae type 1.
The presence of large numbers of IS-elements in the Shigella genomes is likely the major cause of many of the genome rearrangements. IS1 dominates in Sf301, Ss046 and Sb227, and is associated with DNA rearrangements at many different loci in these three genomes. These events could not be random events which must have occurred in accordance with the whole genome property or promoted (or restrained) by other genetic loci. Previously ISIN, also known as iso-IS1, was found only in S.dysenteriae serotype 1 (55). However, we found that Sb227 and Ss046, and the EHEC strain EDL933 all have a single copy of ISIN next to yiiX, a gene encoding a hypothetical protein. The uropathogenic E.coli (UPEC) strain CFT073 has three partial copies but none is at this locus. Since Sd197 also has a copy of ISIN next to yiiX, this locus is likely to be the original site for ISIN acquisition except the UPEC strain. Thus, either the expansion of ISIN is permitted only in S.dysenteriae type 1 or ISIN has transmitted into the other genomes fairly recently.
The fact that ISSbo6 is restricted in SHI-1, SHI-2 and ipaH islands in Sb227 is interesting. It indicates that those pathogenicity islands were acquired earlier than the IS-elements, which probably is mainly distributed among microbes within the Indian subcontinent.
Inversions, probably IS-mediated, are another mechanism that has reshaped the genomes. One of the conserved genetic traits, namely the CG bias strand composition or GC-skew, among the enteric bacteria, is distorted by inversions in the Shigella genomes (Figure 1). This can be significant to gene expression as GC-skew is a reflection of the biased mutational traits in codon positions in the leading and lagging strands under natural selection (22). Shigella colonizes and proliferates in the cell cytosol, a niche unique amongst the enteric bacteria, and thus is likely to be under different selective pressure to change the expression of many genes compared with other enteric bacteria. Inversions and, additionally, translocations effectively lead to preferred leading or lagging strand, orientation and distance to the replication origin for the optimal expression of these genes.
Gene decay or reductive evolution is noted to be an important evolutionary mechanism for the obligate intracellular pathogens, such as Mycobacterium leprae (56). Shigella bacteria, being facultative intracellular pathogens, also employ such a mechanism. The Sd197 genome is obviously smaller than that of MG1655 (Table 1), and the other three sequenced genomes display a net loss of genetic material [excluding the IS sequences which account for 7–12% of the genomes (Table 1)]. Besides deletions, formation of pseudogenes also plays an important part in gene decay, leading to many important characteristics in favour of Shigella pathogenesis.
For many pathogenic bacteria, flagella are responsible for chemotaxis and play a role in tissue invasion (57). Conversely, mammalian hosts detect the conserved domain on flagellin monomers through the Toll-like receptor (TLR)-5, which triggers pro-inflammatory and adaptive immune responses (57). Shigella spends most of the time intracellularly during infection and is very mobile within the cells by polymerising actin using IcsA. Therefore, flagella synthesis is inactivated in all genomes via deletions as well as pseudogenes (Supplementary Table S2). This not only conserves energy but also allows evasion of TLR-5 mediated innate and adaptive immunity.
Adherence to the host cell surface via fimbriae is generally assumed to be important for bacteria to establish an infection. However, Shigella infective doses can be very low despite all the fimbrial genes orthologous to those of EHEC being inactivated. The YadA-like proteins are the only Shigella adhesins identified so far and are intact only in Ss046. Perhaps, the efficient invasion mechanism through Ipa proteins and the TTSS has overcome the need for fimbriae and other adherence factors, which has led to the inactivation of the fimbrial genes as well as the yadA genes. While it is important to investigate the significance of the intact YadA in Ss046 for virulence, sequencing or performing northern blotting on more epidemiologic S.sonnei strains will indicate whether or not the yadA gene is prone to gene decay.
In summary, the Shigellas have evolved from different strains of E.coli and have become highly specific human pathogens through extensive convergent evolution involving gain and loss of functions. A similar scenario is also observed in Typhi and Paratyphi A of S.enterica; through convergent evolution, mainly involving gene decay, these pathogens have become human restricted, with similar virulence properties (58). This study has provided valuable information for further investigation of the pathogenicity, epidemiology and virulence of one of the important human pathogens, and provides some insight into how these pathogens have evolved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
The authors thank P. J. Sansonetti and K. Turner for critical reading of the manuscript. This work was funded by the National Basic Research Priorities Program (grant no. 2005CB522904) and the High Technology Research and Development Program (grant no. 2001AA223011) from the Ministry of Science and Technology of China. Funding to pay the Open Access publication charges for this article was provided by MSTC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hale T.L. Genetic basis of virulence in Shigella species. Microbiol. Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff K.L., Winickoff J.P., Ivanoff B., Clemens J.D., Swerdlow D.L., Sansonetti P.J., Adak G.K., Levine M.M. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 1999;77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 3.Sansonetti P.J. Slaying the Hydra all at once or head by head? Nature Med. 1998;4:499–500. doi: 10.1038/nm0598supp-499. [DOI] [PubMed] [Google Scholar]
- 4.Cheasty T., Rowe B. Antigenic relationships between the enteroinvasive Escherichia coli O antigens O28ac, O112ac, O124, O136, O143, O144, O152, and O164 and Shigella O antigens. J. Clin. Microbiol. 1983;17:681–684. doi: 10.1128/jcm.17.4.681-684.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan R., Lumb B., Ryan D., Reeves P.R. Molecular evolution of large virulence plasmid in Shigella clones and enteroinvasive Escherichia coli. Infect. Immun. 2001;69:6303–6309. doi: 10.1128/IAI.69.10.6303-6309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sansonetti P.J. Microbes and microbial toxins: paradigms for microbial-mucosal interactions III. Shigellosis: from symptoms to molecular pathogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G319–G323. doi: 10.1152/ajpgi.2001.280.3.G319. [DOI] [PubMed] [Google Scholar]
- 7.Bernardini M.L., Mounier J., d'Hauteville H., Coquis-Rondon M., Sansonetti P.J. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl Acad. Sci. USA. 1989;86:3867–3871. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sansonetti P.J., Hale T.L., Dammin G.J., Kapfer C., Collins H.H., Jr, Formal S.B. Alterations in the pathogenicity of Escherichia coli K-12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect. Immun. 1983;39:1392–1402. doi: 10.1128/iai.39.3.1392-1402.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajakumar K., Sasakawa C., Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect. Immun. 1997;65:4606–4614. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moss J.E., Cardozo T.J., Zychlinsky A., Groisman E.A. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 1999;33:74–83. doi: 10.1046/j.1365-2958.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- 11.Jin Q., Yuan Z., Xu J., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., et al. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J., Goldberg M.B., Burland V., Venkatesan M.M., Deng W., Fournier G., Mayhew G.F., Plunkett G., III, Rose D.J., Darling A., et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 2003;71:2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pupo G.M., Lan R., Reeves P.R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl Acad. Sci. USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 15.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 16.Yang J., Wang J., Yao Z.J., Jin Q., Shen Y., Chen R. GenomeComp: a visualization tool for microbial genome comparison. J. Microbiol. Methods. 2003;54:423–426. doi: 10.1016/s0167-7012(03)00094-0. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Chen L., Yu J., Sun L., Jin Q. ShiBASE: an integrated database for comparative genomics of Shigella. Nucleic Acids Res. 2006;34:xxx–xxx. doi: 10.1093/nar/gkj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sayeed S., Reaves L., Radnedge L., Austin S. The stability region of the large virulence plasmid of Shigella flexneri encodes an efficient postsegregational killing system. J. Bacteriol. 2000;182:2416–2421. doi: 10.1128/jb.182.9.2416-2421.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahassi E.M., O'Dea M.H., Allali N., Messens J., Gellert M., Couturier M. Interactions of CcdB with DNA gyrase. Inactivation of Gyra, poisoning of the gyrase–DNA complex, and the antidote action of CcdA. J. Biol. Chem. 1999;274:10936–10944. doi: 10.1074/jbc.274.16.10936. [DOI] [PubMed] [Google Scholar]
- 21.Lan R., Stevenson G., Reeves P.R. Comparison of two major forms of the Shigella virulence plasmid pINV: positive selection is a major force driving the divergence. Infect. Immun. 2003;71:6298–6306. doi: 10.1128/IAI.71.11.6298-6306.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 23.Shu S., Setianingrum E., Zhao L., Li Z., Xu H., Kawamura Y., Ezaki T. I-CeuI fragment analysis of the Shigella species: evidence for large-scale chromosome rearrangement in S.dysenteriae and S.flexneri. FEMS Microbiol. Lett. 2000;182:93–98. doi: 10.1111/j.1574-6968.2000.tb08880.x. [DOI] [PubMed] [Google Scholar]
- 24.Schneider D., Duperchy E., Coursange E., Lenski R.E., Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tillier E.R., Collins R.A. Genome rearrangement by replication-directed translocation. Nature Genet. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 26.Nakata N., Tobe T., Fukuda I., Suzuki T., Komatsu K., Yoshikawa M., Sasakawa C. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol. 1993;9:459–468. doi: 10.1111/j.1365-2958.1993.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 27.Maurelli A.T., Fernandez R.E., Bloch C.A., Rode C.K., Fasano A. ‘Black holes’ and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl Acad. Sci. USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perna N.T., Mayhew G.F., Posfai G., Elliott S., Donnenberg M.S., Kaper J.B., Blattner F.R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shepherd J.G., Wang L., Reeves P.R. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from P.shigelloides in a recent event. Infect. Immun. 2000;68:6056–6061. doi: 10.1128/iai.68.10.6056-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjelloun-Touimi Z., Sansonetti P.J., Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol. Microbiol. 1995;17:123–135. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 31.Henderson I.R., Czeczulin J., Eslava C., Noriega F., Nataro J.P. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 1999;67:5587–5596. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al Hasani K., Henderson I.R., Sakellaris H., Rajakumar K., Grant T., Nataro J.P., Robins-Browne R., Adler B. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect. Immun. 2000;68:2457–2463. doi: 10.1128/iai.68.5.2457-2463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fasano A., Noriega F.R., Maneval D.R., Jr, Chanasongcram S., Russell R., Guandalini S., Levine M.M. Shigella enterotoxin 1: an enterotoxin of Shigella flexneri 2a active in rabbit small intestine in vivo and in vitro. J. Clin. Invest. 1995;95:2853–2861. doi: 10.1172/JCI117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purdy G.E., Payne S.M. The SHI-3 iron transport island of Shigella boydii 0–1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 2001;183:4176–4182. doi: 10.1128/JB.183.14.4176-4182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyckoff E.E., Duncan D., Torres A.G., Mills M., Maase K., Payne S.M. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 1998;28:1139–1152. doi: 10.1046/j.1365-2958.1998.00873.x. [DOI] [PubMed] [Google Scholar]
- 36.Baumler A.J., Norris T.L., Lasco T., Voight W., Reissbrodt R., Rabsch W., Heffron F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Prada C.M., Hoover D.L., Tall B.D., Hartman A.B., Kopelowitz J., Venkatesan M.M. Shigella flexneri IpaH(7.8) facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun. 2000;68:3608–3619. doi: 10.1128/iai.68.6.3608-3619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyotome T., Suzuki T., Kuwae A., Nonaka T., Fukuda H., Imajoh-Ohmi S., Toyofuku T., Hori M., Sasakawa C. Shigella protein IpaH(9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 2001;276:32071–32079. doi: 10.1074/jbc.M101882200. [DOI] [PubMed] [Google Scholar]
- 39.Tauschek M., Gorrell R.J., Strugnell R.A., Robins-Browne R.M. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandkvist M., Michel L.O., Hough L.P., Morales V.M., Bagdasarian M., Koomey M., DiRita V.J., Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonough M.A., Butterton J.R. Spontaneous tandem amplification and deletion of the shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 1999;34:1058–1069. doi: 10.1046/j.1365-2958.1999.01669.x. [DOI] [PubMed] [Google Scholar]
- 42.Perna N.T., Plunkett G., III, Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 43.Eitel J., Dersch P. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect. Immun. 2002;70:4880–4891. doi: 10.1128/IAI.70.9.4880-4891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid Y., Grassl G.A., Buhler O.T., Skurnik M., Autenrieth I.B., Bohn E. Yersinia enterocolitica adhesin A induces production of interleukin-8 in epithelial cells. Infect. Immun. 2004;72:6780–6789. doi: 10.1128/IAI.72.12.6780-6789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lan R., Alles M.C., Donohoe K., Martinez M.B., Reeves P.R. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 2004;72:5080–5088. doi: 10.1128/IAI.72.9.5080-5088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukiya S., Mizoguchi H., Tobe T., Mori H. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 2004;186:3911–3921. doi: 10.1128/JB.186.12.3911-3921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fontaine A., Arondel J., Sansonetti P.J. Role of Shiga toxin in the pathogenesis of bacillary dysentery, studied by using a Tox- mutant of Shigella dysenteriae 1. Infect. Immun. 1988;56:3099–3109. doi: 10.1128/iai.56.12.3099-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strauch E., Lurz R., Beutin L. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 2001;69:7588–7595. doi: 10.1128/IAI.69.12.7588-7595.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Recktenwald J., Schmidt H. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage phiP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 2002;70:1896–1908. doi: 10.1128/IAI.70.4.1896-1908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escobar-Paramo P., Clermont O., Blanc-Potard A.B., Bui H., Le Bouguenec C., Denamur E. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 2004;21:1085–1094. doi: 10.1093/molbev/msh118. [DOI] [PubMed] [Google Scholar]
- 51.Mills M., Payne S.M. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 1997;65:5358–5363. doi: 10.1128/iai.65.12.5358-5363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skaar E.P., Humayun M., Bae T., DeBord K.L., Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 53.Lawlor K.M., Daskaleros P.A., Robinson R.E., Payne S.M. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect. Immun. 1987;55:594–599. doi: 10.1128/iai.55.3.594-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keusch G.T., Bennish M.L. Shigellosis. In: Evans A.S., Brachman P.S., editors. Bacterial Infections of Humans. NY: Plenum Publishing Co.; 1998. pp. 631–656. [Google Scholar]
- 55.Ohtsubo H., Nyman K., Doroszkiewicz W., Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981;292:640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- 56.Cole S.T., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 57.Ramos H.C., Rumbo M., Sirard J.C. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 2004;12:509–517. doi: 10.1016/j.tim.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 58.McClelland M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nature Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete genome sequences have been deposited in the GenBank. The accession numbers for chromosomes and virulence plasmids are Sd197, CP000034 and CP000035; Sb227, CP000036 and CP000037; Ss046, CP000038 and CP000039. In addition, genome annotation and comparative analysis can be obtained at ShiBASE (http://www.mgc.ac.cn/ShiBASE/) (18).