Abstract
Adaptive regulation of gene expression in response to environmental changes is a general property of bacterial pathogens. By screening an ordered transposon mutagenesis library of Mycobacterium tuberculosis, we have identified three mutants containing a transposon in the coding sequence or in the 5′ regions of genes coding for two-component signal transduction systems (trcS, regX3, prrA). The intracellular multiplication capacity of the three mutants was investigated in mouse bone marrow-derived macrophages. Only the prrA mutant showed a defect in intracellular growth during the early phase of infection, and this defect was fully reverted when the mutant was complemented with prrA-prrB wild-type copies. The mutant phenotype was transient, as after 1 week this strain recovered full growth capacity to reach levels similar to that of the wild type at day 9. Moreover, a transient induction of prrA promoter activity was observed during the initial phase of macrophage infection, as shown by a prrA promoter-gfp fusion in M. bovis BCG infecting the mouse macrophages. The concordant transience of the prrA mutant phenotype and prrA promoter activity indicates that the PrrA-PrrB two-component system is involved in the environmental adaptation of M. tuberculosis, specifically in an early phase of the intracellular growth, and that, similar to other facultative intracellular parasites, M. tuberculosis can use genes temporarily required at different stages in the course of macrophage infection.
Most bacterial pathogens modulate their gene expression in response to the diverse environmental changes encountered during infection. This property may be especially advantageous for intracellular pathogens to adapt to inner compartments of host cells. The capacity of Mycobacterium tuberculosis to survive and multiply intracellularly likely plays a key role in making this bacterium one of the most successful human pathogens (for a review, see reference 3). Several studies, mainly based on RNA analysis or on the use of green fluorescent protein, have led to the identification of M. tuberculosis genes regulated in response to phagocytosis, (e.g., see references 7, 16, 24, 27, and 29). However, regulatory systems controlling intracellular gene expression have not been identified yet.
In bacteria, two-component systems provide a conserved mechanism for the adaptive regulation of gene expression in response to a wide variety of environmental signals. These systems involve a histidine kinase sensor protein and a cognate response regulator, which classically controls transcription of target genes (20). They regulate important and diverse physiological or metabolic processes such as stress responses, nutrient utilization, quorum sensing, or cell division (12). Moreover, they frequently play a critical role in virulence, by coordinating expression of genes involved in pathogenesis (e.g., see reference 5). Some of them have been shown to be required for entry, survival, or multiplication inside host cells, such as for Salmonella spp. (e.g., see references 8, 14, and 15), Yersinia (19) and Brucella spp. (25).
The genome of M. tuberculosis has the potential to encode 11 complete two-component systems (1). Their function and the signals sensed by these systems are not known, and so far only four of them, MtrA-MtrB (Rv3246c-Rv3245c), SenX3-RegX3 (Rv0490-Rv0491), TrcS-TrcR (Rv1032c-1033c), and DevR-DevS (Rv3132c-3133c), have been partially characterized (2, 9, 11, 26, 28). Expression of the Mycobacterium bovis BCG mtrA gene has been shown to be upregulated in murine macrophages (4, 28). In contrast, in M. tuberculosis this gene is not regulated in macrophages and was found to be indispensable for this organism (30). Recently, the expression of another M. tuberculosis two-component system operon, designated prrA-prrB, was found, by selective capture of transcribed sequences, to be induced in human primary macrophages (7). However, the implication of these genes in intracellular growth of M. tuberculosis has not been elucidated up to now.
In an effort to study the function of M. tuberculosis two-component systems, we have screened an ordered transposon mutagenesis library (13) for mutants deficient in these systems. We describe here the identification of three mutants affected in the genes coding for Rv0491 (RegX3 response regulator), Rv0903c (PrrA response regulator), and Rv1032c (TrcS histidine kinase), respectively. The consequences of these mutations for survival and multiplication in murine macrophages were investigated, and the intracellular expression profiles of the corresponding genes were analyzed.
MATERIALS AND METHODS
Bacterial strains.
M. tuberculosis Mt103 is a wild-type strain which was isolated from a tuberculosis patient (13). The BCG Pasteur strain (isolate 1173P2; World Health Organization, Stockholm, Sweden) was used for the electroporation of the pFluo plasmids. All mycobacterial strains were grown in liquid Sauton medium (23), in Middlebrook 7H9 medium (Difco) supplemented with 0.05% Tween 80 and ADC (0.2% dextrose, 0.5% bovine serum albumin fraction V, 0.0003% beef catalase), or on solid Middlebrook 7H10 medium (Difco) supplemented with OADC (0.005% oleic acid, 0.2% dextrose, 0.5% bovine serum albumin fraction V, 0.085% NaCl, 0.0003% beef catalase). Kanamycin and hygromycin were added when required at a concentration of 20 and 50 μg ml−1, respectively. All cloning steps were carried out in Escherichia coli XL1-blue (Stratagene, La Jolla, Calif.).
Identification of M. tuberculosis mutants deficient in two-component systems.
The M. tuberculosis mutants were isolated by PCR screening of an ordered transposon mutagenesis library (13) as follows. PCR amplifications were carried out on a GenAmp PCR system 9600 (Perkin-Elmer), using the OP primer specific for the inverted repeats of IS1096 and directed outwards from the transposon (13) and primers 5′-CCTCGCGACTCGGTTGCAACGCT-3′ (prrA up) or 5′-CGTGATGAGGGCGTCCTGGTCGTTC-3′ (prrA down), 5′-CCTGGGTCGGACACGGCGTGTT-3′ (trcR up) or 5′-GTCGTGCAGCACGGCAATGAGGTT-3′ (trcR down), and 5′-GGGTGCTGGGCGACCAAACTCTG-3′ (regX3 up) or 5′-CGGCGCCGACGAGCAGCTTT-3′ (regX3 down) for prrB-prrA, trcS-trcR, and senX3-regX3, respectively. PCR fragments were purified by using the Geneclean II kit (Bio 101) and directly sequenced using an automated DNA sequencer (model 373; Applied Biosystems), a dye deoxy terminator cycle sequencing kit (Applied Biosystems) and the oligonucleotide prrA up, trcR down, or regX3 1465 (5′-GCAGGCGCGAGAGCCCGAAC-3′).
DNA extraction and Southern blot analysis.
DNA was extracted from the mycobacteria as described previously (21), except that no d-cycloserine was added to the bacterial culture. After enzyme digestion of the DNA and electrophoresis under standard conditions, the agarose gels were incubated in 0.25 N HCl for 15 min, and Southern blotting was carried out overnight in 0.4 N NaOH, using positively charged nylon membranes (Boehringer). Membranes were rinsed four times in 6× SSC (0.9 M NaCl, 0.09 M C6H5Na3O7 · 2H2O, pH 7.0). The Megaprime random-primed labeling kit (Amersham) and 5 μCi of [α-32P]dCTP (6,000 mCi/mmol) were used to label probes. The entire Tn5368 transposon was isolated as an NcoI-NotI fragment from pPR32 (21) and used as the transposon probe. The senX3-regX3 probe was obtained as an EcoRI-BamHI fragment from pRegX3Bc1 (26), while the prrA and trcS probes were DNA fragments obtained by PCR using the pairs of oligonucleotides 5′-AAAAGATCTATGGGCGGCATGGACACTGGTGTGA-3′ and 5′-AAAAAGCTTATTCTGCATACGCAGCACGAATCCGACT-3′ and 5′-AAGGATCCGGAATCGTCGGGCACAC-3′ and 5′-AAAAAGCTTCAGGCGGTAGTGGCGATCTGCTGC-3′, respectively. Prehybridization and hybridization were carried out at 68°C using 6× SSC, 5× Denhardt's reagent (containing [per liter] 1 g of Ficoll Type 400, 1 g of polyvinylpyrrolidone, and 1 g of bovine serum albumin), 0.5% sodium dodecyl sulfate (SDS), and salmon sperm DNA (100 μg ml−1). Serial washes were performed under the following conditions: two washes in 2× SSC-0.5% SDS at room temperature for 5 min, two washes in 0.2× SSC-0.1% SDS at room temperature for 15 min, two washes in 0.2× SSC-0.1% SDS at 42°C for 15 min, and two washes in 0.1× SSC-0.1% SDS at 68°C for 15 min. X-Omat AR X-ray film (Kodak) was exposed overnight to the blots at −80°C.
Plasmid constructions.
The prrB-prrA, trcR-trcS, and senX3-regX3 operons were amplified by PCR from M. tuberculosis Mt103 genomic DNA using the following pairs of oligonucleotides: 5′-GCTCTAGATGAGGTCAGGGCCGCAGGGCTAAC-3′ and 5′-CCGCTCGAGGCTACCGCCGGATCGGGTTG-3′, 5′-GGACTAGTCTCTCAGGCGGTAGTGGCGATCTG-3′ and 5′-CCCACAACCTGGTCACGATCAG-3′, and 5′-GGATCCAATTGTTTGAGATCCCACCTGC-3′ and 5′-CTAGCCCTCGAGTTTGTAGCCCAG-3′, respectively. The senX3-regX3 PCR product was digested with BamHI (the BamHI site is underlined) and cloned into the pBSh-D shuttle vector (22), previously digested with BamHI and EcoRV. This gave rise to pBSh-D-SRX3. The prrA-prrB and trcR-trcS products were first introduced into the EcoRV site of pZero-2 (Invitrogen). The resulting plasmids were then digested with NotI, blunt ended, and then digested with BamHI, and the inserts were cloned into pBSh-D, resulting in pBSh-D-902/3c and pBSh-D-1032/3c, respectively.
The 5′ regions of prrA, trcR, and senX3 were amplified by PCR from pBSh-D-902/3c, pBSh-D-1032/3c, and pBSh-D-SRX3 using the following pairs of oligonucleotides: 5′-AAGGATCCTTACACCCGAGGTGAGGTCACAC-3′ and 5′-AAAGATCTGCTACCGCCGGATCGGGTTG-3′, 5′-TTGGATCCTCAGATGGCTTGCCGCGGACGTTGAC-3′ and 5′-GAAGATCTCCCACAACCTGGTCACGATCAG-3′, and 5′-AGATCTTTACGAGAACACAGTCACAAGGAA-3′ and 5′-GGATCCTGGTGCGCGGTGGTACGTATC-3′, respectively. These sequences included in-frame stop codons (italicized) and BamHI or BglII sites (underlined). The PCR products digested with BamHI or BglII were cloned into the BamHI and ScaI sites of pJFX2 (27), yielding pFluo1, pFluo2, and pFluo5, respectively. All constructs were verified by sequencing.
Preparation and infection of mouse bone marrow-derived macrophages.
Bone marrow cells were prepared and infected as previously described (13), with minor modifications. Bone marrow cells were flushed from the femurs of 7- to 8-week-old BALB/c mice and suspended in Dulbecco modified Eagle medium (DMEM) with Glutamax II and a low glucose (1 g liter−1) concentration (Gibco BRL) and enriched with 10% heat-inactivated fetal calf serum (Gibco BRL) and macrophage colony-stimulating factor (10 ng ml−1; R&D Systems). For the infection assays, the macrophages were seeded in 24-well cell culture clusters (Techo Plastic Products) (2 × 105 cells per well in a 1-ml volume) and allowed to differentiate for 6 to 8 days. Aliquots (500 μl) of mycobacterial suspensions were centrifuged at 4,000 × g for 15 min and then resuspended in 500 μl of Sauton medium. Bacterial aggregates were sedimented by centrifugation at 20 × g for 10 min. The bacterial supernatant was recovered, and bacterial concentrations were adjusted to 2 × 104 bacteria ml−1 with DMEM. An aliquot of each M. tuberculosis suspension used to infect the macrophages was plated onto Middlebrook 7H10 agar to establish the number of live bacteria in the inoculum.
The infection assay was as follows. The culture medium of each macrophage-containing well was removed, and 1 ml of the mycobacterial suspension was added to obtain a multiplicity of infection (MOI) of 0.1:1. Control wells containing noninfected macrophages were filled with 1 ml of fresh culture medium. Infected and noninfected cultures were incubated at 37°C in a 5% CO2 atmosphere for 18 h. Infection was terminated by removing the overlying medium and washing each well three times with 1 ml of DMEM (Gibco BRL) before adding 1 ml of fresh culture medium per well. At days 1, 3, 6, and 9, the numbers of intracellular CFU were evaluated for four independent wells containing infected macrophages for each of the three mutant strains and the wild-type M. tuberculosis strains. For this purpose, macrophage monolayers were washed three times in DMEM buffer and then lysed in 200 μl of phosphate-buffered saline (PBS) containing 0.1% Triton X-100 (Sigma). Different dilutions of the lysates were plated onto 7H10, 7H10-kanamycin, or 7H10-hygromycin agar to perform counts of Mt103 and the mutant and complemented mutant strains. The experiment was carried out three times independently.
Mouse infection.
For intravenous infection experiments, 8-week-old female BALB/c mice were infected in the thin-layer vein with 106 viable units of parental or mutant strains. At indicated times, mice were sacrificed, and individual lungs, liver, and spleen were removed and homogenized. Serial dilutions were plated onto 7H10 medium for colony counting. For aerosol infection, C57BL/6J mice purchased from The Jackson Laboratory (Bar Harbor, Maine) were maintained under specific-pathogen-free conditions on sterile bedding, with water and mouse chow administered ad libitum. Mice were infected using a Glas-Col (Terre Haute, Ind.) aerosol generator such that bacteria were deposited in the lungs of each animal. The numbers of viable bacteria in target organs were determined at various time points by plating serial dilutions of lung homogenates on nutrient Middlebrook 7H11 agar for colony counting. Mice were killed by CO2 asphyxiation, and the lungs were aseptically excised. Each of these organs was individually homogenized in physiological saline solution, and serial dilutions of the organ homogenate were plated on nutrient 7H11 agar. For all experiments, a determination of the infecting dose was performed on lungs of mice infected for 1 day.
Flow cytometry.
Bone marrow-derived macrophages were prepared and infected as described above at an MOI of 1:1 by BCG containing pFluo1, pFluo2, pFluo5, pJFX2 (negative control), or pFXJ4 (positive control) grown in minimal Sauton medium. After 4 h of infection, extracellular bacteria were removed by washing three times with DMEM, and incubation was continued at 37°C under 5% CO2. At different time points after infection, the macrophages were washed three times with PBS and incubated with 50 μl of trypsin (2.5 g liter−1 [Gibco BRL]) at 37°C for 5 min. Cells were then resuspended in 200 μl of PBS and analyzed with a FACSCalibur (Becton Dickinson). The flow cytometer was calibrated to measure only the fluorescence of the infected macrophage cell population and not of the free bacilli, by using side and forward scatter parameters of a noninfected macrophage suspension as a standard reference. This reference cell suspension was also used to eliminate the effects of macrophage autofluorescence.
RESULTS
Identification of mutations in two-component system genes in an ordered transposon mutagenesis library of M. tuberculosis.
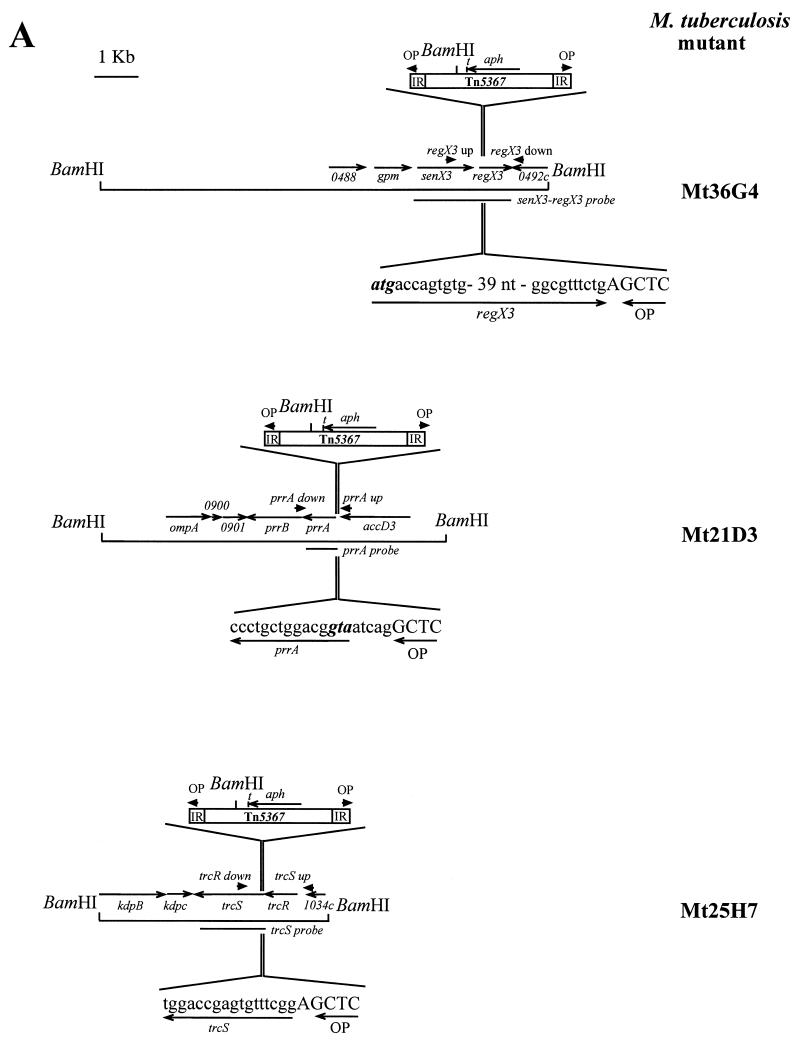
The 6,912 M. tuberculosis transposition mutants assembled in 72 pools of 96 mutants each were screened by PCR using a strategy similar to that previously described (13). Primer OP, specific for the inverted repeats at both extremities of IS1096 and directed outward from the transposon, was used together with primer prrA up, trcR up, or regX3 up. These primers are located 183, 244, and 667 bp upstream of the prrA, trcR, and regX3 start codons, respectively, in the M. tuberculosis H37Rv genome (Fig. 1A). PCR amplifications were performed with the 72 pools of chromosomal DNA, and the PCR products with a size consistent with an insertion of the transposon within or near the prrA-prrB, trcR-trcS, and senX3-regX3 operons were identified. Amplicons with a size of about 220 and 1,000 bp were obtained for pools 21 and 25 with primers OP-prrA up and OP-trcR up, respectively, while three pools (pools 29, 36, and 37) yielded products of about 750 bp with primers OP-regX3 up. These sizes are consistent with insertions near the start codons of prrA and trcS and in the 5′ part of the regX3 coding region, respectively. To confirm the location of the insertion sites, PCR amplifications were performed on the same pools using primer OP together with primers prrA down, trcR down, and regX3 down, located on the opposite side of the predicted insertion sites. Unique amplification products with the expected sizes were obtained for each of the relevant pools (1,000 bp for pool 21; 400 bp for pool 25; and 850 bp for pools 29, 36, and 37). Sequences of the PCR products obtained with primers OP-prrA up, OP-trcR down, and OP-regX3 up indicated that the transposon was inserted 5 nucleotides upstream of the predicted prrA start codon, after nucleotide 12 of the trcS open reading frame (ORF) and after nucleotide 57 of the regX3 ORF, respectively (Fig. 1A). The chromosomal DNA of each of the 96 mutants of pools 21, 25, and 36 was individually analyzed using primer OP with primers prrA up, trcR down, and regX3 up, respectively. One amplification product of the expected size was obtained from a single DNA sample for pools 21 and 25, corresponding to mutants Mt21D3 and Mt25H7, respectively. Three different samples of pool 36 gave the same PCR fragment with the expected size. One of the three (Mt36G4) was selected for further analysis.
FIG. 1.
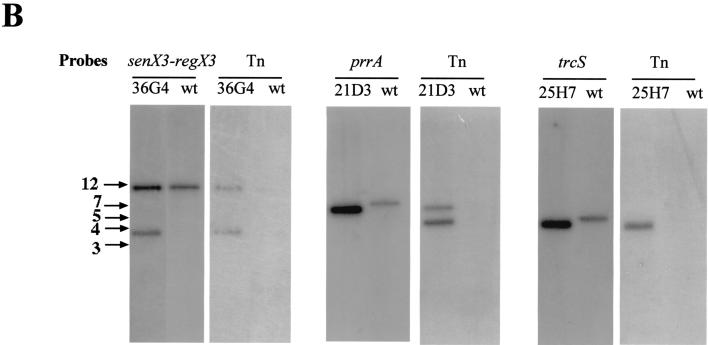
Identification by PCR of mutations in two-component system genes among an M. tuberculosis transposon mutagenesis library. (A) The genes mutated by Tn5367 insertion are shown together with flanking genes and chromosomal BamHI sites. Arrowheads labeled OP or regX3, prrA, and trcS up or down indicate the positions of the primers used for the PCR screening. IR and t designate the inverted repeats and the trp transcriptional terminator present in the transposon, respectively. The sequence of the Tn5367 insertion sites in the three mutants is indicated. (B) Southern blot analysis of the mutants. Genomic DNA was digested with BamHI and probed with fragments of the target genes (shown in Fig. 1A) or with the entire transposon. Arrows on the left indicate the positions of DNA size markers (sizes are expressed in kilobases). wt, wild type.
DNA from the wild-type M. tuberculosis parent strain Mt103 and mutants Mt21D3, Mt25H7, and Mt36G4 was restricted with BamHI, which cuts once in the transposon, and analyzed by Southern blot hybridization using various probes corresponding to the transposon or to the different target genes (Fig. 1B). Hybridization of DNA from Mt21D3 with a prrA probe, from Mt25H7 with a trcS probe, and from Mt36G4 with a senX3-regX3 probe showed a shift in size of a BamHI fragment between the wild-type and each mutant DNA, confirming the transposon insertion in the probed genes. Hybridization with the transposon probe revealed two signals for Mt21D3 and Mt36G4 and one signal for Mt25H7, which from the position of the insertion identified by sequencing was concluded to be a doublet. In each case the size of one of the fragments corresponded to that of the fragment obtained with the target gene probe, further confirming that these regions were disrupted by a transposon insertion in each of the three mutants orientated as shown in Fig. 1A.
Survival and multiplication of Mt21D3, Mt25H7, and Mt36G4 within macrophages.
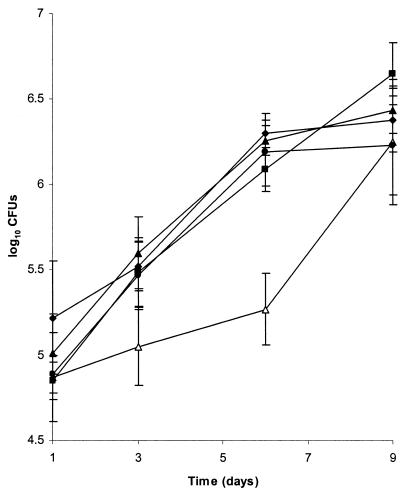
Mutants Mt21D3, Mt25H7, and Mt36G4 and the parental strain Mt103 showed similar growth rates in Middlebrook 7H9 medium-OADC and in minimal synthetic Sauton medium, and none of the mutants exhibited an extended lag phase, even when stationary-phase cultures were resuspended in fresh medium (data not shown). To investigate the effects of the transposon insertions on intracellular multiplication of the three mutants, we compared their growth rates with that of the wild-type strain within mouse bone marrow-derived macrophages. As shown in Fig. 2, mutants Mt25H7 and Mt36G4 had intracellular growth rates similar to that of the wild-type strain Mt103. In contrast, in three independent experiments, Mt21D3 showed a significant impairment in growth during the first days of infection, reaching an approximately 10-fold difference in CFU at day 6 compared to Mt103. However, after this initial period, Mt21D3 recovered full growth capacity to reach plateau levels of CFU similar to that of the wild type at day 9. The experiment could not be continued beyond day 9, because the infected macrophages started to undergo extensive lysis, as evidenced by microscopic observations. By using a recently constructed single-copy shuttle vector (22), we introduced the wild-type prrA-prrB genes into Mt21D3. When bone marrow-derived macrophages were infected with the complemented strain, named Mt21D3F, the intracellular growth kinetics were essentially identical to that of Mt103 (Fig. 2), indicating that wild-type prrA-prrB fully complemented the 21D3 mutation. Quantitative analysis of the Mt21D3F colonies recovered from infected macrophages indicated that the complementing plasmid was stably maintained during intracellular multiplication (data not shown).
FIG. 2.
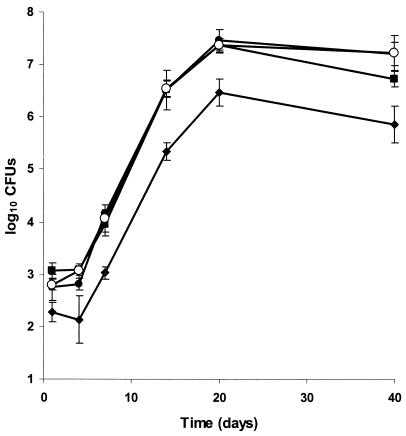
Growth of Mt21D3, Mt21D3F, Mt25H7, Mt36G4, and Mt103 in mouse bone marrow-derived macrophages. Macrophages were infected at an MOI of 0.1:1 with 104 CFU of Mt21D3 (open triangles), Mt21D3F (filled triangles), Mt25H7 (circles), Mt36G4 (diamonds), or Mt103 (squares). At days 1, 3, 6, and 9, intracellular bacteria were recovered and plated for counting. For each time point and each strain, results are expressed as means ± standard deviations (error bars) of CFU counts of quadruplicate wells.
Analysis of prrA-prrB, trcR-trcS, and senX3-regX3 gene expression.
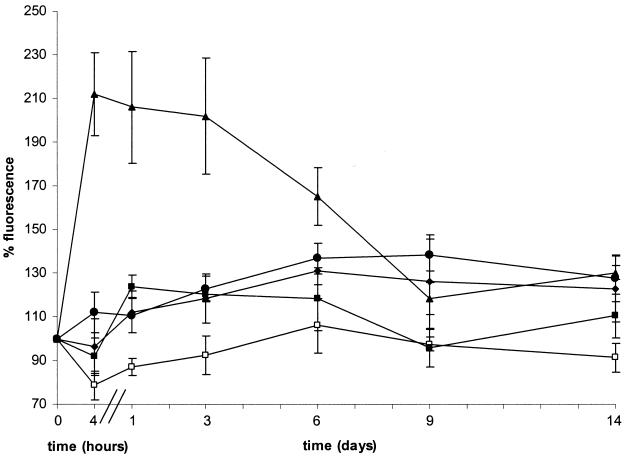
To investigate a potential relationship between the above phenotypes and intracellular expression profiles of the corresponding genes, we constructed transcriptional fusions of the 5′ regions of prrA, of trcR, or of senX3 with a gfp reporter gene. These constructs were then introduced into M. bovis BCG, and the recombinant BCG strains were analyzed by flow cytometry. Under axenic conditions no fluorescence above background levels was detected for recombinant BCG containing prrA-gfp, indicating that prrA is not expressed under these conditions (Fig. 3). However, as soon as 4 h after infection of mouse bone marrow-derived macrophages, fluorescence intensity increased about twofold above background levels. This fluorescence level was maintained during the first 3 days of infection and then gradually decreased to reach background levels again at day 9. By comparison, fluorescence levels seven times higher than background were detected under axenic conditions with recombinant BCG producing green fluorescent protein under the control of promoter Pbla. This fluorescence level was maintained throughout the entire duration of macrophage infection. No fluorescence induction was observed when recombinant BCG containing prrA-gfp was incubated with fresh tissue culture medium or with culture supernatants from macrophages infected or not with BCG (not shown).
FIG. 3.
Activity of the prrA, trcS, and senX3 promoters. Bone marrow-derived macrophages were infected at an MOI of 1:1 with BCG containing pFluo1 (prrA [filled triangles]), pFluo2 (trcS [filled circles]), pFluo5 (senX3 [filled diamonds]), pJFX2 (negative control [open squares]), or pFXJ4 (positive control [filled squares]). After 4 h and 1, 3, 6, 9, and 14 days, the mean fluorescence intensities of infected macrophages were analyzed as described in Materials and Methods. The relative fluorescence (representing means ± standard deviations [error bars] from four independent assays) is expressed as a percentage of the mean fluorescence at time zero (set at 100%) corresponding to recombinant BCG strains in minimal Sauton medium. The mean fluorescences at time zero were 38, 43, 119, 48, and 325 arbitrary units for BCG containing pFluo1, pFluo2, pFluo5, pJFX2, and pFXJ4, respectively.
Unlike what was observed for prrA-gfp, BCG containing senX3-gfp showed significant fluorescence under axenic culture conditions (three times higher than background levels), consistent with our earlier results obtained with a lacZ reporter gene fusion (26). The levels of fluorescence did not change significantly upon phagocytosis and remained high during the entire experiment, indicating that the expression of senX3-regX3 is not modulated in the intracellular compartments of bone marrow-derived macrophages. Finally, very low fluorescence levels were detected for recombinant BCG containing trcR-gfp, regardless of whether the cells were grown under axenic conditions or after infection of macrophages.
Survival and multiplication of Mt21D3, Mt25H7, and Mt36G4 in mice.
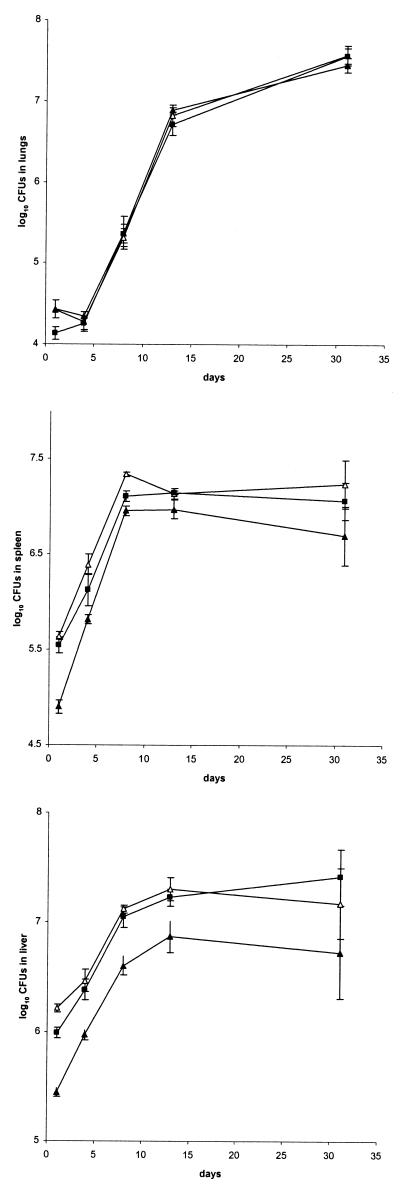
Finally, we investigated the consequences of the transposon insertions in the three M. tuberculosis mutants on their multiplication capacity within the lungs, by aerosol infection of BALB/c mice with 103 CFU of each of the three mutants or of Mt103. The mycobacterial loads of the lungs were analyzed up to 40 days after infection. As shown in Fig. 4, all three mutants showed growth kinetics similar to that of Mt103. Mutant Mt21D3 was further tested by infecting BALB/c mice intravenously with 106 CFU of Mt21D3 or Mt103. Analysis of the mycobacterial loads in the lungs, the spleen, and the liver revealed no difference in the growth rates in any of the organs between the mutant and the wild-type strain, up to 31 days after infection (Fig. 5).
FIG. 4.
Growth of Mt21D3, Mt25H7, Mt36G4, and Mt103 in the lungs of aerosol-infected C57BL/6J mice. For each time point and each strain—Mt21D3 (open circles), Mt25H7 (filled circles), Mt36G4 (diamonds), and Mt103 (squares)—the results are expressed as means ± standard deviations (error bars) of CFU counts for at least three mice.
FIG. 5.
Growth of Mt21D3 and Mt103 in the lungs (upper panel), spleens (middle panel), and livers (lower panel) of BALB/c mice infected intravenously. For each time point and each strain—Mt21D3 (open triangles), Mt21D3F (filled triangles), and Mt103 (squares)—results are expressed as means ± standard deviations (error bars) of CFU counts for at least three mice.
DISCUSSION
In this study, we have systematically screened a recently constructed ordered transposition library for mutants deficient in two-component signal transduction systems (13). Ten out of the eleven two-component system operons present in the M. tuberculosis genome were targeted (data not shown), and three different mutants were identified. One of them contains an insertion within trcS, which codes for a sensor. The two other mutants contain insertions within regX3 and 5 nucleotides upstream of the prrA coding sequence, respectively. Each of these two genes encodes a response regulator. All three two-component system operons are present in BCG, as evidenced by PCR amplification from chromosomal BCG DNA (F. Ewann, unpublished observations).
prrA and prrB located 14 nucleotides downstream probably belong to the same operon (1). The gene downstream of prrB, which codes for a putative membrane protein, is oriented in the opposite direction. The gene upstream of prrA, encoding a subunit of an acetyl coenzyme A carboxylase involved in lipid metabolism, is separated from prrA-prrB by 132 bp. In mutant Mt21D3, the transposon located 5 nucleotides upstream of the prrA ORF (Fig. 1A) presumably prevents transcription of the prrA-prrB operon, as a transcriptional terminator is present at the 3′ end of the aph gene in the transposon (17). While Mt21D3 was unaffected in its in vitro growth, it was impaired in its intracellular multiplication capacity during the first days of mouse macrophage infection, reaching an approximately 10-fold difference in CFU at day 6 compared to the parent strain. This phenotype was completely reverted when the mutant was complemented with a wild-type prrA-prrB copy, which constitutes independent evidence for a functional defect of prrA-prrB in Mt21D3 and rules out a polar effect on downstream gene expression. Interestingly, after the initial days of infection, the mutant eventually recovered its growth capacity to reach a maximal level of infection similar to that of the wild-type strain. This indicates a transient effect of the mutation, which is overcome in later stages of infection. Concordant with this transient phenotype, expression of prrA was rapidly, but also transiently, induced during infection of macrophages, as evidenced by the use of a BCG strain containing a transcriptional prrA-gfp fusion. As the green fluorescent protein used in this study is a rather stable molecule, the lack of continued increase in fluorescence after the initial rapid induction suggests that prrA is expressed only shortly after infection and then rapidly switched off. These results are consistent with those obtained by Graham and Clark-Curtiss (7), who were able to isolate the prrA cDNA from M. tuberculosis phagocytosed by human macrophages but not from M. tuberculosis grown in broth culture. Altogether the results indicate that the two-component system encoded by prrA-prrB is involved specifically in an early step in intracellular multiplication of M. tuberculosis. Interestingly, such transient defects in intracellular multiplication have been recently reported for transposon mutants in various genes of another facultative intracellular pulmonary pathogen, Legionella pneumophila (6, 10). These mutants showed an initial lag in multiplication within macrophages but were also able to multiply as well as the wild-type strain after prolonged incubation. Taken together with these findings, the results described here suggest a rather complex course of macrophage infection by such pathogens, comprising different stages during which these bacteria use genes required in a timely fashion.
Transcription of other M. tuberculosis genes, such as those encoding sigma factors or proteins involved in cation transport or lipid and cell-wall metabolism or invasion, is also induced during the first days of human macrophage infection (7). One of these genes, encoding isocitrate lyase, has recently been shown to be required for persistence of M. tuberculosis in mice (18). The availability of the Mt21D3 mutant will now allow us to test whether induction upon phagocytosis of any of these genes is controlled by prrA-prrB.
Despite its growth defect in the early phase of macrophage infection ex vivo, mutant Mt21D3 was unaffected in its capacity to multiply in the organs of mice infected intravenously or by aerosol, compared to the wild-type strain. It is likely that the mouse model of acute infection used here is not sensitive enough to detect a role in the early phases of macrophage infection.
Unlike the expression of prrA-prrB, the expression of the genes encoding the two other two-component systems did not appear to be regulated within the macrophages, nor did their inactivation affect intracellular growth of M. tuberculosis within bone marrow-derived macrophages or in mice during the acute phase of infection. These results indicate that trcS-trcR and senX3-regX3 do not play a major role in the initial phases of infection in these models.
Acknowledgments
Christophe Guilhot is gratefully acknowledged for very helpful discussion and advice, Alain Baulard is thanked for critical reading of the manuscript, James Triccas is thanked for the gift of pFXJ plamids, and Jean Rauzier is thanked for DNA sequencing.
This work was supported by INSERM, Institut Pasteur de Lille, through a grant from the Ministère de l'Education Nationale, de la Recherche et de la Technologie and from the Indo-French Center for the Promotion of Advanced Research. F.E. holds a fellowship of the Région Nord-Pas-de-Calais. K.P. holds a fellowship of the Ministère de la Recherche. P.S. is a Chercheur du Centre National de Recherche Scientifique.
Editor: R. N. Moore
REFERENCES
- 1.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 2.Dasgupta, N., V. Kapur, K. K. Singh, T. K. Das, S. Sachdeva, K. Jyothisri, and J. S. Tyagi. 2000. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber. Lung Dis. 80:141-159. [DOI] [PubMed] [Google Scholar]
- 3.Deretic, V., and R. A. Fratti. 1999. Mycobacterium tuberculosis phagosome. Mol. Microbiol. 31:1603-1609. [DOI] [PubMed] [Google Scholar]
- 4.Dhandayuthapani, S., L. E. Via, C. A. Thomas, P. M. Horowitz, D. Deretic, and V. Deretic. 1995. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol. Microbiol. 17:901-912. [DOI] [PubMed] [Google Scholar]
- 5.Dziejman, M., and J. J. Mekalanos. 1995. Two-component signal transduction and its role in the expression of bacterial virulence factors, p. 305-317. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 6.Edelstein, P. H., M. A. C. Edelstein, F. Higa, and S. Falkow. 1999. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. USA 96:8190-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisman, E. A., E. Chiao, C. J. Lipps, and F. Heffron. 1989. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl. Acad. Sci. USA 86:7077-7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haydel, S. E., N. E. Dunlap, and W. H. Benjamin. 1999. In vitro evidence of two-component system phosphorylation between the Mycobacterium tuberculosis TrcR/TrcS proteins. Microb. Pathog. 26:195-206. [DOI] [PubMed] [Google Scholar]
- 10.Higa, F., and P. H. Edelstein. 2001. Potential virulence role of the Legionella pneumophila ptsP ortholog. Infect. Immun. 69:4782-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himpens, S., C. Locht, and P. Supply. 2000. Molecular characterization of the mycobacterial SenX3-RegX3 two-component system: evidence for autoregulation. Microbiology 146:3091-3098. [DOI] [PubMed] [Google Scholar]
- 12.Hoch, J. A., and T. J. Silhavy (ed.). 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 13.Jackson, M., C. Raynaud, M. A. Laneelle, C. Guilhot, C. Laurent-Winter, D. Ensergueix, B. Gicquel, and M. Daffe. 1999. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol. Microbiol. 31:1573-1587. [DOI] [PubMed] [Google Scholar]
- 14.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas, R. L., and C. A. Lee. 2000. Unraveling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 16.Mariani, F., G. Cappelli, G. Riccardi, and V. Colizzi. 2000. Mycobacterium tuberculosis H37Rv comparative gene-expression analysis in synthetic medium and human macrophage. Gene 253:281-291. [DOI] [PubMed] [Google Scholar]
- 17.McAdam, R. A., T. R. Weisbrod, J. Martin, J. D. Scuderi, A. M. Brown, J. D. Cirillo, B. R. Bloom, and W. R. Jacobs. 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect. Immun. 63:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 19.Oyston, P. C., N. Dorrell, K. Williams, S. R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 21.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picardeau, M., C. Le Dantec, and V. Vincent. 2000. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology 146:305-313. [DOI] [PubMed] [Google Scholar]
- 23.Sauton, B. 1912. Sur la nutrition minérale du bacille tuberculeux. C. R. Acad. Sci. Ser. III Sci. Vie 155:860-863. [Google Scholar]
- 24.Smith, I., O. Dussurget, G. M. Rodriguez, J. Timm, M. Gomez, J. Dubnau, B. Gold, and R. Manganelli. 1998. Extra and intracellular expression of Mycobacterium tuberculosis genes. Tuber. Lung Dis. 79:91-97. [DOI] [PubMed] [Google Scholar]
- 25.Sola-Landa, A., J. Pizarro-Cerda, M. J. Grillo, E. Moreno, I. Moriyon, J. M. Blasco, J. P. Gorvel, and I. Lopez-Goni. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 26.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 27.Triccas, J. A., F. X. Berthet, V. Pelicic, and B. Gicquel. 1999. Use of fluorescence induction and sucrose counterselection to identify Mycobacterium tuberculosis genes expressed within host cells. Microbiology 145:2923-2930. [DOI] [PubMed] [Google Scholar]
- 28.Via, L. E., R. Curcic, M. H. Mudd, S. Dhandayuthapani, R. J. Ulmer, and V. Deretic. 1996. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J. Bacteriol. 178:3314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson, R. J., L. E. Desjardin, N. Islam, B. M. Gibson, R. A. Kanost, K. A. Wilkinson, D. Poelman, K. D. Eisenach, and Z. Toossi. 2001. An increase in expression of a Mycobacterium tuberculosis mycolyl transferase gene (fbpB) occurs early after infection of human monocytes. Mol. Microbiol. 39:813-821. [DOI] [PubMed] [Google Scholar]
- 30.Zahrt, T. C., and V. Deretic. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]