Abstract
The plasmid-encoded AIDA (adhesin involved in diffuse adherence) autotransporter protein derived from diffuse-adhering clinical Escherichia coli isolate 2787 and the TibA (enterotoxigenic invasion locus B) protein encoded by the chromosomal tib locus of enterotoxigenic E. coli (ETEC) strain H10407 are posttranslationally modified by carbohydrate substituents. Analysis of the AIDA-I adhesin showed that the modification involved heptose residues. AIDA-I is modified by the heptosyltransferase activity of the product of the aah gene, which is located directly upstream of adhesin-encoding gene aidA. The carbohydrate modification of the TibA adhesin/invasin is mediated by the TibC protein but has not been elucidated. Based on the sequence similarities between TibC and AAH (autotransporter adhesin heptosyltransferase) and between the TibA and the AIDA proteins we hypothesized that the AIDA system and the Tib system encoded by the tib locus are structurally and functionally related. Here we show that (i) TibC proteins derived from different ETEC strains appear to be highly conserved, (ii) recombinant TibC proteins can substitute for the AAH heptosyltransferase in introducing the heptosyl modification to AIDA-I, (iii) this modification is functional in restoring the adhesive function of AIDA-I, (iv) a single amino acid substitution at position 358 completely abolishes this activity, and (v) antibodies directed at the functionally active AIDA-I recognize a protein resembling modified TibA in ETEC strains. In summary, we conclude that, like AAH, TibC represents an example of a novel class of heptosyltransferases specifically transferring heptose residues onto multiple sites of a protein backbone. A potential consensus sequence for the modification site is suggested.
The plasmid-encoded AIDA (adhesin involved in diffuse adherence) autotransporter system was originally identified in clinical isolate 2787 (O126:H27) (2). Adhesin AIDA-I confers the diffuse adherence phenotype of the wild-type strain to recipient Escherichia coli K-12 strains (4). The protein is synthesized as a 130-kDa preproprotein harboring a long signal peptide of 49 amino acids (3), which is cleaved during transport through the inner membrane. C-terminal processing during or after translocation of the adhesin through the outer membrane generates adhesin AIDA-I and 47.5-kDa C-terminal fragment AIDAc (25, 26). Translocator AIDAc is a β-barrel integral outer membrane protein and mediates the transport of AIDA-I to the surfaces of the bacteria (12, 13). Even after cleavage AIDA-I remains associated with the outer membrane. The adhesin function of the AIDA-I protein is dependent on the activity of the AAH (autotransporter adhesin heptosyltransferase) protein, which catalyzes the modification of AIDA-I with on average 19 heptose residues (5). For this the AAH protein uses the activated ADP-glycero-manno-heptose precursor of the lipopolysaccharide (LPS) biosynthetic pathway. This modification has been shown to be necessary for the adhesin activity of AIDA-I (5). The aah gene is located directly upstream of the AIDA-encoding aidA gene. In all functional members of the AIDA adhesin family the aah gene and the aidA gene are always associated (21).
The tib locus (enterotoxigenic invasion locus B), originally identified in enterotoxigenic Escherichia coli (ETEC) strain H10407, confers adherence and invasion properties to E. coli K12 strains (10, 11). The tib locus encodes four proteins TibA to -D. TibA is synthesized as a 100-kDa precursor and seems to be the adhesin/invasin for human intestinal epithelial cells (17). The amino acid sequence of TibA shows homology to that of AIDA-I in the N-terminal part and to that of the β-barrel structure of AIDAc in the C-terminal part. However, in contrast to what is found for AIDA, only the signal sequence is cleaved in TibA and no additional C-terminal processing was detected. Therefore, TibA remains covalently connected to the surface of the bacteria (16). Interestingly, the functional form of TibA is modified by the addition of carbohydrates very much reminiscent of AIDA-I (16). This modification is mediated by the TibC protein (J. G. Mammarappallil, B. W. Hronek, L. D. Mettenburg, and E. A. Elsinghorst, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. B57, 2001), which exhibits a high sequence similarity to the AAH protein.
Because of the apparent homology of the two systems, we hypothesized that also in the tib locus the TibC protein represents a heptosyltransferase. Therefore, we were interested to investigate whether this novel putative heptosyltransferase is specific for the tib locus of enterotoxigenic E. coli (ETEC) or whether it might be able to replace the AAH heptosyltransferase of the AIDA system, generating a functional AIDA-I adhesin. Here we report on the functional substitution of the AAH heptosyltransferase and the modification of AIDA-I by several TibC-homologous proteins derived from different ETEC strains.
MATERIALS AND METHODS
Bacteria and plasmids.
E. coli K-12 strains C600 (F− thi-1 thr-1 leuB6 lacY1 tonA21 supE44 λ−) (1) and BL21(DE3) (F− ompT hsdSB [rB− mB−] gal dcm T7 gene1) (24) were used. E. coli clinical isolates of different origins were employed (Table 1). T7 expression vector pET-20b(+) (Novagen, Bad Soden, Germany) has a pBR322 origin and mediates ampicillin resistance, and vector pGP1-2 has a P15A origin and mediates kanamycin resistance (29). Plasmids pIB264 (2) and pIB9 (5) are pBR322 derivatives and confer ampicillin resistance. Plasmid pIB264 harbors the genes for the expression of the authentic AIDA system (aah and aidA). In pIB9 (5) the aah gene has been deleted. Bacteria were grown in standard I medium (Merck, Darmstadt, Germany). For maintenance of plasmids the medium was supplemented with the appropriate antibiotic (100 μg of ampicillin/ml and/or 30 μg of kanamycin/ml).
TABLE 1.
E. coli clinical isolates used in this study
| Strain | Serotype | Reference or sourcea |
|---|---|---|
| DAEC 2787 | O126:H27 | 2 |
| ETEC G1253 | O147:H19:K88 | HK |
| ETEC 117/86 | O6:H− | HK |
| ETEC 147/1 | O128:H− | HK |
| ETEC 149 | O149:H88 | HK |
| ETEC 164/82 | O148:H28 | HK |
| ETEC 297/87 | O25:H42 | HK |
| ETEC 28.4/97 | n.d.b | HK |
| ETEC E2539-C1 | n.d. | CB |
| ETEC TX-1 | n.d. | CB |
| ETEC EDL1493 | n.d. | CB |
| ETEC 440TL | n.d. | CB |
| ETEC 469ST | n.d. | CB |
HK, H. Karch, Münster, Germany; CB, C. le Bouguénec, Paris, France.
n.d., not determined.
PCR analysis, cloning, and sequencing.
For chromosomal DNA preparation exponentially growing bacteria were collected by centrifugation, resuspended in water, incubated at 100°C for 15 min, and again centrifuged. The supernatant was used as a DNA template in the PCR employing Taq polymerase (Roche Diagnostics, Mannheim, Germany). For the detection and amplification of the tibC gene PCR conditions were set as follows: 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C for 30 cycles. Sequences of oligonucleotides used as primers are listed in Table 2. Plasmids were constructed by using established methods described by Sambrook et al. (22). Plasmid transfer to bacteria was achieved by following the protocol of Mandel and Higa (18). For in vitro mutagenesis the QuickChange site-directed mutagenesis kit (Stratagene, Amsterdam, The Netherlands) was used. DNA sequences were determined by MWG Biotech AG (Ebersberg, Germany). Sequence analysis was performed with the HUSAR program (Heidelberg Unix Sequence Analysis Resources) of the Deutsches Krebsforschungszentrum (Heidelberg, Germany).
TABLE 2.
Sequences of oligonucleotide primers used in this study
| Primer | Sequencec | Positiona | Tmb (°C) |
|---|---|---|---|
| CM1 | CTGCAGGCTACATTCTGGG | 569-587 | 58.8 |
| CM2 | GTAAAGTCCTCTGCACCCCA | 833-814 | 59.4 |
| CM5 | GGATATATCATATGTCAACGCTGAAGAATAC | 1-20 | 62.9 |
| CM6 | CTATTAACAGAATTCGCCTTGAC | 599-577d | 57.1 |
| CM7 | CCACCACGATTTTCTGTGGTGCCCGAGAC | 1053-1081 | 78.0e |
| CM8 | GTCTCGGGCACCACAGAAAATCGTGGTGG | 1081-1053 | 78.0e |
Position in the sequence of tibC of ETEC H10407, accession no. AF131891, except where indicated otherwise.
Tm, melting temperature. Values were provided by the manufacturer.
Nucleotides in boldface are not complementary to the template DNA. Nucleotides corresponding to NdeI (CM5) and EcoRI (CM6) restriction sites are underlined.
Calculated as described in the QuickChange site-directed mutagenesis kit.
Induction and analysis of protein expression by denaturing gel electrophoresis and Western blotting.
Recombinant gene expression in E. coli BL21(DE3) was induced at an optical density at 600 nm of 0.6 to 0.8 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h.
For the preparation of whole-cell lysates bacteria were suspended in 62.5 mM Tris-HCl (pH 6.8)-20% glycerol-3% sodium dodecyl sulfate (SDS)-8% β-mercaptoethanol and incubated for 10 min at 100°C.
After protein separation by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting was performed in 25 mM Tris-HCl (pH 7.2) essentially as described previously (6, 30). The nitrocellulose membrane was blocked with 5% skim milk-phosphate-buffered saline (PBS) for 1 h at room temperature. The different antisera were applied in 0.1% bovine serum albumin (BSA)-PBS. After 1 h of incubation the nitrocellulose was washed three times for 10 min each with 0.06% Brij 35-PBS and incubated with a 1:5,000 dilution of alkaline phosphatase (AP)-conjugated second antibody (Jackson ImmunoResearch Laboratories, West Grove, Pa.) in 0.1% BSA-PBS. Following repeated washings with 0.06% Brij 35-PBS, the bound antibody was visualized by incubation with nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate as the substrate in AP buffer (100 mM Tris-HCl [pH 9.5], 1 mM MgCl2).
The following antisera were used: (i) antiserum raised against C600(pIB4) exhaustively preadsorbed against C600(pBR322), which recognizes primarily the protein backbone of the AIDA-I adhesin (4); (ii) antiserum raised against the isolated and purified AIDAc protein carrying an additional signal sequence (26); and (iii) antiserum raised against solubilized AIDA-I adhesin recognizing the modification (5).
Adhesion assay.
Bacterial adherence to HeLa cells was monitored essentially as described by Cravioto et al. (7) with modifications. For each assay about 108 bacteria grown overnight at 37°C with aeration in standard I medium (Merck) were incubated for 5 min in 1 ml of PBS containing 0.5% d-mannose. The bacterial suspension was added to HeLa cell monolayers on coverslips, just before the monolayers reached confluence. After 1 h of incubation at 37°C, the cells were washed extensively with PBS to remove any nonadherent bacteria. The cells were fixed in 70% methanol, stained with Giemsa (10% solution in water) for better contrast, and evaluated for adhering bacteria by light microscopy.
Detection of a glycan modification.
After separation by SDS-PAGE and Western blotting, glycoproteins were detected by employing the DIG glycan detection kit (Roche Diagnostics). Oxidation of carbohydrate residues was performed in 10 mM sodium metaperiodate in 100 mM sodium acetate buffer (pH 5.5) for 20 min. After being washed with PBS, the filter was incubated with digoxigenin (DIG)-3-O-succinyl-ɛ-aminocaproic acid-hydrazide for 1 h followed by a washing with PBS and incubation with blocking reagent. DIG-labeled proteins were detected with horseradish peroxidase-labeled anti-DIG antibodies. For visualization of bound antibodies the membranes were developed with Super Signal West Pico chemiluminescent substrate (Pierce Chemical Co., Rockford, Ill.) and subsequently exposed to X-ray film.
RESULTS
Sequence comparison of the AIDA and the tib locus.
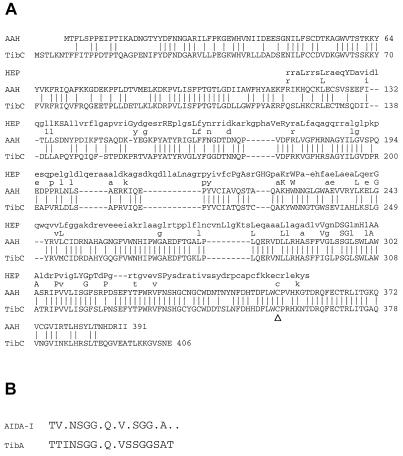
Comparison of the amino acid sequences of AAH and TibC protein showed 76.0% similarity according to the best-fit algorithm of Devereux et al. (8). The amino acid sequences of the two proteins were highly homologous, exhibiting 69.2% identical residues. The highest variation is seen in the N and C termini (Fig. 1A).
FIG. 1.
Comparative sequence analysis of AAH and TibC. (A) Homology between the amino acid sequences of the AAH and the TibC proteins and the consensus sequence of heptosyltransferases (HEP). Letters between the consensus sequence of heptosyltransferases and the AAH sequence, identical amino acids (uppercase letter, strong consensus positions; lowercase letter, best identity at a weak consensus position); lines between the sequence of the AAH protein and the TibC protein, match lines (exact matches); Δ, mutated cysteine residue leading to a complete loss of activity in the recombinant TibC of ETEC strain TX-1. (B) Comparison of the consensus sequences of the N-terminal repeat regions of AIDA-I and TibA.
A search of Hidden Markov models (14) to detect potential structural homologues of the AAH and TibC proteins resulted in the identification of the consensus sequence of heptosyltransferases involved in the biosynthesis of the inner core structures of LPS (Fig. 1A). This supported the possibility that, like the aah gene in the AIDA system, the tibC gene might encode a heptosyltransferase. Prior to the recognition of the AAH protein of the AIDA system as a heptosyltransferase (5), the heptosyltransferases identified were shown to specifically target carbohydrate structures in, e.g., the synthesis of the inner core of LPS molecules and the modification of S-layer carbohydrates.
Further comparative analysis revealed additional homologies between AIDA-I and TibA. This sequence homology is most significant in the N-terminal region, where in both proteins repeats of a distinct amino acid sequence motif have been identified. The consensus sequence derived from these repetitive motifs is given in Fig. 1B. Therefore, not only the modifying but also the modified proteins exhibit a significant sequence homology.
PCR analysis of ETEC strains.
To identify the tib locus in further ETEC strains, we examined 12 ETEC strains derived from two different strain collections for the presence of tibC by PCR analysis as described in Materials and Methods. As a control we used AIDA prototype diffuse-adhering E. coli (DAEC) strain 2787 and recombinant strain C600(pIB264), which both express the AIDA system, and E. coli C600. Total DNA was used as the template with primers CM1 and CM2, which were designed to be complementary to the tibC gene of ETEC H10407 in a region where it is highly homologous to the aah gene. Amplification by PCR should result in a DNA fragment of 265 bp. With total DNA from E. coli C600 as the template no DNA fragment of the expected size could be obtained, whereas PCR amplification of total DNA derived from strains C600(pIB264) and DAEC 2787 generated a small amount of the expected fragment. In 4 of the 12 ETEC strains analyzed by PCR a fragment indicative of the presence of the tibC gene could be amplified. This also showed that the tib locus is apparently not a common denominator of ETEC strains, as it was found in only one-third of the strains investigated in this study.
Cloning of the tibC gene from different ETEC strains.
Total DNA of the four tibC-positive ETEC strains was used as a template in PCRs with primers CM5 and CM6 (flanking the tibC gene in ETEC H10407) as described in Materials and Methods. The amplified 1.8-kb DNA fragment contained the tibC gene and 600 bp of tibA. Specific amplification was achieved in all four strains. The fragment was restricted with NdeI and EcoRI and ligated with vector pET-20b(+) restricted with the same enzymes. In the resulting plasmids (pCM-I series) the tibC gene is under the control of the T7 promoter. Therefore, to test the function of the cloned genes, it was necessary to reclone the appropriate DNA fragment in pGP1-2. This vector contains a P15A origin and is therefore compatible with the plasmids derived from pBR322 (ColE1 origin), such as pIB9. The pCM-I plasmids were restricted with BglII and EcoRI. The 1.9-kb fragment containing the T7 promoter and the tibC gene was isolated from an agarose gel, ligated with vector pGP1-2 restricted with BamHI and EcoRI, and transformed in E. coli C600. The resulting plasmids were analyzed by restriction analysis and denoted pCM-II.
Expression of the TibC protein.
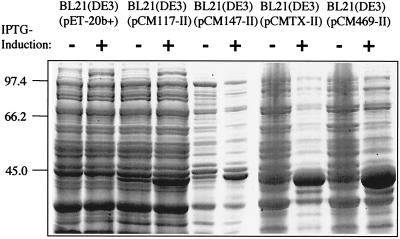
To analyze the recombinant TibC protein, the pCM-II plasmids were transformed into E. coli strain BL21(DE3). Total cellular proteins were separated by SDS-PAGE and stained with Coomassie blue. In all samples an additional protein band was detected after induction with IPTG (Fig. 2). It corresponds to a protein with a molecular mass of about 45 kDa, as expected for the TibC protein. Bacteria grown in the absence of IPTG showed small amounts of the protein, whereas in bacteria grown in the presence of IPTG a larger amount of the protein could be detected. The amount of expressed TibC protein varies among the four recombinant strains.
FIG. 2.
Expression of recombinant TibC protein in E. coli BL21(DE3). Total cellular proteins of different recombinant E. coli strains were separated by SDS-10% PAGE. The plasmids contain a tibC gene derived from the particular ETEC strain indicated in the plasmid name. Proteins were detected by staining with Coomassie blue. The total cellular extracts were prepared from recombinant bacteria without (−) and with (+) prior induction of T7 polymerase synthesis with IPTG for 3 h. Molecular sizes of the marker proteins are given in kilodaltons.
Modification of AIDA-I by the TibC protein.
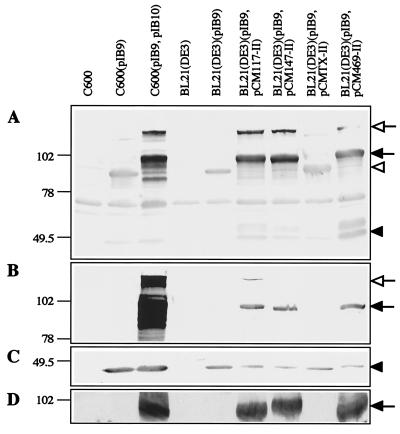
The recombinant pCM-II plasmids harboring the tibC gene derived from the different wild-type ETEC strains were transformed into BL21(DE3)(pIB9). For expression of tibC the recombinant strains were grown in the presence of IPTG for 3 h. Samples of total cellular proteins were analyzed by Western blotting and glycan detection (Fig. 3). Carbohydrate-modified AIDA-I was identified by (i) the shift in electrophoretic mobility compared to that for the unmodified form synthesized in BL21(DE3)(pIB9) (Fig. 3A), (ii) recognition in Western blots with different antisera (anti-AIDA recognizes the modification, and anti-C600[pIB4] recognizes the protein backbone of the adhesin) (Fig. 3A and B), and (iii) the reaction in the glycan detection assay (Fig. 3D). For the strains harboring the pCM117-II, pCM147-II, and pCM469-II plasmids, modified AIDA-I was identified in the total cellular proteins by all three criteria (Fig. 3A, B, and D). Interestingly, in the strain harboring pCMTX-II only unmodified AIDA could be detected (Fig. 3). The protein extracts were analyzed by Western blotting with antiserum directed against AIDAc, and were found to contain comparable amounts of recombinant AIDA protein (Fig. 3C).
FIG. 3.
Western blots and glycan detection of total cellular proteins extracted from different E. coli strains. Proteins were separated by SDS-10% PAGE. AIDA-specific proteins were identified with preadsorbed antiserum raised against C600(pIB4) (A), anti-AIDA-I serum (B), and anti-AIDAc serum (C). (D) Glycoproteins were labeled with DIG-hydrazide after periodate oxidation and visualized with an anti-DIG antibody. Molecular sizes of the marker proteins are given in kilodaltons. Open arrow, proprotein; solid arrow, AIDA-I; open arrowhead, unmodified AIDA-I; solid arrowhead, AIDAc
Adherence properties of different recombinant strains.
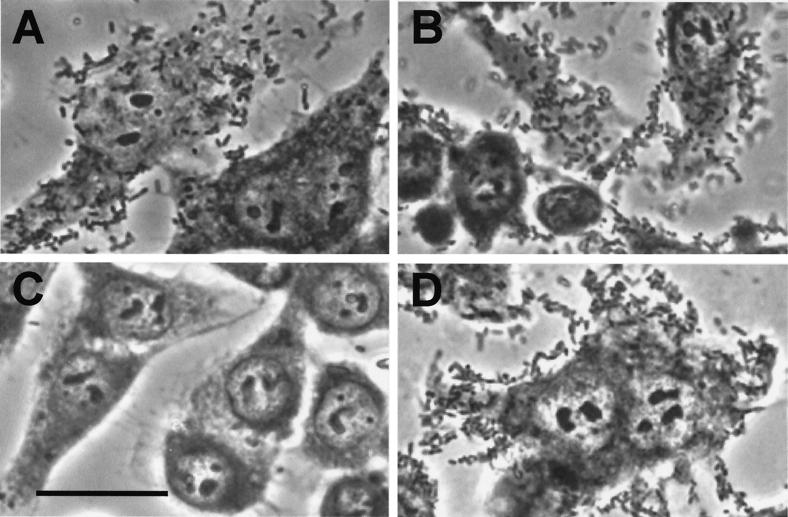
In the original system only heptose-modified AIDA is able to mediate adherence to HeLa cells (5), whereas the translocation of AIDA to the bacterial surface is not influenced (3). To test if the modification in AIDA-I catalyzed by recombinant TibC is sufficient to confer the original phenotype, we analyzed the recombinant strains in an adhesion assay. The three strains synthesizing a modified AIDA-I exhibited the same pattern of diffuse adherence as wild-type strain DAEC 2787 (Fig. 4A, B, and D). As expected, the strain harboring pCMTX-II, which synthesized only the unmodified AIDA-I adhesin, is not able to attach to HeLa cells (Fig. 4C).
FIG. 4.
Adherence properties of E. coli strains expressing a recombinant tibC gene. (A) BL21(DE3)(pIB9, pCM117-II); (B) BL21(DE3)(pIB9, pCM147-II); (C) BL21(DE3)(pIB9, pCMTX-II); (D) BL21(DE3)(pIB9, pCM469-II). HeLa cell monolayers were incubated with 108 bacteria in PBS (37°C) in the presence of 0.5% d-mannose. After extensive washing with PBS, cells and bacteria were stained with Giemsa for better contrast and assayed for adherence by light microscopy. Bar, 25 μm.
Sequence comparison of the four cloned tibC genes.
To elucidate why TibC derived from strain TX-1 is not able to modify AIDA-I despite being available in rather large amounts (Fig. 2), the DNA sequences of the different recombinant tibC genes were determined. Comparison of the new TibC sequences with the known sequence encoded by the tibC gene from strain H10704 (16) showed a near 100% identity with only a few single-amino-acid exchanges. This indicated a very high conservation among strains derived from different collections. The differences between the published sequences of tibC and its product, TibC, from ETEC H10407 (16) and each of the novel versions of tibC and TibC are listed in Table 3. The amino acid exchanges at position 90 and 348 seemed to have no influence on the activity of the TibC protein, whereas the exchange at position 358 (Fig. 1A), as found in the TibC protein encoded by pCMTX-II, leads to a complete loss of the glycosyltransferase activity (Fig. 3 and 4).
TABLE 3.
Comparison of the amino acid sequences of the recombinant TibC proteins and associated nucleotide sequencesa
| Wild-type ETEC strain | Difference in:
|
|
|---|---|---|
| Nucleotides | Amino acids | |
| 117/86 | G144 to T | —b |
| G268 to A | E90 to K | |
| C621 to T | — | |
| 147/1 | G268 to A | E90 to K |
| C621 to T | — | |
| T1043 to C | L348 to P | |
| 469ST | — | — |
| TX-1 | T1072 to C | C358 to R |
In vitro mutagenesis of the cloned tibC gene of ETEC strain TX-1.
The loss of glycosyltransferase activity of the recombinant TibC derived from ETEC strain TX-1 seems to be caused by the single-amino-acid exchange at position 358. To investigate whether this is actually the case, we performed an in vitro mutagenesis with plasmid pCMTX-I to reintroduce the cysteine residue, which is conserved in all other TibC sequences, at position 358 (primers CM7 and CM8; 30 s at 95°C, 1 min at 65°C, and 12 min at 72°C for 12 cycles). A recombinant strain harboring a plasmid carrying a tibC sequence encoding an Arg358Cys mutation (pCMTX-III) was analyzed for synthesis of the TibC protein. The protein was detected in about the same amount as in strains with the original plasmid. The TibC protein was then assessed for its ability to modify AIDA-I. The Arg358Cys amino acid exchange fully restored the activity. The AIDA-I adhesin was modified as with the other TibC proteins, and the recombinant strain exhibited the same diffuse pattern of adherence to HeLa cells as that previously described (data not shown).
Examination of the tibC-harboring ETEC strains.
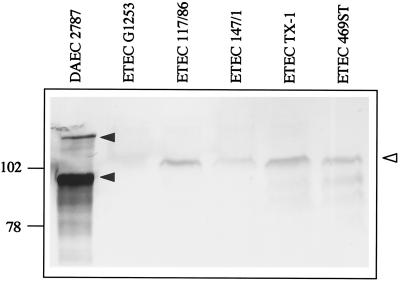
Western blot analysis of total cellular proteins of ETEC strains with anti-AIDA serum recognized protein bands corresponding in size to the modified TibA protein in all four strains (Fig. 5). As the anti-AIDA serum recognizes the modification of the AIDA adhesin, this result indicates that in all four ETEC strains including TX-1 TibC exhibits glycosyltransferase activity and TibA is modified. Furthermore, recognition of the modification in TibA by antibodies directed against the modification of AIDA-I suggests that the modifications by carbohydrate moieties of AIDA-I and TibA are very similar if not identical.
FIG. 5.
Western blot of total cellular proteins extracted from different E. coli strains. ETEC strains 117/86, 147/1, TX-1, and 469ST harbor the tib locus, whereas ETEC isolate G1253 does not. DAEC 2787 expressing the AIDA system serves as a positive control. Proteins were separated by SDS-10% PAGE. Modified proteins were identified with anti-AIDA-I serum recognizing the heptosyl modification. Solid arrowheads, AIDA proprotein and modified AIDA-I adhesin; open arrowhead, modified TibA protein. Molecular sizes of the marker proteins are given in kilodaltons.
To confirm this result the DNA sequence of a newly amplified tibC-specific DNA fragment of ETEC TX-1 was determined. In this newly amplified sequence a nucleotide exchange at position 1072 was not detected and the tibC sequence of the TX-1 strain was the same as that of tibC of H10407. These results show that TibC from strain TX-1 has no mutation and modifies the TibA protein whereas the recombinant TibC obtained by PCR amplification showed an amino acid exchange and had lost its modifying function. These findings can be readily explained by a mistake of the Taq polymerase during amplification of the tibC-specific DNA fragment.
DISCUSSION
Protein glycosylation has for a long time been thought to be restricted to eukaryotes as prokaryotes have been considered to lack the necessary machinery for protein glycosylation (19, 20). In recent years, however, protein glycosylation has been demonstrated and recognized as a rather frequent posttranslational modification in archaea and also in eubacteria. As important human pathogens, such as Campylobacter jejuni (28), Chlamydia trachomatis (15, 27), Neisseria meningitidis (23), and Mycobacterium tuberculosis (9) express glycosylated proteins, the investigation of a putative function of bacterial glycosylation in pathogenesis has developed into an actively expanding area of research.
Plasmid-encoded autotransporter adhesin AIDA was identified in a clinical isolate of E. coli and was subsequently characterized in our laboratory (2-5, 12, 26). Recently, we demonstrated that the AIDA-I adhesin is posttranslationally modified with, on average, 19 heptose residues, and we further showed that this modification is essential for the adhesive function of the protein (5). We identified the modifying enzyme as the product of the aah gene, which has been found to be always associated with a functional AIDA system (5, 21). The AAH protein (autotransporter adhesin heptosyltransferase) is a cytoplasmic heptosyltransferase which utilizes activated ADP-glycero-manno-heptose residues pinched from the pathway of LPS synthesis to specifically modify the AIDA protein.
TibA, a chromosomally encoded glycosylated autotransporter adhesin/invasin, was described by Elsinghorst and collegues (11, 16, 17). Recently, it was reported (Mammarappallil et al., Abstr. 101st Gen. Meet. Am. Soc. Microbiol.) that the glycosylation of TibA is apparently mediated by the product of the tibC gene. Sequence comparison of the tibC gene and the aah gene, which had been shown to encode a heptosyltransferase (5), demonstrated significant similarity. In addition, homology to the consensus sequence for the known heptosyltransferases involved in LPS core synthesis could be identified in the products of both genes (5, 16). AIDA-I and TibA seem to be functionally and structurally related proteins, as both have repetitive sequence motifs in their N termini (5, 16). Therefore, we hypothesized that TibC also might represent a heptosyltransferase, and we were interested to see whether TibC might be able to substitute for AAH in the AIDA system and as such restore the adhesin activity of the AIDA-I autotransporter adhesin.
For the identification of the tibC gene by PCR we investigated 12 ETEC isolates derived from two different strain collections. Four strains harboring the tibC gene were identified; the genes were amplified by PCR and subsequently cloned and expressed under the control of a T7 promoter (Fig. 2). This indicates that the tibC gene might be present in only a fraction of ETEC strains (11) and is not a conserved gene. Reconstitution of a recombinant strain which expresses only the AIDA autotransporter and which lacks the aah heptosyltransferase gene with three of the four cloned tibC genes derived from different ETEC strains restored glycosylation and the activity of the AIDA-I adhesin. Glycosylation was demonstrated by the shift in electrophoretic mobility and reactivity in the glycan detection assay (Fig. 3). Furthermore, three out of four recombinant TibC proteins also restored the diffuse-adherence phenotype mediated by the AIDA-I adhesin (Fig. 4). However, the recombinant tibC gene derived from ETEC strain TX-1 did not restore the activity of AIDA-I, although the protein was expressed in sufficient amounts (Fig. 2). Further analysis of the recombinant DNA sequences detected a near identity to the published tibC sequence of ETEC strain H10407, leading to only a few dispersed single-amino-acid exchanges. In the recombinant tibC gene of strain TX-1, however, we identified a nucleotide exchange at position 1072 that resulted in amino acid exchange Cys358Arg. As in the four known TibC sequences a cysteine residue is conserved at this position, and it appeared that this single mutation might be responsible for the lack of activity. To prove that this is really the case, we reintroduced a cysteine at this position (Arg358Cys; pCMTX-III) by site-specific mutagenesis. The introduction of the conserved cysteine residue fully restored the activity of TibC. This indicates either that the cysteine residue is directly involved in the catalytic activity of the glycosyltransferase or, more likely, that this cysteine is part of an essential disulfide loop needed for the correct folding of the enzyme. Interestingly, this cysteine residue is the only Cys of the 10 to 12 cysteines shared by AAH and TibC which is also present in the heptosyltransferase consensus sequence (Fig. 1A). Other single-amino-acid exchanges, compared to the prototype H10407 TibC sequence, identified in two further recombinant TibC proteins apparently did not influence their activity (Table 3).
Interestingly, in the wild-type ETEC strains including TX-1, a glycosylated protein corresponding to the modified TibA protein (104 kDa) could be identified (Fig. 5) by the modification-specific anti-AIDA antiserum (5). This clearly demonstrated that in the wild-type ETEC TX-1 strain TibC is active and, furthermore, suggested that the inactivating mutation might have been introduced by the Taq polymerase during PCR amplification. Reamplification and sequencing of tibC derived from ETEC strain TX-1 resulted in an active TibC protein carrying the expected cysteine residue at position 358.
Recognition of TibA in all four tib-positive ETEC strains by the anti-AIDA-I antiserum (Fig. 5) demonstrated that the glycosylation of TibA is related if not identical to the modification with heptose residues recently identified in AIDA-I (5). In conclusion, our results strongly indicate that, like the AAH protein, the TibC protein functions as a protein-specific heptosyltransferase. With respect to the homology of the repetitive sequences of the N terminus of AIDA-I and TibA (Fig. 1), it is tempting to speculate that one of the serine residues (boldface) of consensus sequence TV-NSGG-Q-V-SGG-A--, derived from the AIDA-I repetitive motif, might represent the actual modification site. However, this has to be confirmed by further studies.
In summary, we show that the TibC glycosyltransferase, identified as the product of the chromosomal tib loci of certain ETEC strains, is able to functionally replace the autotransporter adhesin heptosyltransferase (AAH) of the plasmid-encoded AIDA system in fully restoring activity to the AIDA-I adhesin depending on the modification of AIDA with heptose residues. Therefore, the AAH and the TibC proteins represent the first examples of a novel class of heptosyltransferases which target proteins.
Acknowledgments
We thank H. Karch (Hygiene Institut, Universitätsklinikum Münster) and C. le Bougenec (Institut Pasteur, Paris) for the kind gift of ETEC strains.
This study was supported in part by the Deutsche Forschungsgemeinschaft (DFG SCHM 770/7-2; SFB293 B5).
C. Moormann and I. Benz contributed equally to this work.
Editor: V. J. DiRita
REFERENCES
- 1.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 4.Benz, I., and M. A. Schmidt. 1992. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27). Infect. Immun. 60:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 6.Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 7.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:389-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobos, K. M., K. Swiderek, K.-H. Khoo, P. J. Brennan, and J. T. Belisle. 1995. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect. Immun. 63:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsinghorst, E. A., and D. J. Kopecko. 1992. Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect. Immun. 60:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konieczny, M. P. J., I. Benz, B. Hollinderbäumer, C. Beinke, M. Niederweis, and M. A. Schmidt. 2001. Modular organization of the AIDA autotransporter translocator: the N-terminal β1-domain is surface-exposed and stabilizes the transmembrane β2-domain. Antonie Leeuwenhoek 80:19-34. [DOI] [PubMed] [Google Scholar]
- 13.Konieczny, M. P. J., M. Suhr, A. Noll, I. B. Autenrieth, and M. A. Schmidt. 2000. Cell surface presentation of recombinant (poly-)peptides including functional T cell epitopes by the AIDA autotransporter system. FEMS Immunol. Med. Microbiol. 27:321-332. [DOI] [PubMed] [Google Scholar]
- 14.Krogh, A., M. Brown, I. S. Mian, K. Sjolander, and D. Haussler. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235:1501-1531. [DOI] [PubMed] [Google Scholar]
- 15.Kuo, C.-C., N. Takahashi, A. F. Swanson, Y. Ozeki, and S.-I. Hakomori. 1996. An N-linked high-mannose type oligosaccharide, expressed at the outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J. Clin. Investig. 98:2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindenthal, C., and E. A. Elsinghorst. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandel, M., and A. Higa. 1970. Calcium dependent bacteriophage DNA infection. J. Mol. Biol. 53:159-162. [DOI] [PubMed] [Google Scholar]
- 19.Messner, P. 1997. Bacterial glycoproteins. Glycoconj. J. 14:3-11. [DOI] [PubMed] [Google Scholar]
- 20.Moens, S., and J. Vanderleyden. 1997. Glycoproteins in prokaryotes. Arch. Microbiol. 168:169-175. [DOI] [PubMed] [Google Scholar]
- 21.Niewerth, U., T. Voss, A. Frey, C. le Bouguénec, G. Baljer, S. Franke, and M. A. Schmidt. 2001. The AIDA autotransporter system is associated with F18 and Stx2e in Escherichia coli isolates derived from pigs suffering from edema disease and postweaning diarrhea. Clin. Diagn. Lab. Immunol. 8:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Stimson, E., M. Virji, K. Makepeace, A. Dell, H. R. Morris, G. Payne, J. R. Saunders, M. P. Jennings, S. Barker, M. Panico, I. Blech, and E. R. Moxon. 1995. Meningococcal pili: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17:1201-1214. [DOI] [PubMed] [Google Scholar]
- 24.Studier, F. W., and B. A. Moffatt. 1987. T7 lysozyme inhibits transcription by T7 polymerase. Cell 49:221-227. [DOI] [PubMed] [Google Scholar]
- 25.Suhr, M. 1998. Prozessierung des AIDA-I Vorläuferproteins: Topologie von AIDAc in der äuβeren Membran und Möglichkeiten der Präsentation heterologer Antigene. Ph.D. thesis. University of Münster, Münster, Germany.
- 26.Suhr, M., I. Benz, and M. A. Schmidt. 1996. Processing of the AIDA-I precursor: removal of AIDAc and evidence for the outer membrane anchoring as a β-barrel structure. Mol. Microbiol. 22:31-42. [DOI] [PubMed] [Google Scholar]
- 27.Swanson, A. F., and C.-C. Kuo. 1994. Binding of glycan of the major outer membrane protein of Chlamydia trachomatis to HeLa cells. Infect. Immun. 62:24-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 29.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]