Abstract
Previous studies suggested that PspC is important in adherence and colonization within the nasopharynx. In this study, we conducted mutational studies to further identify the role PspC plays in the pathogenesis of pneumococci. pspC and/or pspA was insertionally inactivated in a serotype 2 Streptococcus pneumoniae strain and in a serotype 19 S. pneumoniae strain. In the mouse colonization model, pneumococcal strains with mutations in pspC were significantly attenuated in their abilities to colonize. In a mouse pneumonia model, strains with mutations in pspC were unable to infect or multiply within the lung. Using reverse transcriptase PCR we were able to demonstrate that pspC is actively transcribed in vivo, when the bacteria are growing in the nasal cavity and in the lungs. In the bacteremia model, a strain mutated for pspC alone behaved like the wild type, but the absence of both pspC and pspA caused accelerated clearance of the bacteria. Intranasal immunization with PspC with cholera toxin subunit B as an adjuvant protected against intranasal challenge. Evidence was also obtained that revertants that spontaneously acquired PspC expression could multiply and colonize the nasal tissue. This latter finding strongly indicates that pneumococci are actively metabolizing and growing while in the nasopharynx.
PspC (59 to 105 kDa) is a paralog of PspA, a virulence factor found on all pneumococci (11, 13). Both molecules contain similar structural domains, including a coiled-coil α helix, followed by a proline-rich region and a choline-binding domain (11, 43). While the α-helical regions of PspA and PspC are distinct, the proline-rich region and choline-binding domains are indistinguishable. PspC is found on about 75% of pneumococci (11, 22). The molecule is also termed CbpA and/or SpsA based on its ability to bind choline and/or the secretory component of immunoglobulin A (IgA), respectively (22, 33). PspC interacts with the complement pathway through its ability to bind complement component C3 (12) and factor H (17, 25). It has been previously demonstrated that PspC acts as a cell surface adhesin and plays a major role in nasopharyngeal colonization in an infant rat model and that it is important in binding to cytokine-activated epithelial cells (33). It has been proposed that pneumococci migrate across the mucosal barrier by using the polymeric Ig (pIg) machinery and that this process is mediated through the interaction between PspC and the pIg receptor (pIgR).
Streptococcus pneumoniae varies spontaneously between opaque and transparent phenotypes at frequencies of 10−3 to 10−6 per generation (38, 39). It was noted that regardless of the phenotype injected into the mouse, the opaque phase generally predominates among the bacteria recovered from the blood. The transparent phase is more commonly associated with bacteria in the nasopharynx (39). This general finding of the tropism of these different phases for carriage and bacteremia has been confirmed by our laboratory with several additional strains of S. pneumoniae (D. E. Briles, A. Virolainen, and R. Fulghum, unpublished data). The difference in colony morphology is also associated with differential protein expression (31, 38). PspC is expressed preferentially in transparent as opposed to opaque colonies (33).
PspA, the paralog of PspC, is the protein primarily responsible for binding human lactoferrin to the pneumococcal cell surface (20, 21). Studies have also demonstrated that PspA interferes with deposition of the complement on the surface of the bacteria (37). PspA is a protection-eliciting immunogen. Immunization studies with PspA have demonstrated protection against intravenous, intraperitoneal, and intranasal routes of infection (40, 42). When mice are immunized with PspC fragments containing proline-rich and/or choline-binding regions, antibodies are elicited that cross-react with PspA and can protect against invasive disease (11).
S. pneumoniae strain D39 and its nonencapsulated derivatives are widely used in pneumococcal laboratories throughout the world. D39 is a serotype 2 strain and is lethal for mice (3, 5). The 50% lethal dose in CBA/N mice infected intravenously is about 10 CFU. When mutations in pspA were introduced into D39, the 50% lethal dose was reduced by a small but statistically significant amount (30). At a dose above the minimum lethal dose of both strains, the PspA-positive strain caused rapid death significantly sooner than the corresponding PspA-negative strain. On the other hand, when PspA was mutated in the serotype 3 strain WU2, the strain became virtually avirulent (9, 30). WU2 does not contain the structural gene for pspC, and no detectable PspC protein is present on Western blots. These findings led us to speculate that perhaps PspC, a paralog of PspA, might largely compensate for the virulence role of PspA in strains where PspA had been genetically deleted. Therefore, in these studies we constructed mutants of D39 (serotype 2) and EF3030 (serotype 19) that lacked PspA, PspC, or both PspA and PspC.
Our results indicated that a pspA-pspC double mutant of D39 was more attenuated in a mouse sepsis model than in either a pspA or a pspC mutant alone. Our findings provide strong in vivo support for the growing evidence (11, 22, 33) that PspC plays a major role at mucosal surfaces. We have also demonstrated for the first time the importance of PspC in pneumococcal pneumonia. We showed that pspC was transcribed in vivo, consistent with the expectation that it is produced in vivo. The initial studies conducted by Rosenow et al., by Hammerschmidt et al., and in our laboratory suggest that PspC may contribute to the pathogenesis of S. pneumoniae (11, 22, 23). Furthermore, our studies indicate that PspC can serve as a mucosal immunogen and can elicit protection against nasopharyngeal colonization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The pneumococcal strains used in this study are listed in Table 1. S. pneumoniae D39, a serotype 2 strain, is a clinical isolate (3). EF3030, a serotype 19F strain, was isolated from a patient with otitis media (2). S. pneumoniae was grown in Todd-Hewitt broth containing 0.5% yeast extract and plated on blood agar plates with 3% sheep erythrocytes in a candle jar (Difco Laboratories, Detroit, Mich.). Tetracycline (15 μg/ml) or erythromycin (0.3 μg/ml) was added to the blood agar plates when antibiotics were used for the growth of S. pneumoniae. Escherichia coli was grown in Luria-Bertani broth or agar with 50 μg of ampicillin/ml, 15 μg of tetracycline/ml, or 300 μg of erythromycin/ml.
TABLE 1.
Strains used in this study
| Strain | Derivation | Relevant phenotype | Reference(s) |
|---|---|---|---|
| D39 | Clinical isolate, 1914b | PspA+ PspC+ | 3 |
| WG44.1 | R×1 | PspA− PspC+ Emr | 43 |
| JY53 | D39 | PspA− PspC+ Emr | 43 |
| TRE108 | D39 | PspA+ PspC− Emr | This study |
| TRE118 | D39 | PspA+ PspC− Emr | 20 |
| TRE121/136a | D39 | PspA− PspC− Emr, Tcr | 20, this study |
| EF3030 | Clinical isolate, 1992b | PspA+ PspC+ | 2 |
| TRE124 | EF3030 | PspA+ PspC− Emr | This study |
| TRE125 | EF3030 | PspA+ PspC− Tcr | This study |
| BG7322 | Clinical isolate | PspA+ PspC+ | 8 |
| PLN-A | D39 | PspA+ PspC+ Ply− | 7 |
Multiple numbers indicate independently derived mutants.
Year of isolation.
Mice.
Six- to 12-week-old CBA/CAHN-XID/J (CBA/N) or BALB/cByJ mice were obtained from Jackson Laboratories (Bar Harbor, Maine). CBA/N mice lack the ability to produce antibodies to polysaccharides and lack serum antibodies to the phosphocholine determinant of pneumococcal teichoic acids. Mice were maintained in a specific-pathogen-free environment.
Cloning and insertion duplication mutagenesis.
A DNA fragment from pspC was amplified by PCR with oligonucleotides PB11 and ABW26, corresponding to bp 461 to 480 and bp 731 to 749, respectively, in pspC from strain D39 (Table 2) (Briles et al., unpublished). The amplified PCR product was cloned into the pTOPO cloning vector (Invitrogen, Carlsbad, Calif.) according to the manufacturer’s protocol. Inserts from the pTOPO plasmid were digested with EcoRI, and the pneumococcal DNA was subcloned into the appropriate suicide vector.
TABLE 2.
Primers used in this study
| Gene | Primer | Sequence |
|---|---|---|
| pspC | ABW26 | 5′-ACTTCTCAAAAGCTGCGTC-3′ |
| PB10 | 5′-GGAATTCACAGAGAACGAGGGAAGTAC-3′ | |
| PB11 | 5′-CCGAATTCCGCGACAGAGAACGAGGGAAGTAC-3′ | |
| pspA | LSM12 | 5′-CCGGATCCAGCGTCGCTATCTTAGGGGCTGGTT-3′ |
| SKH57 | 5′-TCGTCATATTTCCTCTGAGC-3′ | |
| endA | PB16 | 5′-ATTGAAGCTCCTAGTCAAGCATTGG-3′ |
| PB17 | 5′-ACTTTGCCCAGGCTGTCTGAAC-3′ |
Two plasmids were used for insertion duplication mutagenesis. Both plasmids contain an E. coli origin of replication and an antibiotic marker selectable in both E. coli and S. pneumoniae. The plasmid pSF143 (36) contains a tetracycline resistance gene, and pJY4164 contains an erythromycin resistance gene. The plasmid pJY4164 is a derivative of pMU1327 that has the streptococcal origin of replication deleted (1, 44). Neither plasmid can replicate autonomously in S. pneumoniae, and antibiotic-resistant transformants are obtained when the plasmid integrates by recombination into the homologous region of the host chromosome.
Pneumococcal transformations.
Pneumococci were transformed as described by Yother et al. (45), with the following modifications. Synthetically synthesized competence factor was obtained from Zymed Laboratories (San Francisco, Calif.) (23). Cultures were induced with 500 ng of competence factor/ml at 37°C for 12 min prior to the addition of purified plasmid. Mutations in pspA and pspC were made independently, with one gene containing an antibiotic resistance marker and the other gene containing the alternate antibiotic resistance marker. To construct a strain that contained mutations in both PspA and PspC, genomic DNA from the PspC-null mutant was used to transform a strain carrying the PspA-null mutation. Transformants were selected for resistance to both erythromycin and tetracycline. Lack of expression of PspA and/or PspC was confirmed by Western blot analysis as described by Brooks-Walter et al. (11).
Genomic DNA from antibiotic-resistant transformants was isolated by chloroform-isoamyl alcohol extraction. DNA was digested with restriction enzymes, and Southern hybridizations were performed essentially as described in McDaniel et al. (29), with the following modifications. (i) Hybridization was performed at 42°C in hybridization buffer with 50% formamide and 2× SSC (pH 7.5) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). (ii) The high stringency wash was conducted at 65°C in 0.01× SSC and 0.1% sodium dodecyl sulfate. The full-length Rx1pspA probe consists of nucleotides 1 to 1999 of the published Rx1pspA sequence (43). The 288-bp fragment of pspC from D39 used to make the mutation in pspC was labeled to serve as a pspC-specific probe. Stringency was determined by the formula tm = 81.5°C + 16.6 log M + 0.41(% GC) − 600/n − 0.62(% formamide), where tm is the melting temperature and M is the molar concentration of cations and n is the probe length in base pairs. By this calculation, our high stringency wash allows a 5% mismatch. Probes were labeled by digoxigenin with the Genius System according to the manufacturer's protocols (Boehringer-Mannheim, Indianapolis, Ind.).
Virulence experiments.
S. pneumoniae was grown to an optical density at 600 nm of 0.5. Bacterial cultures were pelleted by centrifugation and frozen in aliquots of Todd-Hewitt broth with 10% glycerol. The bacterial count was determined by thawing an aliquot, serially diluting the bacteria, and plating the bacteria onto blood agar plates. Pneumococci used for inoculations were diluted in lactated Ringer's solution to the desired CFU level prior to infections.
In the mouse bacteremia model, BALB/cByJ mice were infected intravenously with 106 CFU of bacteria in 0.2 ml of lactated Ringer's solution. Using a 75-μl heparinized microhematocrit capillary tube (Fisher Scientific, Pittsburgh, Pa.), mice were bled retro-orbitally at 1 min, 1 h, and 4 h. The collected blood was serially diluted and plated onto blood agar plates to enumerate the bacteria. The experiment was terminated after 21 days (504 h).
Intranasal infections were performed as described by Wu et al. (41). CBA/N mice were infected intranasally with 1 × 107 bacteria in 10 μl of lactated Ringer's solution. Infected mice were bled and sacrificed, and their nasal cavities were washed with 50 μl of Ringer's solution as previously described by Wu et al. (41). The nasal wash was serially diluted and double-plated onto both blood agar with gentamicin and blood agar with gentamicin and optochin.
Lung infections (35) with EF3030 and its derivatives were performed by anesthetizing CBA/N mice with Metofane during infection. Suspensions of 40 μl of lactated Ringer's solution containing 5 × 106 bacteria were introduced into the nares of the mice. After 6 days, the nasal cavity was washed as described above and the lungs were removed from the chest cavity. The lobes of the lungs were placed into 2 ml of Ringer's solution in a stomacher bag, homogenized, serially diluted, and plated. The viable counts were determined after overnight incubation.
RT-PCR.
Bacteria were collected from pooled nasal washes (strain D39, intranasal infection, ∼103 bacteria) or from pooled nasal washes and lungs (strain EF3030, lung infection, ∼5 × 104 bacteria). RNA was extracted immediately from the samples by using the FastRNA kit Blue (Bio101, Vista, Calif.). The RNA samples were treated with DNase I. Transcripts of the different genes were detected by reverse transcriptase PCR (RT-PCR) with gene-specific primers (pspC, PB10/ABW26; endA, PB16/PB17; pspA, LSM12/SKH57) listed in Table 2. As a control for the presence of contaminating DNA, all samples were run as duplicates and, in one set, the RT was inactivated by incubation at 95°C for 10 min before proceeding with the PCR.
Intranasal immunization.
PspC from D39 (amino acids 255 to 445) was expressed as a fusion protein with a PelB leader sequence and a six-His-tagged C-terminal extension in the pET20b expression system (Novagen, Madison, Wis.). Expression was induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in the expression strain E. coli BL21(DE3), and the expressed protein was purified by affinity chromatography with nondenaturing conditions on a nickel column according to the manufacturer's protocol. Isolated protein was quantitated by using a protein assay (Bio-Rad, Hercules, Calif.).
Mice were immunized intranasally once a week for 3 weeks with 2 μg of purified PspC in 10 μl of Ringer's solution. The first two immunizations contained 4 μg of cholera toxin B subunit (CTB) (40). Three weeks after the last immunization, mice were challenged with 6 × 106 CFU of a pneumolysin mutant of D39 or serotype 6B strain BG7322 (7). One week later, mice were sacrificed and their nasal cavities were washed to enumerate the number of CFU per nose. Twenty-four hours prior to challenge, 50 μl of carbachol (100 μg/ml) was injected intraperitoneally to increase salivary secretions. Using a pipette, 50 μl of saliva was collected from the oral cavity. Mice were also bled retro-orbitally into capillary tubes (75 μl). The blood was immediately mixed with 0.5 ml of 1% bovine serum albumin in phosphate-buffered saline. This mixture was centrifuged for 5 min, and the supernatant was removed and stored at −20°C. Direct enzyme-linked immunosorbent assays were conducted as described by Wu et al. (40). Antibody reactivity to PspC was determined and depicted as the titer giving 33% of maximum binding by a standard curve in each assay.
RESULTS
Confirmation of mutants by Southern blot analysis.
Insertion duplication mutagenesis was used to construct pspC mutants. An internal portion of the pspC gene was cloned into a plasmid that encodes an antibiotic resistance marker but cannot replicate autonomously in S. pneumoniae. During insertion duplication mutagenesis, recombinant plasmids integrate within the gene homologous to cloned DNA fragments (30). Transformants were selected on the basis of resistance to antibiotics (erythromycin or tetracycline), a property encoded by the integrated plasmid.
Transformation is an inherently mutational process, and spontaneous mutations may occur during recombination (19). For both pspC and pspA-pspC double mutants, at least two independent strains were made to control for secondary, unlinked mutations that may affect the phenotype of the resulting mutant. The plasmid used for insertion duplication mutagenesis was also inserted downstream of the pspC gene to take into account the importance of a possible transcriptional unit or a gene linked to pspC. In the case of pspC mutations, both plasmids pSF143 and pJY4164 containing pspC inserts were introduced separately to produce independent mutants of D39. These strains and others used in the study are listed in Table 1.
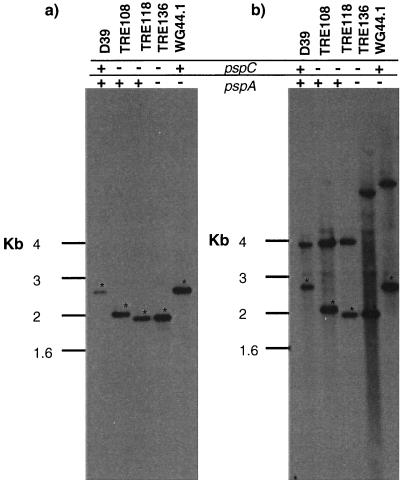
At high stringency, a pspC-specific probe identified a 2.8-kb fragment in HindIII-EcoRI-digested chromosomal DNA from strain D39 (Fig. 1). Insertion of a nonreplicating plasmid into the chromosome introduced a second EcoRI restriction site from the plasmid into the pspC gene. Consequently, the DNA fragment that hybridizes with the pspC-specific probe in TRE108, TRE118 (mutated in pspC), and TRE136 (mutated in both pspC and pspA) is approximately 2.0 kb, compared to 2.8 kb for nonmutated pspC from strain D39. While TRE108 carries an erythromycin resistance marker in pspC, TRE118 was mutated in pspC with plasmid pSF143 carrying a tetracycline resistance marker. Thus, although both strains are mutated in pspC, they show differences in their restriction patterns. TRE136 also carries a tetracycline resistance marker in pspC, and therefore, it shows a restriction pattern similar to that of TRE118 with respect to pspC. TRE136 and WG44.1, both mutated in pspA, show an altered restriction pattern for pspA compared to that of D39, TRE108, or TRE118 (wild type for pspA).
FIG. 1.
Southern blots of chromosomal DNA from D39 transformants. Chromosomal DNA was run in duplicate and developed with a pspC-specific probe (A) or with a full-length pspA probe (B). The DNA fragment containing a mutated gene is shifted in strains where the plasmid has been inserted. An asterisk indicates the fragment containing the pspC gene.
Cell lysates from the insertion duplication mutants were analyzed on Western blots. Polyclonal serum to PspC did not detect wild-type or truncated PspC in the lysates of these insertion duplication mutants. These mutations were found to have no effect on growth rates in vitro (data not shown).
Intranasal carriage of pneumococci in the mouse model.
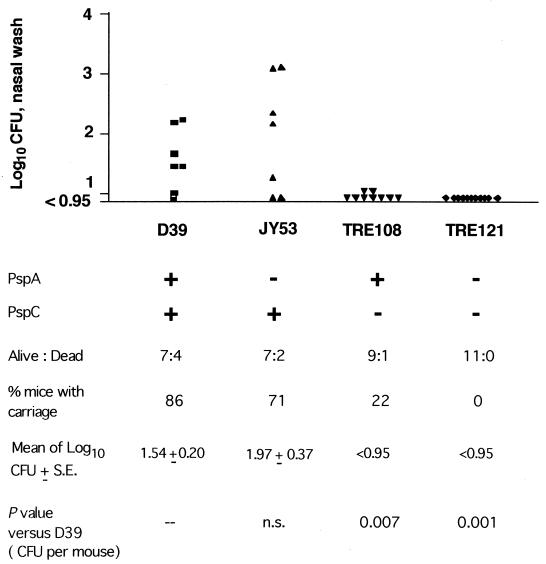
Nonanesthetized CBA/N mice were inoculated intranasally with 10 μl containing 1 × 107 CFU of either S. pneumoniae D39 or the isogenic strains mutated in PspC and/or PspA (Fig. 2). Four of 11 mice died after intranasal inoculation with wild-type D39, and 6 of the 7 remaining mice were colonized intranasally with D39. Following intranasal inoculation with JY53 (strain D39, mutant for PspA), 2 of the 9 mice died and the number of CFU colonizing the nasal tissue was slightly higher than for wild-type D39. Only 1 of the 10 mice infected with TRE108 (strain D39, mutant for PspC) died, and carriage was virtually absent in the remaining mice. Pneumococci were recovered from only 2 of the mice and only at a minimal level. Carriage was completely eliminated with TRE121 (double mutant of D39, lacking both PspA and PspC). None of the mice infected with this mutant died. A second set of independently generated mutants, TRE118 (lacking PspC) and TRE136 (lacking both PspA and PspC) also did not colonize the mouse nasopharynx (log CFU < 0.95).
FIG. 2.
Effect of mutation of pspA and pspC on intranasal carriage of D39. CBA/N mice were infected with 1 × 107 CFU of D39 or its isogenic mutants. Mice were sacrificed 7 days postinfection, and their nasal cavities were washed with 50 μl of Ringer's solution. The nasal washes were plated to enumerate the bacteria. P values versus D39 were calculated by using the Wilcoxon two-sample rank test. S.E., standard error.
Capsular serotype 19 pneumococci colonize the nasopharynx at a higher level than is usually observed for D39 (D. E. Briles and R. Fulghum, unpublished data). Mice inoculated in the nose with wild-type strain EF3030 carried an average of 103.5 CFU per nose, whereas isogenic strains lacking PspC showed no ability to be carried. Identical results were obtained with each of the independent mutants. Three out of 10 mice that were infected with the strain of EF3030 lacking PspC (TRE125) showed some colonization in the nasopharynx, with the nasal washes containing a few hundred bacteria. However, the pneumococci isolated from these three mice were identified as revertants because they were antibiotic sensitive and because their pspC gene was identical to that of the wild type by PCR and Southern analysis (data not shown). EF3030 strains mutated in PspC that remained antibiotic resistant did not colonize the nose. In the above experiments, as well as in the following infection models, strains with the plasmid introduced downstream of pspC (controls for downstream polar effects) behaved similarly to the wild-type strains (data not shown).
Lung infections in the mouse model.
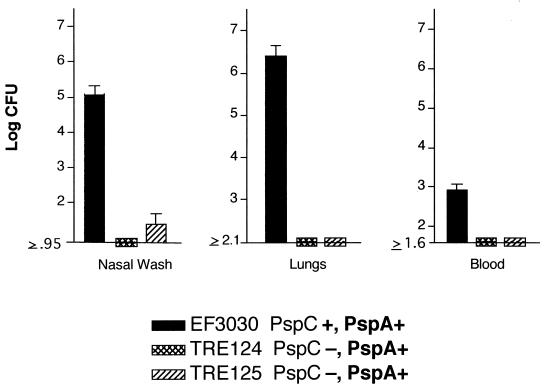
If mice are anesthetized and infected intranasally with about 106 CFU of strain EF3030 in 40 μl of Ringer's solution, pneumococci are aspirated into the lungs and a pneumonia-like disease develops. The bacteria multiply in the lung tissue for over a week. Under these conditions, EF3030 infects the lungs and colonizes the nasal cavity but is found only transiently and in low numbers in the blood. If anesthetized mice are infected, however, with doses as high as 107 or more, they become septic and die within a few days (D. E. Briles, unpublished data). Three groups of mice were intranasally infected with EF3030 or an isogenic strain lacking PspC. Six days after challenge, the mice were sacrificed and the nasal wash and lung tissue were serially diluted and plated to determine the number of bacteria. Both of the strains bearing the pspC-null mutation were severely hampered in their abilities to cause pulmonary infection, and pneumococci containing an insertionally inactivated pspC gene were not recovered in the lung tissue (Fig. 3).
FIG. 3.
Effect of pspC mutations on lung infection in CBA/N mice. Groups of six anesthetized CBA/N mice were challenged intranasally with 5 × 106 CFU of wild-type EF3030 and two isogenic PspC-negative strains. Mice were sacrificed 6 days after challenge, and nasal wash, lung tissue, and blood samples were plated on blood agar plates to determine the number of bacteria.
Bacteremic infections in the mouse model.
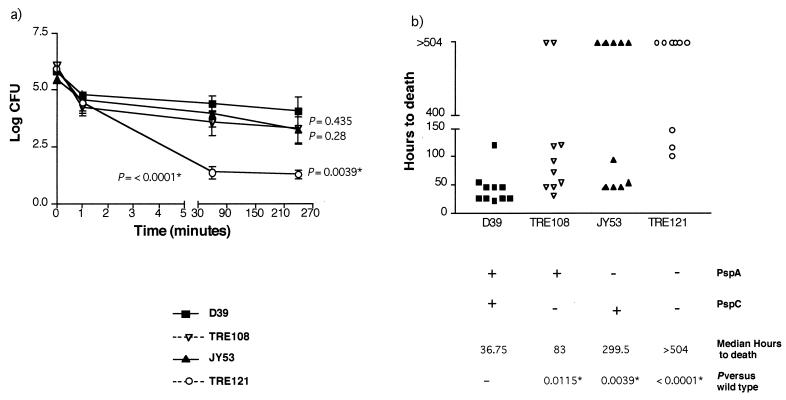
Previous studies have indicated that strains lacking PspA are attenuated in virulence and are generally cleared faster from the blood than the corresponding wild-type strain (10, 30, 37). We wanted to determine if the absence of PspC altered the blood clearance of the pneumococci and the survival of the mice. When BALB/c mice were intravenously infected with the parent strain, D39, and its isogenic mutant, TRE108 (lacking PspC), it was observed that TRE108 levels (in CFU) were only marginally lower than those of D39 in the first 4 h (see Fig. 5a). With respect to time to death, however, TRE108 killed significantly more slowly than D39 (Fig. 4b). With JY53 (lacking PspA), a trend towards a drop in CFU levels was observed after 4 h, although these numbers were not significantly different from those of wild-type D39 (Fig. 4a). In previous studies with CBA/N mice, JY53 was shown to have a statistically more-rapid clearance than D39 (30). The double knockout, TRE121, showed greatly enhanced clearance from the blood and lower virulence. It showed much more-rapid blood clearance than the wild type or either of the single mutants. Five of the 10 mice had completely cleared the bacteria by 1 h. In the absence of both PspA and PspC, there was also a statistically significant increase in the survival time of the mice from 36.75 h to greater than 504 h (Fig. 4b). This increase in survival time was much greater than that seen with either of the two single mutants with respect to time to death. The delay in death for mice infected with TRE121 was also significantly different from that for those infected with TRE108 (P = 0.0174), indicating a role for PspA. However, the survival times of mice infected with TRE121 were not significantly different from those of mice infected with JY53 (P = 0.245). Thus, it appeared that the pspA-pspC double mutant was less virulent than the pspA or pspC single mutant alone.
FIG. 5.
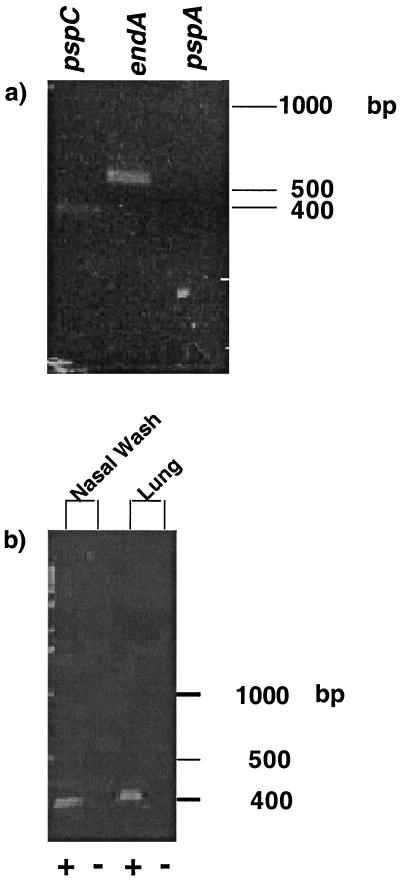
Detection of transcripts by RT-PCR. RNA was extracted from strain D39 in the nasal washes (a) or from strain EF3030 in the nasal washes and lung homogenates (b) of CBA/N mice. RNA was treated with DNase I. Transcripts were detected by RT-PCR with gene-specific primers. Transcripts detected were for pspC, endA, and pspA (a) and pspC (b). The plus and minus signs in panel b indicate reactions carried out in the presence (test) or absence (control) of RT, respectively. Similar controls were also used for the experiments whose results are shown in panel a.
FIG. 4.
Effect of mutations in pspC and pspA in a mouse bacteremia model. Groups of five mice (BALB/cByJ) were challenged intravenously with 1 × 106 CFU of wild-type pneumococci (D39), pspA-negative D39 (JY53), pspC-negative D39 (TRE108), and pspA- and pspC-negative D39 (TRE121). The presence of bacteria in the blood was determined at 1 min, 1 h, and 4 h (a). Subsequently, the number of hours to death was determined in each case (b). P values versus the wild type were calculated by using the Wilcoxon two-sample rank test. Mice that did not die after 21 days (504 h) were assigned a time to death of 504 h for statistical purposes. In each graph, values that are significantly different from D39 are indicated by asterisks.
Detection of the pspC transcript in vivo.
Previously published data (33) as well as the above-described experiments indicated that PspC can play a role in the early stages of infection. To determine if pspC was transcribed during nasal colonization and pneumonia, we used RT-PCR and were able to detect the pspC transcript in the nasal washes of mice infected intranasally with strain D39 (Fig. 5a). We were also able to detect the transcript for the gene encoding endonuclease A (endA), a constitutive gene that served as our positive control. However, we were unable to detect the transcript for pspA. This result substantiates our previous finding that the absence of pspA does not reduce the ability of the pneumococci to colonize the nasopharynx. Thus, pspA may not be required at this location. A similar experiment performed on samples from the nasal washes and lungs of mice infected with EF3030 (pneumonia model) confirmed that pspC is transcribed in vivo (Fig. 5b).
Intranasal immunization with PspC.
To examine protection against carriage, we immunized mice with PspC cloned from strain D39. D39 is the homologous challenge strain for the immunizing recombinant PspC. Although D39 does not carry well in mice, it is quite invasive. The pneumolysin-negative strain of D39, PLN-A, is less invasive (4), but as reported here, it is much better at establishing nasal carriage than D39. For this reason, the challenge strain used for this study is the pneumolysin-negative mutant of D39, PLN-A.
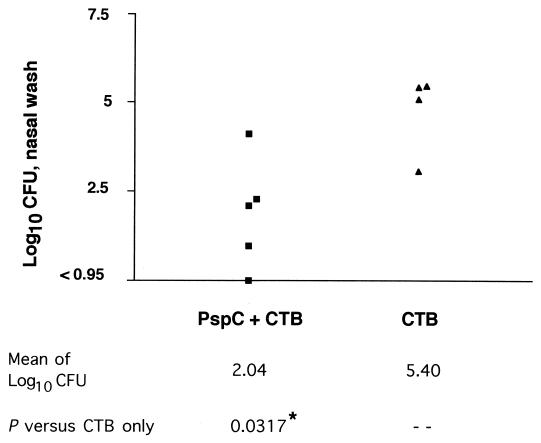
When the mice were challenged with PLN-A, the numbers of bacteria recovered from the nasal washes of the immunized mice were significantly lower than those from the control mice (Fig. 6). We also challenged mice intranasally with BG7322, a capsule-type 6B strain that has a clade B PspC (same type as D39 and as that used by Brooks-Walter et al. [11]). The number of BG7322 pneumococci recovered in the nasal washes of the five mice immunized with PspC was 102.26 ± 0.60 (geometric mean CFU and standard error) compared to 103.56 ± 0.51 in the control mice. The trend with BG7322 colonization was towards protection but was not statistically significant. Saliva and serum collected 24 h prior to challenge were analyzed to determine the amount of IgA antibody present in mucosal secretions and total antibody present in the serum. The serum from immunized mice had an antibody (Ig) log titer of 3.03, whereas the serum from control mice had an antibody log titer of <1.66, the limit of detection. We were unable to detect salivary IgA antibody specific to PspC in either the immunized or the control mice.
FIG. 6.
Intranasal immunization with PspC. CBA/N mice were immunized with 2 μg of purified PspC and 4 μg of CTB or CTB alone once a week for 3 weeks. Three weeks after the last immunization, the mice were challenged intranasally with PLN-A (pneumolysin mutant of D39). Mice were sacrificed after 7 days, the nasal cavities were washed with Ringer's solution, and the nasal wash samples were plated to determine the CFU counts. P values were calculated by using the one-tailed Wilcoxon test. One mouse in the CTB group died and was assigned a nasal wash log CFU value of 6 for statistical purposes.
DISCUSSION
These studies demonstrate that PspC is important for maintaining the pneumococci in the ecological niche for nasal carriage. We have examined the effect of PspC and PspA mutations in serotype 2 and serotype 19 strains. Utilizing the mouse pneumonia, carriage, and bacteremia models, we can conclude that PspC is a fundamental determinant in mucosal colonization and pulmonary infection acquired through a mucosal route.
Carriage in the nasopharynx is the first step of the disease process. Host-cell surface carbohydrates containing N-acetylgalactosamine β-1-4 galactose, N-acetylglucosamine β-1-4 galactose, and protein structures like the platelet-activating factor receptor, are all important in the adherence of pneumococci, as has been observed with many pulmonary pathogens (2, 14). In vitro studies conducted by Rosenow et al. (33) proposed that PspC is a pneumococcal adhesin. PspC was shown to bind immobilized lacto-N-neotetraose, and the addition of lacto-N-neotetraose intranasally prevented colonization of the nasopharynx (24, 33). Serotype 6B and serotype 2 strains mutated in pspC were attenuated in their abilities to colonize the nasopharynx in the infant rat model (33). These findings are in agreement with this study showing that PspC is necessary for optimal carriage in the adult mouse model.
Studies by Zhang et al. have indicated that pneumococci exploit the ability of PspC to interact with pIgR to migrate across the mucosal barrier (46). While pIgR is highly expressed in the nasopharynx and the upper respiratory tract, its level is marginal in the lower respiratory tract, which is the site of manifestation of pneumonia. It has been proposed that strains mutant for PspC are unable to colonize the nasopharynx primarily due to the lack of the PspC-pIgR interaction (46). However, our results indicate that PspC is also important for infection of the lungs. We eliminated the possibility that these effects are due to another gene by utilizing at least two independent mutants as well as by using strains where the plasmid was introduced downstream of pspC, to account for polar mutations. It is likely that PspC has to interact with a factor other than pIgR in the lungs to maintain and propagate a successful infection. PspC has been shown to be an important inflammatory stimulus (27), and maybe it is this lack of stimulus that abolishes the ability of a pspC mutant strain to initiate pneumonia.
While many of the components required for adherence and colonization have been determined, the physiological interaction of the bacteria and the host is poorly understood. Several pneumococcal factors have been implicated to play an important role at this stage of infection (15, 18, 28, 33, 34). However, it has been difficult scientifically to address the growth kinetics of bacteria in the host because colonization is a dynamic process. Bacteria adhere to the nasopharynx and remain within this niche unless a breach in the host defenses is present and the bacteria are able to cause a more-invasive infection.
Bacteria must also maintain some level of replication in the nasopharynx; however, excessive growth or invasion could cause an aggravated immune response, resulting in clearance of the bacteria from the mucosal surface. Evidence for this hypothesis is found in the present study. D39 carries very poorly in mice and commonly cannot be recovered from carriage at all. In the present study we observed that mutations in both pspA and ply enhanced the carriage of this strain. Both of these genes have been previously shown to be important for in vivo virulence of S. pneumoniae (7, 30, 37). Thus, it would appear that reducing the invasion of the D39 pneumococci in CBA/N mice enhanced their ability to carry in the nasal tissue. It is therefore possible that inflammation induced by successful invasion of deeper tissues causes a loss of carriage.
We have isolated PspC revertants from the nasal wash, indicating that a small number of pneumococci, in the absence of antibiotic pressure, have eliminated the suicide vector and reconstituted the pspC gene. These PspC revertants then go on to multiply and colonize the nasopharynx. These results indicate that pneumococci are very metabolically active and rapidly divide within the nasopharynx. Our results also demonstrate that pspC is actively transcribed when the bacteria are growing in vivo. These findings emphasize that there is a selective pressure for expression of a functional PspC in vivo.
While PspC appeared to be important for the establishment of carriage and pneumonia, it did not appear to be an important virulence factor during bacteremia by itself. However, the loss of both PspA and PspC impairs the ability of the pneumococci to cause bacteremia. This observation supports our initial hypothesis that in at least some strain backgrounds, the activities of PspA and PspC are able to complement each other. Such synergy has been previously observed between PspC and another virulence factor, pneumolysin (6). Interestingly, all three of these virulence proteins have been shown to interact with the complement pathway. PspA interacts with the alternate complement pathway and has anticomplementary activity (37). PspC can bind to factor H of the alternate complement pathway (25). Pneumolysin can activate the classical complement pathway in the absence of anti-pneumolysin antibodies, thereby depleting complement locally (32). One could thus speculate that the synergy seen between these proteins could be because they may each be able to interfere with complement-dependent opsonization in vivo.
It has previously been shown that PspC can elicit protection against sepsis (11) and that a mixture of choline-binding proteins including PspC can protect against carriage (33). As a consequence of our findings that demonstrate an essential role of PspC in carriage, it was anticipated that PspC might also confer protective immunity. This study demonstrates that immunization with PspC elicited systemic mucosal antibody and reduced the level of carriage in the nasopharynx. PspC-specific antibody was also present in the serum. We were unable to detect increased levels of IgA in the saliva, but these results do not rule out a potential role for IgA antibody to PspC.
Protection against carriage will prevent pneumococcal infections and impede the distribution of the pathogen within the population. In the case of the polysaccharide-protein conjugate vaccines under development, studies are still ongoing. The data released to date, however, indicate that in some cases antibody reactive to the capsular polysaccharide can partially reduce carriage of specific capsular types but that the increased carriage of capsular types not represented in the vaccine compensated for this loss (16, 26).
Reduction of carriage rates of pneumococci should result in herd immunity and provide protection against disease, even among children who have not yet been immunized and among immunodeficient individuals for whom a vaccine that protects only against sepsis may be of marginal benefit. The findings of this study suggest that PspC may be an excellent candidate for a mucosal pneumococcal vaccine.
Acknowledgments
We thank Orlanda Thomas, Yvette Hale, and Robert Fulghum for technical assistance.
This work was supported by National Institute of Allergy and Infectious Diseases (National Institutes of Health) grant R01 AI21548 and National Heart, Lung, and Blood Institute grant P60 HL56418. A.B.-W. was funded by the University of Alabama Comprehensive Minority Faculty Development Fellowship.
P.B. and A.B.-W. contributed equally to this work.
Editor: E. I. Tuomanen
REFERENCES
- 1.Achen, M. G., B. E. Davidson, and A. J. Hillier. 1986. Construction of plasmid vectors for the detection of streptococcal promoters. Gene 45:45-49. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, B., J. Dahmen, T. Frejd, H. Leffler, G. Magnusson, G. Norri, and C. S. Eden. 1983. Identification of an active disaccharide unit of a glycoconjugate receptor for pneumococci attaching to human pharyngeal epithelial cells. J. Exp. Med. 158:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benton, K. A., M. P. Everson, and D. E. Briles. 1995. A pneumolysin-negative mutant of Streptococcus pneumoniae causes chronic bacteremia rather than acute sepsis in mice. Infect. Immun. 63:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benton, K. A., J. C. Paton, and D. E. Briles. 1997. Differences in virulence of mice among Streptococcus pneumoniae strains of capsular types 2, 3, 4, 5, and 6 are not attributable to differences in pneumolysin production. Infect. Immun. 65:1237-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry, A., and J. Paton. 2000. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of genes encoding pneumolysin and other putative pneumococcal virulence proteins. Infect. Immun. 68:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, A. M., J. Yother, D. E. Briles, D. Hansman, and J. C. Paton. 1989. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect. Immun. 57:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briles, D. E., M. J. Crain, B. M. Gray, C. Forman, and J. Yother. 1992. A strong association between capsular type and mouse virulence among human isolates of Streptococcus pneumoniae. Infect. Immun. 60:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briles, D. E., S. K. Hollingshead, E. Swiatlo, A. Brooks-Walter, A. Szalai, A. Virolainen, L. S. McDaniel, K. A. Benton, P. White, K. Prellner, A. Hermansson, P. C. Aerts, H. Van Dijk, and M. J. Crain. 1997. PspA and PspC: their potential for use as pneumococcal vaccines. Microb. Drug Resist. 3:401-408. [DOI] [PubMed] [Google Scholar]
- 10.Briles, D. E., J. Yother, and L. S. McDaniel. 1988. Role of pneumococcal surface protein A in the virulence of Streptococcus pneumoniae. Rev. Infect. Dis. 10:S372-S374. [DOI] [PubMed] [Google Scholar]
- 11.Brooks-Walter, A., D. E. Briles, and S. K. Hollingshead. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect. Immun. 67:6533-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, Q., D. Finkel, and M. Hostetter. 2000. Novel purification scheme and functions for a C3-binding protein from Streptococcus pneumoniae. Biochemistry 39:5450-5457. [DOI] [PubMed] [Google Scholar]
- 13.Crain, M. J., W. D. Waltman, J. S. Turner, J. Yother, D. E. Talkington, L. M. McDaniel, B. M. Gray, and D. E. Briles. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect. Immun. 58:3293-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cundell, D. R., N. P. Gerard, C. Gerard, I. Idanpaan-Heikkila, and E. I. Tuomanen. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435-438. [DOI] [PubMed] [Google Scholar]
- 15.Cundell, D. R., J. N. Weiser, J. Shen, A. Young, and E. I. Tuomanen. 1995. Relationship between colonial morphology and adherence of Streptococcus pneumoniae. Infect. Immun. 63:757-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagan, R., M. Muallem, R. Malemed, O. Leroy, and P. Yagupsky. 1997. Reduction of pneumococcal nasopharyngeal carriage in early infancy after immunization with tetravalent pneumococcal vaccines conjugated to either tetanus toxoid or diphtheria toxoid. Pediatr. Infect. Dis. J. 16:1060-1064. [DOI] [PubMed] [Google Scholar]
- 17.Dave, S., A. Brooks-Walter, M. K. Pangburn, and L. S. McDaniel. 2001. PspC, a pneumococcal surface protein, binds human factor H. Infect. Immun. 69:3435-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grist, R. W., and L. O. Butler. 1983. Effect of transforming DNA on growth and frequency of mutation of Streptococcus pneumoniae. J. Bacteriol. 153:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakansson, A., H. Roche, S. Mirza, L. S. McDaniel, A. Brooks-Walter, and D. E. Briles. 2001. Characterization of binding of human lactoferrin to pneumococcal surface protein A. Infect. Immun. 69:3372-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt, S., G. Bethe, P. Remanen, and G. S. Chhatwal. 1999. Identification of pneumococcal surface protein A as a lactoferrin-binding protein of Streptococcus pneumoniae. Infect. Immun. 67:1683-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammerschmidt, S., S. R. Talay, P. Brandtzaeg, and G. S. Chhatwal. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol. 25:1113-1124. [DOI] [PubMed] [Google Scholar]
- 23.Haverstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Idanpaan-Heikkila, I., P. M. Simon, D. Zopf, T. Vullo, P. Cahill, K. Sokol, and E. Tuomanen. 1997. Oligosaccharides interfere with the establishment and progression of experimental pneumococcal pneumonia. J. Infect. Dis. 176:704-712. [DOI] [PubMed] [Google Scholar]
- 25.Janulczyk, R., F. Iannelli, A. G. Sjoholm, G. Pozzi, and L. Bjorck. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J. Biol. Chem. 275:37257-37263. [DOI] [PubMed] [Google Scholar]
- 26.Lipsitch, M. 1999. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg. Infect. Dis. 5:336-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madsen, M., Y. Lebenthal, Q. Cheng, B. L. Smith, and M. K. Hostetter. 2000. A pneumococcal protein that elicits interleukin-8 from pulmonary epithelial cells. J. Infect. Dis. 181:1330-1336. [DOI] [PubMed] [Google Scholar]
- 28.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDaniel, L. S., J. S. Sheffield, E. Swiatlo, J. Yother, M. J. Crain, and D. E. Briles. 1992. Molecular localization of variable and conserved regions of pspA, and identification of additional pspA homologous sequences in Streptococcus pneumoniae. Microb. Pathog. 13:261-269. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel, L. S., J. Yother, M. Vijayakumar, L. McGarry, W. R. Guild, and D. E. Briles. 1987. Use of insertional inactivation to facilitate studies of biological properties of pneumococcal surface protein A (PspA). J. Exp. Med. 165:381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overweg, K., C. D. Pericone, G. G. Verhoef, J. N. Weiser, H. D. Meiring, A. P. De Jong, R. De Groot, and P. W. Hermans. 2000. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect. Immun. 68:4604-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paton, J. C. 1996. The contribution of pneumolysin to the pathogenicity of Streptococcus pneumoniae. Trends Microbiol. 4:103-106. [DOI] [PubMed] [Google Scholar]
- 33.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25:819-829. [DOI] [PubMed] [Google Scholar]
- 34.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19:803-813. [DOI] [PubMed] [Google Scholar]
- 35.Takashima, K., K. Tateda, T. Matsumoto, J. Iizawa, M. Nakao, and K. Yamaguchi. 1997. Role of tumor necrosis factor alpha in pathogenesis of pneumococcal pneumonia in mice. Infect. Immun. 65:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 37.Tu, A.-H. T., R. L. Fulgham, M. A. McCory, D. E. Briles, and A. J. Szalai. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect. Immun. 67:4720-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiser, J. N. 1998. Phase variation in colony opacity by Streptococcus pneumoniae. Microb. Drug Resist. 4:129-135. [DOI] [PubMed] [Google Scholar]
- 39.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62:2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, H.-Y., M. Nahm, Y. Guo, M. Russell, and D. E. Briles. 1997. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage and infection with Streptococcus pneumoniae. J. Infect. Dis. 175:839-846. [DOI] [PubMed] [Google Scholar]
- 41.Wu, H.-Y., A. Virolainen, B. Mathews, J. King, M. Russell, and D. E. Briles. 1997. Establishment of a Streptococcus pneumoniae nasopharyngeal model of pneumococcal carriage in adult mice. Microb. Pathog. 23:127-137. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, M., L. S. McDaniel, K. Kawabata, D. E. Briles, R. J. Jackson, J. R. McGhee, and H. Kiyono. 1997. Oral immunization with PspA elicits protective humoral immunity against Streptococcus pneumoniae infection. Infect. Immun. 65:640-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yother, J., G. L. Handsome, and D. E. Briles. 1992. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J. Bacteriol. 174:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yother, J., L. S. McDaniel, and D. E. Briles. 1986. Transformation of encapsulated Streptococcus pneumoniae. J. Bacteriol. 168:1463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, J. R., K. E. Mostov, M. E. Lamm, M. Nanno, S. Shimida, M. Ohwaki, and E. Tuomanen. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827-837. [DOI] [PubMed] [Google Scholar]