Abstract
PCR-based subtractive genome hybridization produced clones harboring inserts present in Brazilian purpuric fever (BPF) prototype strain F3031 but absent in noninvasive Haemophilus influenzae biogroup aegyptius isolate F1947. Some of these inserts have no matches in the GenBank database, while others are similar to genes encoding either known or hypothetical proteins. One insert represents a 2.3-kb locus with similarity to a Thermotoga maritima hypothetical protein, while another is part of a 7.6-kb locus that contains predicted genes encoding hypothetical, phage-related, and carotovoricin Er-like proteins. The presence of DNA related to these loci is variable among BPF isolates and nontypeable H. influenzae strains, while neither of them was detected in strains of types a to f. The data indicate that BPF-causing strain F3031 harbors unique chromosomal regions, most of which appear to be acquired from unrelated microbial sources.
Haemophilus influenzae biogroup aegyptius was identified in the mid 1980s as the etiological agent of Brazilian purpuric fever (BPF), a frequently fatal invasive pediatric disease (6). Originally, a clone was isolated from patients with BPF in Sao Paulo State, Brazil (7). However, after outbreaks in other regions of Brazil (12, 34) and in Australia (19, 37) and a case of a child from Connecticut with an infection consistent with BPF (35), it is clear that a single H. influenzae biogroup aegyptius strain or clone is not the sole agent responsible for BPF. These observations led to the hypothesis that BPF-causing strains harbor DNA that is absent in noninvasive isolates, which may encode the factors that transformed a benign microorganism into an aggressive pathogen. This hypothesis was tested by using a genomewide approach based on PCR-based subtractive genome hybridization.
Generation of a subtracted genomic library
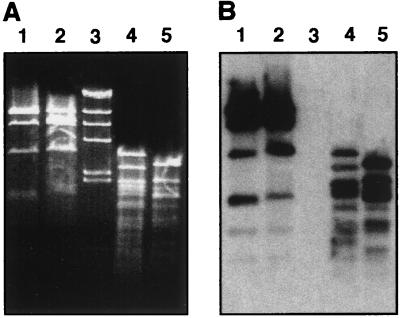
We first addressed an important concern: both strains contain a 24-MDa plasmid (7). The significance of this is that differences in plasmid content between the F3031 and F1947 strains could result in misleading subtractive hybridization results. Restriction and Southern blot analyses (25) showed that the plasmids from F3031 and F1947 have similar AccI restriction profiles and cross hybridize, although they have some differences in their RsaI patterns (Fig. 1A and B). Nevertheless, the plasmid contents of these two strains are similar and should not affect the subtractive hybridization process.
FIG. 1.
Agarose gel electrophoresis and Southern hybridization of plasmid DNA. (A) The 24-MDa plasmid pF3031 from strains F3031 (lanes 2 and 5) and F1947 (lanes 1 and 4) was digested with AccI (lanes 1 and 2) and RsaI (lanes 4 and 5), and restriction fragments were separated by electrophoresis in an ethidium bromide-stained gel. Lane 3 contained HindIII-digested λ DNA. (B) The DNA fragments were blotted onto nitrocellulose and probed with 32P-labeled pF3031.
A library enriched in DNA unique to F3031 was made with the Clontech PCR-Select bacterial genome subtractive kit by using F3031 and F1947 total DNAs. After confirming that such a library was obtained, a secondary PCR with nested primers was conducted and the amplicons were cloned with a TA system (Invitrogen) and Escherichia coli DH5α or TOP10F′ (Table 1) competent cells. Colony hybridization (25) with 32P-labeled, RsaI-digested F1947 and F3031 genomic DNA showed that approximately 27% of the clones contained DNA unique to F3031, a value comparable to that described by the kit manufacturer and reported previously in a similar analysis of Helicobacter pylori strains (2). This apparently low percentage of unique clones could be due to a base substitution(s) and restriction fragment length polymorphisms in DNA common to both strains that can decrease subtraction efficiency (2). The presence of F3031 DNA fragments containing patches of sequences homologous to F1947 is another reason for the apparently low subtraction efficiency. Nevertheless, the analyses described below show that more than half of the subtracted clones examined contain DNA unique to BPF-associated strain F3031.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| H. influenzae biogroup aegyptiusa | ||
| F3031 | Invasive BPF isolate, prototype strain | 7 |
| F1947 | Noninvasive non-BPF isolate | 7 |
| F3029 | Invasive BPF isolate | 7 |
| F3033 | Invasive BPF isolate | 7 |
| F3037 | Invasive BPF isolate | 7 |
| F4380 | Invasive BPF isolate from Australia | 37 |
| Connecticut | Invasive BPF isolate from United States | CDCb via A. Lesse |
| Valparaiso | Invasive BPF isolate from Brazil | CDC via A. Lesse |
| H. influenzae | ||
| DL42 | Type b | E. Hansen |
| DL63 | Type b | E. Hansen |
| Eagan | Type b | S. Goodgal |
| TN106 | Nontypeable | E. Hansen |
| 165-NP | Nontypeable | L. Bakaletz |
| 86-028NP | Nontypeable | L. Bakaletz |
| 1128MEE | Nontypeable | L. Bakaletz |
| 1728MEE | Nontypeable | L. Bakaletz |
| AMC 36-A-3 | Type a | ATCCc |
| AMC 36-A-5 | Type c | ATCC |
| AMC 36-A-6 | Type d | ATCC |
| AMC 36-A-7 | Type e | ATCC |
| AMC 36-A-8 | Type f | ATCC |
| E. coli | ||
| TOP10F′ | Host cloning strain | Invitrogen |
| DH5α | Host cloning strain | 25 |
| Plasmids | ||
| pUC18 | BamHI- and alkaline phosphatase-treated cloning vector | Pharmacia |
| pCR2.1 | TA cloning vector | Invitrogen |
| pF3031 | 24-MDa H. influenzae biogroup aegyptius plasmid | This study |
All of the H. influenzae biogroup aegyptius strains listed, except F4380, Connecticut, and Valparaiso, are clinical isolates from Brazil and contain 24-MDa plasmid pF3031.
CDC, Centers for Disease Control and Prevention.
ATCC, American Type Culture Collection.
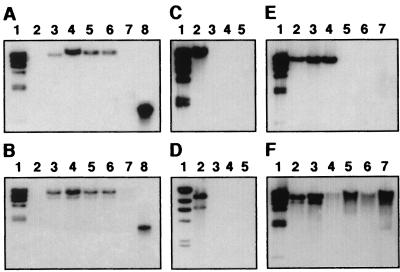
Analysis of 19 clones showed that all of them contained 300- to 1,500-bp inserts that hybridized with the F3031 genomic DNA probe, while only three also hybridized with the F1947 genomic DNA probe. In contrast, probing of the F3031 and F1947 genomic DNAs with each of the 19 clones showed that while 11 of them contained F3031-specific DNA, 8 had inserts common to both strains. These conflicting results could be due to changes in the probe-to-target ratios in the hybridization experiments, as reported in a similar analysis of Salmonella enterica serovar Typhimurium (9). Interestingly, the restriction fragments with homology to both probes displayed different sizes in F3031 and F1947, suggesting the presence of polymorphisms in the alleles contained within these common fragments. Two additional clones (MU33 and MU34) were examined in greater detail. MU33 and MU34 are 900- and 1,100-bp subtracted fragments, respectively, that were located on different F3031 HindIII genomic fragments (Fig. 2A and B, lanes 2) and were not detected in strain F1947 (Fig. 2A and B, lanes 3). Neither of them hybridized with pF3031 (Fig. 2A and B, lanes 4) or with each other (Fig. 2A and B, lanes 5).
FIG. 2.
Southern blot analysis of H. influenzae biogroup aegyptius genomic and plasmid DNAs. Genomic DNAs from F3031 (lane 2) and F1947 (lane 3) and pF3031 (lane 4) were digested with HindIII. EcoRI-digested pMU34 and pMU33 were loaded into lanes A5 and B5, respectively. In both panels, lane 1 contained HindIII-digested λ DNA and lane 6 was empty. PCR amplicons of the pMU33 (lane A7) and pMU34 (lane B7) inserts were used as positive controls. DNA fragments were separated by agarose gel electrophoresis and blotted onto nitrocellulose. Blots were probed with 32P-labeled HindIII-digested λ DNA and 32P-labeled PCR amplicons of MU33 (A) and MU34 (B).
In silico analyses of subtracted DNA common to F3031 and F1947
Table 2 shows that all of the common fragments except B1 encode products with matches in the GenBank database, some of which are related to hypothetical (F5) or bacteriophage (E8) proteins. The product of D3 is similar to the RP4-encoded TraE protein. Hybridization of pD3 with the plasmids present in F3031 and F1947 but not with their chromosomes proved that D3 represents plasmid rather than chromosomal DNA differences. Whether these differences affect gene transfer or the virulence of H. influenzae biogroup aegyptius harboring pF3031-like elements remains to be examined by either testing the virulence of plasmidless isogenic derivatives or comparing the complete nucleotide sequences of the plasmids present in these two H. influenzae biogroup aegyptius strains.
TABLE 2.
Sequence analysis and G+C content of subtracted clones
| Clone | Insert size (bp) | Homolog(s)b | G+C content (%)c | Scored | E valued |
|---|---|---|---|---|---|
| B1a | 960 | No significant match | 38.0 | NAe | NA |
| B2 | 1,129 | Rd HI1494, hypothetical protein | 37.5 | 199 | 6e-50 |
| B3 | 558 | Rd HI1467, hypothetical ABC transport protein | 33.9 | 243 | 1e-63 |
| D2 | 356 | D. radiodurans hypothetical protein | 43.8 | 59.0 | 2e-08 |
| D3a | 1,025 | traE of plasmid RP4 | 38.2 | 262 | 9e-69 |
| D5a | 1,088 | Rd HI0361, hypothetical fecE iron transport gene | 39.3 | 601 | 1e-171 |
| D11 | 506 | Rd HI1265, conserved hypothetical protein | 39.3 | 335 | 2e-91 |
| D13 | 676 | No significant match | 39.6 | NA | NA |
| E3 | 1,253 | Carotovoricin Er | 39.9 | ||
| Tail sheath protein | 152 | 1e-35 | |||
| Tail core protein | 147 | 4e-34 | |||
| E8a | 616 | P22 phage antirepressor protein | 37.8 | 189 | 4e-47 |
| E9a | 444 | H. influenzae hmcD hemocin gene | 22.0 | 178 | 7e-44 |
| E10 | 423 | Rd HI0873, glucose 4,6-dehydratase (rffG) | 37.1 | 178 | 3e-44 |
| F2a | 1,151 | Hypothetical genes | 34.5 | ||
| N. meningitidis MC58 adhesin/invasin | 73 | 8e-12 | |||
| HI0422, ATP-dependent RNA helicase (srmB) | 153 | 5e-36 | |||
| F5a | 1,370 | Rd hypothetical proteins | 35.1 | ||
| HI1273, conserved hypothetical protein | 148 | 3e-34 | |||
| HI1266, hypothetical protein | 193 | 7e-42 | |||
| HI1265, conserved hypothetical protein | 179 | 1e-43 | |||
| F6a | 296 | Rd HI0291/HI0292 hypothetical Hg binding proteins | 35.4 | 76 | 3e-13 |
| F7 | 885 | Carotovoricin Er baseplate protein | 42.4 | 149 | 5e-34 |
| F10 | 763 | H. influenzae immunoglobulin A protease (iga) | 35.9 | 534 | 1e-151 |
| F17 | 590 | Rd hypothetical proteins | 33.4 | ||
| HI1466.1, hypothetical TonB-dependent receptor protein | 228 | 6e-59 | |||
| HI1467, hypothetical ABC transport protein | 132 | 8e-30 | |||
| F20 | 556 | Carotovoricin Er tail protein | 43.3 | 410 | 1e-21 |
| MU33 | 826 | Rd HI1508, Mu-like prophage protein GP36 | 41.9 | 50 | 4e-05 |
| MU34 | 1,384 | Thermotoga maritima hypothetical protein | 29.6 | 77 | 1e-12 |
Clone containing DNA present in H. influenzae biogroup aegyptius strains F3031 and F1947. All clones without a superscript a contain DNA unique to strain F3031.
Identification of homologs to subtracted clones is based on BLASTx analysis. The designation HI followed by a number corresponds to loci identified in the H. influenzae Rd KW20 genome (10).
G+C content was determined with Artemis (http://www.sanger.ac.uk) by using a window of 120 nucleotides.
The score and E value of the BLASTx analysis are shown. Scores of less than 50 were not considered significant matches.
NA, not applicable.
E9 is similar to the H. influenzae type b strain E1a hemocin gene (hmcD) and has a G+C content that is much lower than the 38% reported for H. influenzae strain Rd KW20 (10) (hereafter referred to as Rd). Interestingly, the entire hmc E1a locus has a significantly lower G+C content (20), suggesting that it represents a genomic region acquired by H. influenzae encapsulated and nonencapsulated strains from an unrelated source. A small portion of F2 is similar to srmB of Rd (10), which is related to D-E-A-D box helicases (26), while most of it is related to a Neisseria meningitidis MC58 adhesin (32) and the Moraxella catarrhalis UspA2 and UspA2H proteins. The latter is involved in adhesion of this pathogen to conjunctival epithelial cells (8, 14). Interestingly, BPF and non-BPF strains attach to these cells (30), although the expression and role of these proteins in H. influenzae biogroup aegyptius remain to be tested. F6 has similarity to Rd HI0291 and HI0292 genes encoding putative Hg-binding proteins (10), while D5 is similar to HI0361 (10), which is related to the E. coli FecE (29) and Yersinia pestis YfeB (3) iron transport proteins. We have shown that F3031 expresses an FecE-like protein and other components of a siderophore-independent iron acquisition system (28).
In silico analyses of subtracted DNA unique to F3031
B3 and F17 are related to an ABC transport system of Rd (10) that includes an ATP-binding protein and a TonB-dependent receptor potentially involved in iron acquisition. These two clones have similar G+C contents, which are approximately 4% lower than that of Rd (10). The product of D2 has a G+C content 6% higher than that of Rd and is similar to a Deinococcus radiodurans R1 conserved hypothetical protein (36). The predicted product of D11 is similar to a hypothetical Rd protein (10) also detected in E. coli (5, 24), Pasteurella multocida (18), and Pseudomonas aeruginosa (31). The D13-encoded protein has a low level of similarity to the product of orf277, which flanks the vrl virulence-related locus found more frequently in virulent than in nonvirulent isolates of Dichelobacter nodosus (4). The products of E3, F7, and F20 are related to different components of the phage-tail-like bacteriocin carotovoricin Er produced by the phytopathogen Erwinia carotovora (22). Although some structural features and the genetic mechanism involved in host range specificity changes have been described (22), the role of carotovoricin Er in the virulence of this plant pathogen remains to be elucidated. While the G+C content of E3 is similar to that of Rd, the content of F7 and F20 is significantly higher than 38%. E10 is similar to rffG (HI0873) of Rd (10), which is present in the genome of other gram-negative pathogens and encodes an enzyme required for the biosynthesis of the O7 lipopolysaccharide in E. coli K1 (16). MU33 contains the 3′ end of the HI1508 Rd homolog (10), encoding a Mu-like protein, and the 5′ end of an open reading frame (ORF) with no match in the GenBank database. The inferred product of MU34 is highly similar to a hypothetical protein of Thermotoga maritima MSB8 (21), a thermophilic bacterium that is considered one of the deepest-branching eubacterial species (1). The G+C content of these two subtracted regions is significantly different from that of Rd, suggesting that they were acquired by F3031 from unrelated microorganisms. Although the roles of all of these predicted genes and proteins in the virulence of this BPF clone are unknown, their absence in the F1947 noninvasive strain suggests that they are virulence factor candidates.
The F10 subtracted fragment maps within the 1,380-bp region of the BPF iga gene reported previously (15). The preliminary observation that F3031 and F1947 contain restriction fragments of different sizes that hybridize with Rd iga (10) and the fact that all three strains secrete immunoglobulin A1 protease activity indicate that F10 represents a distinct genetic form of iga rather than a gene unique to F3031.
Detection of MU33 and MU34 in other H. influenzae strains
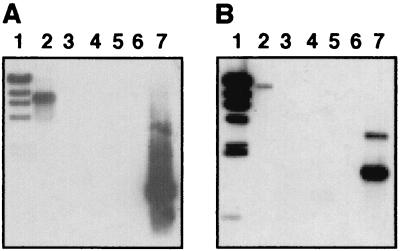
Sequences homologous to MU33 and MU34 were detected in F3029, F3033, and F3037 (Fig. 3A and B, lanes 3 to 6), which express all of the markers assigned to the Brazilian BPF clone (7). Although the Valparaiso and Connecticut BPF strains, which display some of the BPF markers, tested positive for MU33-related DNA, neither of them reacted with the MU34 probe (data not shown). No signals were detected when the F4380 Australian isolate, another BPF strain that does not express all of the markers described in the F3031 prototype strain, was probed with MU33 and MU34 under high-stringency (Fig. 3A and B, lanes 7) or low-stringency (data not shown) conditions. These results show that BPF strains are not identical and support the hypothesis that they have originated from a wider range of sources than originally predicted.
FIG. 3.
Southern blot analysis of H. influenzae biogroup aegyptius and H. influenzae strains. (A and B) Genomic DNAs from H. influenzae biogroup aegyptius strains F1947 (lane 2), F3031 (lane 3), F3029 (lane 4), F3033 (lane 5), F3037 (lane 6), and F4380 (lane 7) were digested with EcoRI. The PCR-amplified inserts of pMU33 (A, lane 8) and pMU34 (B, lane 8) were used as positive controls. (C and D) Genomic DNAs from H. influenzae biogroup aegyptius F3031 (lane 2) and H. influenzae type b strains DL42 (lane 3), DL63 (lane 4), and Eagan (lane 5) were digested with EcoRI. (E and F) Genomic DNAs from H. influenzae biogroup aegyptius F3031 (lane 2) and nontypeable H. influenzae type strains TN106 (lane 3), 165NP (lane 4), 86-028NP (lane 5), 1128MEE (lane 6), and 1728MEE (lane 7) were digested with EcoRI. (A to F) HindIII-digested λ DNA was used as a size marker (lane 1). (A, C, and E) DNA was probed with the radiolabeled, PCR-amplified pMU33 insert and radiolabeled, HindIII-digested λ DNA. (B, D, and F) DNA was probed with the radiolabeled, PCR-amplified pMU34 insert and radiolabeled, HindIII-digested λ DNA.
MU33- and MU34-related DNA was not detected (Fig. 3C and D, lanes 3 to 5, and data not shown) in any of the H. influenzae strains of types a to f tested (Table 1). Two (TN106 and 165NP) of five nontypeable strains (Table 1) contained MU33-related DNA (Fig. 3E, lanes 3 and 4), and all of them tested positive for MU34 (Fig. 3F, lanes 3 to 7). Thus, MU34-related sequences appear to be more common than MU33-like sequences among nontypeable strains, while both seem to be either absent or rare in typeable strains.
Cloning and analysis of the chromosomal region encompassing MU33
Screening of an F3031 library, which was made by cloning 4- to 6-kb partially digested Sau3AI fragments into pUC18, with MU33 as a probe yielded the overlapping clones pMU37 and pMU69. The nucleotide sequence of each clone was determined by using automated procedures (Applied Biosystems) and then assembled (Sequencher 4.1.2, Gene Codes) as a single 7,647-nucleotide contig (hereafter referred to as the BPF33 locus). This locus, which was not detected in F1947 by Southern hybridization, appears to contain seven complete and two partial ORFs, all transcribed in the same direction. BLASTn showed that BPF33 has the sequence 5′-CAACTGAAGATAATACGGTTGAATATGCGGAA-3′, which is also present in P. multocida PM70 (18), and a region 96.6% similar to the sequence 5′-AAAAGCCCAAGCTGAAGCCCAAAAAGCTG-3′ located in the Rd transformation gene cluster (10, 33). Copies of the 5′-AAGTGCGGT-3′ DNA uptake sequence (11) were located in the minus strand between ORFs 4 and 5 and at the ends of ORFs 2, 7, and 8 (Table 3), some of which were part of regions resembling the 29-bp Rd DNA uptake consensus sequence (27). Although the average G+C content of BPF33 is similar to that of Rd (10), six of its ORFs have a G+C content significantly higher than 38% (Table 3). BPF33 encompasses, in addition to MU33, the E3 subtracted region (Table 2) that overlaps ORFs 8 and 9 encoding putative homologs to the carotovoricin Er tail core and tail sheath proteins (Table 3). Accordingly, the inferred products of these two ORFs are similar to their cognate carotovoricin Er proteins. The predicted products of ORFs 1 and 2 are similar to N. meningitidis hypothetical proteins, while ORF 6 seems to encode an Rd Mu-like protein (Table 3). The inferred products of ORFs 3, 4, 5, and 7 have no significant matches in the GenBank database (Table 3). These data indicate that BPF33, which is present in the F3031 BPF-causing strain but absent in non-BPF strain F1947, is a relatively large locus with a mosaic structure that includes bacteriophage genes and novel genes encoding proteins of unknown function. Moreover, work in progress indicates that BPF33 is not simply a Mu-like element. Rather, it seems to be a large region that contains genes encoding proteins potentially involved in the production of a carotovoricin Er-like bacteriocin.
TABLE 3.
Sequence analysis and G+C content of predicted ORFs located within the BPF33 locus
| ORFa | Size (bp) | Protein size (amino acids/kDa) | Homolog(s)b | G+C content (%)c | Scored | E valued |
|---|---|---|---|---|---|---|
| 1e | 1,113 | 3,371/42.7 | N. meningitidis Z2491 hypothetical protein NMA1850 | 38.2 | 133 | 3 e-30 |
| 2 | 501 | 166/18.8 | N. meningitidis Z2491 hypothetical protein NMA1849 | 41.1 | 59 | 5 e-08 |
| 3 | 1,104 | 367/40.7 | No significant match | 42.9 | NAf | NA |
| 4 | 927 | 308/34.1 | No significant match | 38.5 | NA | NA |
| 5 | 318 | 105/12.1 | No significant match | 38.2 | NA | NA |
| 6 | 435 | 144/16.4 | HI1508, Mu-like protein GP36 | 41.8 | 46 | 4 e-08 |
| 7 | 498 | 165/18.5 | No significant match | 42.0 | NA | NA |
| 8 | 1,386 | 461/50.5 | Carotovoricin Er tail sheath protein | 41.2 | 359 | 7 e-98 |
| 9e | 435 | 145/15.9 | Carotovoricin Er tail core protein | 43.0 | 120 | 7 e-27 |
The numbers represent ORFs in the BPF33 locus.
Identification of homologs to subtracted clones was based on BLASTx analysis. The designation HI or NMA followed by a number corresponds to loci identified in the H. influenzae Rd KW20 (10) and N. meningitidis Z2491 genomes (23), respectively.
G+C content was determined with Artemis (http://www.sanger.ac.uk) by using a window of 120 nucleotides.
The score and E value from the BLASTx analysis are shown. Scores of less than 50 were not considered significant matches.
Partial ORF.
NA, not applicable.
Cloning and analysis of the chromosomal region encompassing MU34
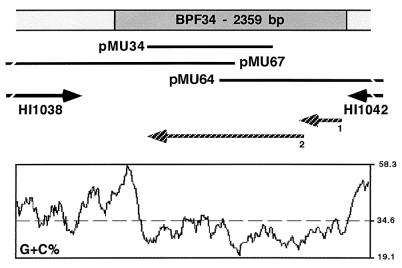
The pMU64 and pMU67 overlapping genomic clones, which were isolated from the genomic library by using MU34 as a probe, were sequenced and assembled as a single contig (Fig. 4). BLASTn showed that this contig contains a 2,359-bp region with no matches in the GenBank database that is flanked on the left and right sides by the HI1038 and HI1042 Rd homologs (10), respectively (Fig. 4). The right-flanking sequence also showed similarity to the H. parainfluenzae HpaI restriction modification genes. Three copies of the DNA uptake signal sequence (11), one in the plus strand and two in the minus strand, were located between the right and left ends of HI1038 and the 2,359-bp region, respectively. DNA hybridization proved that this region, which has a G+C content lower than that of HI1038 and HI1042 (Fig. 4), is present only in F3031, while the HI1038 and HI1042 homologs are present in F3031 and F1947. Therefore, the 2,359-bp region (hereafter referred to as the BPF34 locus) seems to represent a genomic islet unique to the F3031 BPF strain that contains two predicted ORFs (Fig. 4). ORF 1, which is preceded by a putative ribosomal binding site (5′-AGGAAA-3′) and encodes an inferred 146-amino-acid protein, terminates 46 nucleotides within ORF 2. The latter encodes a predicted 532-amino-acid protein. BLASTx analysis revealed that, with the exception of a small stretch, most of BPF34 is highly similar to a 719-amino-acid hypothetical protein in T. maritima strain MSB8 (21). This homology gap could be due to the presence of a real frame shift located close to the right end of BPF34, which could also explain the predicted presence of overlapping ORFs 1 and 2 at this locus.
FIG. 4.
Genetic map and G+C composition of the H. influenzae biogroup aegyptius F3031 BPF34 locus. The radiolabeled insert of subtracted clone pMU34 was used to probe a genomic library of strain F3031 and isolate overlapping clones pMU64 and pMU67. The black bars indicate the DNA inserts in pMU34, pMU64, and pMU67. Clones and predicted ORFs that extend beyond the limits of the BPF34 locus are depicted as solid broken lines and arrows, respectively. Two putative ORFs located within the BPF34 locus are indicated by hatched arrows. The solid broken arrows flanking the BPF34 locus identify F3031 ORFs with sequence similarity to H. influenzae Rd KW20 genes HI1038 and HI1042. In silico analyses were done as described in Table 2.
Conclusions
PCR-based subtraction hybridization proved that BPF-causing strain F3031 contains genomic fragments that are absent in noninvasive strain F1947. Several of these fragments encode putative novel proteins of unknown function, some of which have not been described in H. influenzae and may have been acquired by lateral gene transfer from unrelated bacteria. Although no homologs to well-characterized bacterial virulence genes were identified, some of the DNA unique to F3031 may encode novel virulence traits potentially involved in the pathogenesis of BPF. It was suggested (13) that the BPF-causing strains arose by horizontal gene transfer from N. meningitidis. Interestingly, the inferred products of ORFs 1 and 2 of BPF33 are similar to the NMA1850 and NMA1849 N. meningitidis serogroup A strain Z2491 hypothetical proteins, respectively (23). These homologs were also identified as part of Mu-like phage MuMemB of N. meningitidis serogroup B strain MC58 (17). This region was inserted into a gene encoding an ABC transporter and has a mosaic genetic structure that includes prophage genes and genes coding for hypothetical or unknown functions. Some of these genes encode surface-exposed antigens that could play a role in the virulence of this human pathogen (17). These observations, together with the fact that the BPF33 locus also includes a Mu-gp36 homolog, suggest that horizontal gene transfer has also played a role in the evolution of the BPF invasive clone of H. influenzae biogroup aegyptius.
Nucleotide sequence accession numbers
The nucleotide sequence data in this report have been submitted to the GenBank database and assigned the following accession numbers: B1, AF416103; B2, AF416104; B3, AF4161052; D2, AF416106; D3, AF416107; D5, AF416108; D11, AF416109; D13, AF416110; E3, AF416111; E8, AF416112; E9, AF416113; E10, AF416114; F2, AF416115; F5, AF416116; F6, AF416117; F7, AF416118; F10, AF416119; F17, AF416120; F20, AF416121; MU33, AF416122; MU34, AF416123; BPF33, AF416124; BPF34, AF416125.
Acknowledgments
This work was funded by Miami University research funds and Public Health Service grants R15AI37781-01 and R15AI44776-01A1 from the National Institutes of Health. L. M. Smoot was the recipient of a Sigma Xi grant and a Miami University Dissertation Research Grant. D. D. Franke was the recipient of an Undergraduate Summer Scholarship and an Undergraduate Research Committee grant awarded by Miami University.
We thank D. J. Brenner (Centers for Disease Control and Prevention) and A. Lesse (Veterans Administration Medical Center, Buffalo, N.Y.) for providing the H. influenzae biogroup aegyptius isolates and E. J. Hansen (University of Texas) for providing H. influenzae strains DL42, DL63, and TN106. We thank L. Bakaletz (Ohio State University) for the nontypeable H. influenzae otitis isolates and S. H. Goodgal (University of Pennsylvania) for providing H. influenzae strain Eagan. Additional thanks go to J. M. Musser (NIH) for assistance in sequencing the subtracted clones.
Editor: V. J. DiRita
Footnotes
Present address: Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories National Institute of Allergy and Infectious Disease, National Institutes of Health, Hamilton, MT 59840.
REFERENCES
- 1.Achenbach-Richter, L., R. Gupta, K. O. Stetter, and C. R. Woese. 1987. Were the original eubacteria thermophiles? Syst. Appl. Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearden, S. W., T. M. Staggs, and R. D. Perry. 1998. An ABC transport system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol. 180:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington, S. J., A. S. Huggins, P. A. Johanesen, P. K. Crellin, J. K. Cheung, M. E. Katz, C. L. Wright, V. Haring, and J. I. Rood. 1999. Complete nucleotide sequence of the 27-kilobase virulence related locus (vrl) of Dichelobacter nodosus: evidence for extrachromosomal origin. Infect. Immun. 67:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Brazilian Purpuric Fever Study Group. 1987. Haemophilus aegyptius bacteraemia in Brazilian purpuric fever. Lancet ii:761-763. [PubMed]
- 7.Brenner, D. J., L. W. Mayer, G. M. Carlone, L. H. Harrison, W. F. Bibb, M. C. de Cunto Brandileone, F. O. Sottnek, K. Irino, M. W. Reeves, J. M. Swenson, K. A. Birkness, R. S. Weyant, S. F. Berkley, T. C. Woods, A. G. Steigerwalt, P. A. D. Grimont, R. M. McKinney, D. W. Fleming, L. H. Gheesling, R. C. Cooksey, R. J. Arko, C. V. Broome, and The Brazilian Purpuric Fever Study Group. 1988. Biochemical, genetic, and epidemiological characterization of Haemophilus influenzae biogroup aegyptius. J. Clin. Microbiol. 26:1524-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J.-F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 11.Goodgal, S. H., and M. A. Mitchell. 1990. Sequence and uptake specificity of cloned sonicated fragments of Haemophilus influenzae DNA. J. Bacteriol. 172:5924-5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison, L. H., G. A. da Silva, M. Pittman, D. W. Fleming, A. Vranjac, C. V. Broome, and The Brazilian Purpuric Fever Study Group. 1989. Epidemiology and clinical spectrum of Brazilian purpuric fever. J. Clin. Microbiol. 27:599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroll, J. S., K. E. Wilks, J. L. Farrant, and P. R. Langford. 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95:12381-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomholt, H., and M. Kilian. 1995. Distinct antigenic and genetic properties of the immunoglobulin A1 protease produced by Haemophilus influenzae biogroup aegyptius associated with Brazilian purpuric fever in Brazil. Infect. Immun. 63:4389-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marolda, C. L., and M. A. Valvano. 1995. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 177:5539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masignani, V., M. M. Giuliani, H. Tettelin, M. Comanducci, R. Rappuoli, and V. Scarlato. 2001. Mu-like prophage in serogroup B Neisseria meningitidis coding for surface-exposed antigens. Infect. Immun. 69:2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIntyre, P., G. Wheaton, J. Erlich, and D. Hansman. 1987. Brazilian purpuric fever in Central Australia. Lancet ii:112. [DOI] [PubMed]
- 20.Murley, Y. M., T. D. Edlind, P. A. Plett, and J. J. LiPuma. 1998. Cloning of the haemocin locus of Haemophilus influenzae type b and assessment of the role of haemocin in virulence. Microbiology 144:2531-2538. [DOI] [PubMed] [Google Scholar]
- 21.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, H. A., T. Tomita, M. Hirota, J. Kaneko, T. Hayashi, and Y. Kamio. 2001. DNA inversion in the tail fiber gene alters the host range specificity of carotovoricin Er, a phage-tail-like bacteriocin of phytopathogenic Erwinia carotovora subsp. carotovora Er. J. Bacteriol. 183:6274-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 24.Perna, N. T., G. I. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schmid, S. R., and P. Linder. 1992. D-E-A-D protein family of putative RNA helicases. Mol. Microbiol. 6:283-291. [DOI] [PubMed] [Google Scholar]
- 27.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 28.Smoot, L. M., E. C. Bell, R. L. Paz, K. A. Corbin, D. D. Hall, J. N. Steenbergen, A. C. Harner, and L. A. Actis. 1998. Molecular and genetic analysis of iron uptake proteins in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Front. Biosci. 3:d989-d996. [DOI] [PubMed] [Google Scholar]
- 29.Staudenmaier, H., B. Van Hove, Z. Yaraghi, and V. Braun. 1989. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J. Bacteriol. 171:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St. Geme, J. R., J. R. Gilsdorf, and S. Falkow. 1991. Surface structures and adherence of diverse strains of Haemophilus influenzae biogroup aegyptius. Infect. Immun. 59:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrook-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. M. Lim, K. A. Smith, D. H. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 32.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 33.Tomb, J. F., H. el-Hajj, and H. O. Smith. 1991. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenza Rd. Gene 104:1-10. [DOI] [PubMed] [Google Scholar]
- 34.Tondella, M. L. C., F. D. Quin, and B. A. Perkins. 1995. Brazilian purpuric fever caused by Haemophilus influenzae biogroup aegyptius strains lacking the 3031 plasmid. J. Infect. Dis. 171:209-212. [DOI] [PubMed] [Google Scholar]
- 35.Virata, M., N. E. Rosenstein, J. L. Hadler, N. L. Barrett, M. L. Tondella, L. W. Mayer, R. S. Weyant, B. Hill, and B. A. Perkins. 1998. Suspected Brazilian purpuric fever in a toddler with overwhelming Epstein-Barr virus infection. Clin. Infect. Dis. 27:1238-1240. [DOI] [PubMed] [Google Scholar]
- 36.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, K. W. Minton, R. D. Fleischmann, K. A. Ketchum, K. E. Nelson, S. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilde, B. E., J. W. Pearman, P. B. Campbell, P. B. Swan, and D. L. Gurry. 1989. Brazilian purpuric fever in Western Australia. Med. J. Aust. 150:344-346. [DOI] [PubMed] [Google Scholar]