Abstract
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis identified two conserved, immunogenic Staphylococcus aureus cell wall proteins, of 40 and 87 kDa, expressed under iron-restricted growth conditions in vitro and in vivo. N-terminal sequencing and subsequent genome analysis showed that these proteins are encoded by adjacent monocistronic open reading frames designated frpA and frpB, respectively. Studies with an S. aureus fur mutant confirmed that expression of FrpA and FrpB is regulated by Fur but that there also appears to be differential expression of these proteins in different iron-restricted media in vitro. FrpA and FrpB share some amino acid sequence homology with each other and with a putative S. aureus membrane protein, FrpC. frpC is the first gene of a Fur-regulated operon encoding four proteins of unknown function (FrpC, -D, -G, and -H) and the binding protein (FrpE) and permease (FrpF) of a putative iron transporter. Antisense mutagenesis and bioassays showed that FrpA and FrpB are not required for growth of S. aureus under iron-restricted conditions in vitro and do not appear to be involved in the transport of iron from siderophores or in binding of hemin. Further phenotypic analysis suggested that FrpA may be involved in adhesion of S. aureus to plastic in vitro. Binding of S. aureus to microtiter wells was found to be iron regulated, and iron-restricted S. aureus containing antisense frpA or frpAB but not frpB constructs showed reduced binding compared to vector construct controls.
The increasing incidence of antibiotic resistance among staphylococci necessitates identification of novel bacterial targets for antimicrobial therapy and candidate antigens for prophylaxis and immunotherapy. Given the documented variable expression of staphylococcal proteins in response to environmental growth conditions (19), it is important that culture conditions selected for such studies reflect as closely as possible those found in vivo. The use of in vivo models and growth media which mimic conditions in tissues has recently been documented (30), e.g., iron limitation allows production of bacteria for phenotypic analysis and sera collected from cases of known staphylococcal disease provide a means of identifying staphylococcal antigens expressed under iron-restricted conditions in the host during infection or colonization.
Iron is an essential nutrient, as it is required for many cellular metabolic processes, but free ferric iron is scarce in a human host since it is bound to high-affinity iron binding proteins such as transferrin. Pathogenic bacteria have adapted well to the severe iron-restricted environment encountered in the host; the extremely low availability of iron in mammalian body fluids serves as a major environmental signal to the bacteria to express virulence determinants and they have developed specialized mechanisms for scavenging iron from the host, thereby enabling them to multiply in host tissue.
Staphylococcus aureus has two main strategies for obtaining iron from the host environment: (i) utilization of diffusible siderophores that scavenge iron from the host and transport it into the bacterial cell via a receptor-mediated mechanism and (ii) direct binding of the host iron-binding proteins by specific receptors at the bacterial cell surface, where the iron is removed and transported into the cell via unknown mechanisms. S. aureus has been shown to produce at least three siderophores, staphyloferrins A (23) and B (9, 12) and aureochelin (8), and it also utilizes a wide range of exogenous siderophores (21). Several putative siderophore transporters have been identified in the S. aureus genome sequences, including the SirABC (13) and SstABCD transporters (26), the hydroxamate siderophore transporters FhuD1 and FhuD2 (6, 28, 29), and a putative iron-manganese transporter, SitABC (7).
S. aureus can also use receptor-mediated mechanisms to obtain iron from human iron-binding proteins such as transferrin (24). A 42-kDa cell wall protein expressed during growth in vivo in experimental animal infections and under iron-restricted conditions in vitro has been identified as a transferrin binding protein on Western blots (25). Amino-terminal amino acid sequence analysis of a 42-kDa transferrin binding protein from S. aureus BB identified it as a glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The role of the cell surface GAPDH in transferrin binding and the mechanisms by which the iron is released from the bound transferrin and then transported into the cell have yet to be determined.
All of these high-affinity iron transport mechanisms need to be tightly regulated in order to limit the amount of free iron in the bacterial cytoplasm, as free ferric iron is potentially highly toxic due to its involvement in the reactions that produce highly destructive free radicals. Global regulators such as Fur usually mediate this iron-responsive regulation. Three Fur homologues have been identified in S. aureus, Fur, PerR, and Zur. Fur represses the expression of the high-affinity siderophore transporters and induces expression of the catalase gene in response to iron (14, 33), PerR mediates the oxidative stress response (14, 15), and Zur appears to be involved in zinc-mediated gene regulation (20). Although capsule formation (18) and slime production (4) have been shown to be affected by iron availability, there is presently little information concerning the iron regulation of other S. aureus virulence determinants unconnected with the acquisition of iron.
In this paper we describe the identification and characterization of two S. aureus cell wall proteins expressed under iron-restricted conditions in vitro and in vivo. We describe how the expression of these proteins in vitro is affected by growth conditions and the use of antisense RNA technology to determine their function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. aureus BB (originally isolated from a case of bovine mastitis), RN4220, and RN6390-B were from our laboratory culture collections. The fresh clinical isolates were obtained from the Queen's Medical Centre, Nottingham, United Kingdom, and MJH010 (fur::tet) (14) was a kind gift from M. Horsburgh and S. Foster, Sheffield, United Kingdom. Staphylococci strains were cultured aerobically at 37°C on Luria-Bertani (LB) broth or agar, with chloramphenicol (5 μg/ml) added when required. Staphylococci were routinely cultured under iron-restricted conditions by growing statically for 18 h at 37°C in RPMI 1640 tissue culture medium containing 2 mg of NaHCO3 per ml which had been depleted of iron by batch incubation with 6% (wt/vol) Chelex 100 (Sigma Ltd.) and then supplemented with 10% RPMI 1640 to provide trace elements required for growth (CRPMI) (26). CRPMI cultures were incubated in 5% CO2 in air. Staphylococci were also cultured aerobically under iron-restricted conditions for 18 h at 37°C in iron-deficient Tris-succinate minimal medium (TSM) (28). Although the exact amount of iron in both media has not been determined, previous studies have shown that both promote the expression of iron-regulated proteins (7, 28). Where indicated, the iron-restricted media were supplemented with 10 μM ferric citrate to produce iron-rich growth conditions.
Escherichia coli TOPO10 (Invitrogen) and BL21-DE3 (Strategene) were cultured at 37°C in LB broth or agar containing appropriate antibiotics. Long-term stocks of bacterial strains were stored at −80°C in 10% (vol/vol) glycerol in LB broth.
Human serum.
Following local ethical committee approval, sera were collected from individuals with no known previous history of serious staphylococcal disease and from cases of proven S. aureus septicemia diagnosed at University Hospital, Nottingham, United Kingdom.
Intraperitoneal rat chamber model for in vivo growth of staphylococci.
S. aureus RN6390-B was grown in intraperitoneal chambers implanted in rats as described by Pike et al. (27). Inocula for chambers were grown overnight in LB broth and diluted in sterile phosphate-buffered saline (PBS), pH 7.4, to give initial chamber counts of approximately 106 S. aureus cells per ml. Chambers were sampled at 72 h postinoculation to assess the phenotype of the in vivo-grown recovered bacteria.
DNA preparation and manipulation.
Genomic DNA was prepared from staphylococci by the cetyltrimethylammonium bromide method described by Ausubel et al. (3). E. coli and staphylococcal plasmid DNA was extracted using Qiagen mini and maxi kits according to the manufacturer's instructions, except that staphylococcal cell walls were digested with 100 μg of lysostaphin per ml in P1 buffer at 37°C for 5 min before the addition of P2 buffer. Restriction endonucleases were purchased from Pharmacia, and DNA manipulation enzymes were purchased from Promega. The restriction site-deficient S. aureus strain RN4220 was transformed with E. coli-harvested plasmid DNA using an electroporation method described by Kraemer and Iandolo (16), with minor modifications as described by Morrissey et al. (26). Plasmid DNA extracted from RN4220 was used to transform the wild-type S. aureus strain RN6390-B.
Construction of pS10 plasmids containing the antisense frpA and frpB genes.
A 606-bp fragment of the frpA gene was amplified from S. aureus DNA using the primers FrpAP for (5′-CCGCGCTGCAGTAAGTATCAATCAGAACAACG-3′) and FrpAS rev (5′-GGCGCGCCCGGGGCTTGTTGTTGTAGTTACAGTAGG-3′), while a 491-bp fragment of the frpB gene was amplified using the primers FrpBP for (5′-CACCTGCAGCTACAACATGAACAAACAG-3′) and FrpBS rev (5′-TAACCCGGGCTGTTGAGTTCCATCTTTC-3′). The PCR products were digested with PstI and SmaI, purified, and ligated into the SmaI- and PstI-digested and purified plasmid pS10 (26). The double antisense plasmid pS10frpAB was constructed by ligation of a PstI-digested frpA PCR product amplified using the primers FrpAP for and FrpAP rev (5′-GGCGCGCTGCAGGCTTGTTGTTGTAGTTACAGTAGG-3′) into PstI-digested pS10frpB. The ligations were transformed into E. coli TOPO10 by electroporation and screened by colony PCR using the original primers for the single antisense constructs and the FrpBS rev and FrpAP for primers for the double construct. The parental plasmid pS10 and the antisense plasmids pS10frpA, pS10frpB, and pS10frpAB were transformed first into RN4220 and then into RN6390-B by electroporation. Transformants were selected on LB agar with chloramphenicol (5 μg/ml).
Expression of the recombinant S. aureus FrpA and FrpB proteins in E. coli.
The S. aureus frpA and frpB genes were amplified by PCR using the primers FrpA Xf (5′-GCAACAGAAGCTACGAACG-3′) and FrpA Xr (5′-TTATTAGTTTTTACGTTTTCTAGG-3′) and FrpB Xf (5′-GCGGGCTCCATGGCAAATGGCGAAGCACAAGCAGCAGC-3′) and FrpB Xr (5′-GCCGCAGGATCCACGAGGTAACTGGATACCTCATCC-3′), respectively, and RN6390-B genomic DNA as the template. The resulting frpA PCR fragment was cloned into pBADThio-TOPO (Invitrogen) and transformed into E. coli Top10 according to the manufacturer's instructions. The frpB gene fragment was cloned into pCR2.1 (Invitrogen). The resulting plasmid was digested with NcoI and BamHI, and the 2-kb frpB fragment was gel purified, ligated into NcoI- and BamHI-digested pET30a (Strategene), and transferred initially into E. coli XL1-Blue and subsequently into E. coli BL21 DE3. Protein expression was induced by the addition of 0.002% arabinose (FrpA) or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (FrpB) to LB broth cultures supplemented with kanamycin (50 μg/ml), and the overexpressed recombinant proteins were purified on nickel affinity columns (Invitrogen) according to the manufacturer's instructions.
Proteinase K treatment of whole S. aureus cells.
Bacteria were grown overnight in CRPMI or TSM with or without supplementation with 10 μg of ferric citrate per ml. Pelleted bacteria (3,500 × g for 5 min) were resuspended in PBS to an optical density at 600 nm of 2. Proteinase K (Boehringer Mannheim) was added to 1-ml aliquots of S. aureus suspension to a final concentration of 0.1 mg/ml, and control and proteinase K-treated samples were incubated at 37°C for 1 h. Bacteria were pelleted and washed four times in PBS plus 1 mM phenylmethylsulfonyl fluoride (Sigma).
SDS-PAGE and immunoblotting.
Pelleted S. aureus cells were fractionated by lysostaphin digestion as previously described (26). Cell wall preparations were concentrated using Microcon YM 50 microconcentrators (Millipore). Unless otherwise stated, all samples were solubilized by boiling in Laemmli sample buffer (17) for 5 min. Polypeptides were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 4% (wt/vol) acrylamide stacking and 10% (wt/vol) resolving gel in a Bio-Rad Mini Protean II gel apparatus as previously described (3). For immunoblotting, polypeptides were transferred to a BioTrace NT membrane (Gelman) followed by blocking and incubation with primary antibody (1/1,000 dilution, 1 h) and conjugate (1/5,000 dilution of protein A-alkaline phosphatase, 1 h) as previously described (2, 3). Bound antibody was detected using CDP Star (Boehringer Mannheim) and exposure to Hyperfilm ECL (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Human transferrin affinity blotting.
Concentrated S. aureus cell walls were mixed with an equal volume of modified double-strength Laemmli sample buffer (17) lacking mercaptoethanol and incubated at 37°C for 10 min. Cell wall polypeptides were resolved by SDS-PAGE and transferred to a Biotrace NT membrane as described above. Membranes were blocked for 1 h at room temperature in 2% (wt/vol) skimmed milk (Oxoid) in PBS before incubation with human transferrin horseradish peroxidase or alkaline phosphatase conjugate (Jackson Immunoresearch Labs) or protein A-horseradish peroxidase conjugate (Sigma) (2.5 μg/ml in 2% [wt/vol] skimmed milk in PBS) for 1 h at room temperature. Where indicated, competition assays were performed by inclusion of human holo-transferrin or bovine serum albumin at 1 mg/ml in the reaction mixture. Membranes were then thoroughly washed in PBS plus 0.3% (vol/vol) Tween 20. Bound alkaline phosphatase conjugate was detected as described above and bound peroxidase conjugate was detected using ECL substrate (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Binding of S. aureus cells to plastic microtiter plates.
S. aureus cells grown in CRPMI were harvested and resuspended in 100 mM Tris-150 mM NaCl, pH 7.5, to an optical density at 600 nm of 6. One-hundred-microliter aliquots of cell suspension were added to triplicate wells of flat-bottomed, tissue culture-treated microtiter plates (Costar; Corning, Inc.) and plates were incubated in a humid atmosphere at 37°C for 15 h. Wells were washed four times with 200 μl of PBS and remaining cells were fixed to wells with 200 μl of 1% (vol/vol) formaldehyde in PBS for 1 h at room temperature. Fixative was removed and bound bacteria were stained with a 1/1,000 (vol/vol) dilution of crystal violet solution (R and J Wood, Paisley, Scotland, United Kingdom) in H2O for 5 min at room temperature. Excess stain was removed by washing with water, plates were air dried, and the density of well staining was determined at 620 nm using a Labsystems iEMS plate reader and Genesis software. Quantitative data presented represent the means from two separate experiments.
RESULTS
Identification of two surface-exposed S. aureus cell wall proteins expressed under iron-restricted conditions in vitro and in vivo.
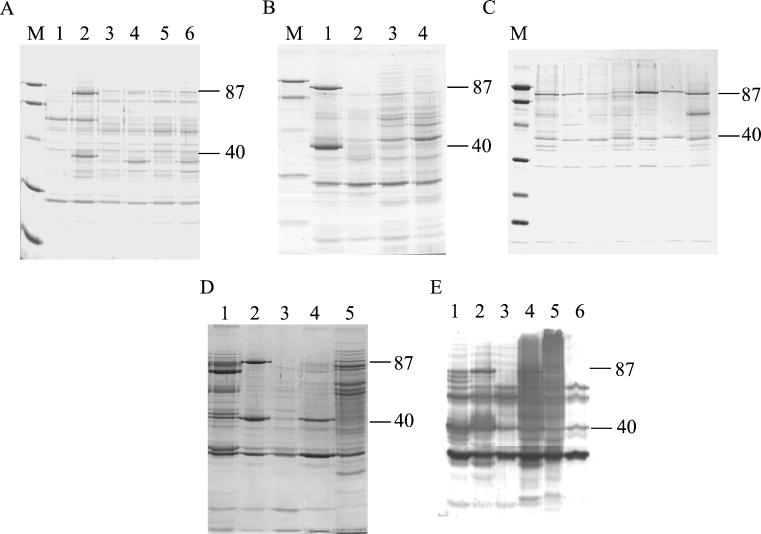
SDS-PAGE analysis of the clinical isolate BB and laboratory strains of S. aureus grown in the defined growth medium CRPMI identified two major iron-regulated cell wall proteins, of approximately 40 and 87 kDa (Fig. 1A). Proteinase K treatment of whole cells of S. aureus RN6390-B demonstrated the surface exposure of the 40- and 87-kDa proteins (Fig. 1B).
FIG. 1.
SDS-PAGE of S. aureus cell wall extracts. (A) Clinical and laboratory strains grown in iron-rich (lanes 1, 3, and 5) and iron-restricted (lanes 2, 4, and 6) CRPMI showing major iron-regulated proteins of 40 and 87 kDa. Lanes: 1 and 2, BB; 3 and 4, 8325-4; 5 and 6, RN6390-B. (B) RN6390-B grown in iron-rich and iron-restricted CRPMI treated with proteinase K showing the surface exposure of the 40- and 87-kDa proteins. Lanes: 1, CRPMI; 2, CRPMI plus proteinase K; 3, iron-rich CRPMI; 4, iron-rich CRPMI plus proteinase K. (C) Fresh clinical isolates grown in CRPMI showing conservation of the 40- and 87-kDa proteins and demonstrating the size variation of both proteins. (D) Differential expression of 40- and 87-kDa proteins. Lanes: 1, RN6390-B recovered from chambers without subculture in vitro; 2 to 5, RN6390-B grown in CRPMI or TSM (lanes 2 and 4) or iron-rich CRPMI or TSM (lanes 3 and 5). (E) Immunoblot of RN6390-B cell wall extracts with pooled human serum. Lanes: 1, cells grown in vivo; 2, CRPMI; 3, iron-rich CRPMI; 4, TSM; 5, iron-rich TSM; 6, LB. The 87- and 40-kDa proteins are indicated. M, size marker.
Interestingly, there appears to be some variability (approximately 1 to 2 kDa) in the sizes of both of these proteins between the clinical isolate BB and the three laboratory strains tested. Analysis of cell wall fractions of seven fresh clinical isolates grown in CRPMI confirmed the conservation of these proteins in S. aureus and the variation in the observed sizes of these proteins among the clinical isolates (Fig. 1C). The 40- and 87-kDa proteins were the predominant iron-regulated cell wall proteins expressed by all S. aureus clinical strains tested when grown in iron-restricted CRPMI.
In contrast, analysis of cell wall protein profiles of RN6390-B grown in a second iron-deficient medium, TSM, indicated that the amount of the 87-kDa protein detected in S. aureus grown in this medium was significantly lower than that seen in CRPMI. Amounts of the 40-kDa protein detected were similar in S. aureus grown in CRPMI or TSM (Fig. 1D).
To investigate whether the 40- and 87-kDa iron-regulated cell wall proteins were expressed in vivo, two approaches were taken. Firstly, cell wall extracts of S. aureus RN6390-B grown for 72 h in rat intraperitoneal implants were analyzed by SDS-PAGE. Figure 1D, lane 1, shows that cell wall extracts of S. aureus grown in vivo contain proteins which comigrate with the 40- and 87-kDa iron-regulated cell wall proteins detected in bacteria grown in iron-restricted CRPMI. The relative amounts of these proteins seen in bacteria recovered from chamber implants were similar to those of the 40- and 87-kDa proteins seen with S. aureus RN6390-B grown in iron-restricted TSM (Fig. 1D).
Secondly, immunoblotting studies with recombinant versions of the 40- and 87-kDa S. aureus cell wall proteins produced in E. coli and human sera collected from S. aureus septicemia patients or individuals with no known previous history of serious staphylococcal disease provided indirect evidence that both proteins are expressed in vivo during infection or colonization by S. aureus. All sera tested contained antibody to these recombinant proteins when tested by immunoblotting (data not shown).
These studies also showed that pooled human septicemia sera contained antibodies to a range of other iron-regulated antigens (Fig. 1E) and highlighted the variability in the total antigenic profile of the cell wall extracts of RN6390-B grown in CRPMI or TSM. Similar immunoblotting studies with S. aureus lysostaphin-insoluble fractions and concentrated culture supernates also showed variation in the antigenic profiles of these cell fractions prepared from S. aureus grown in these media (data not shown).
The 40- and 87-kDa iron-regulated cell wall proteins are encoded by adjacent chromosomal genes.
Amino-terminal sequence analysis was used to identify the genes encoding the 40- and 87-kDa major antigenic cell wall proteins of S. aureus. The amino acid sequences were used to search staphylococcal genome databases (www.ncbi.nlm.nih.gov) to identify the appropriate open reading frames. Adjacent monocistronic open reading frames, with Fur box consensus sequences upstream of the predicted start codons, encode the 40- and 87-kDa proteins. We have designated the gene encoding the 40-kDa protein frpA, for fur-regulated protein A (SA0977), and the one encoding the 87-kDa protein frpB (SA0976). Cell wall anchorage sequences (LPXTG) were found in the C termini of the predicted amino acid sequences. These features of the frpA and frpB genes support the observed cell wall location and iron-regulated nature of the 40-kDa FrpA and 87-kDa FrpB proteins.
The frpA and frpB gene sequences from 8325-4 encode putative polypeptides of 38 kDa with a predicted pI of 9.7 and 72 kDa with a predicted pI of 9.1, respectively. The amino-terminal sequences determined for strains BB and RN6390-B begin 46 and 39 amino acids from the predicted start of the frpA and frpB genes, respectively, indicating that the N-terminal amino acids are signal sequences cleaved from the mature proteins. Consequently, there are significant discrepancies between the observed polypeptide sizes on SDS-PAGE (Fig. 1A) and those for the mature polypeptides as predicted by sequence analysis (approximately 5 to 6 kDa for FrpA and 15 kDa for FrpB).
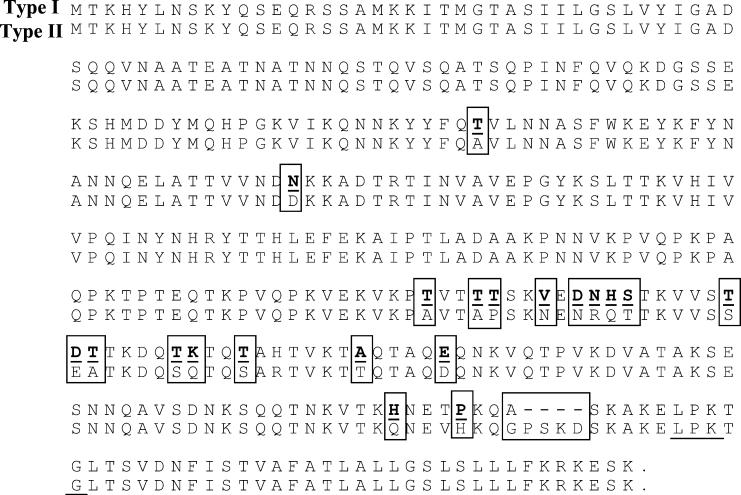
To investigate the reason for the size variability among FrpA and FrpB polypeptides from different S. aureus isolates (Fig. 1A and C), we compared the predicted amino acid sequences of the FrpA and FrpB proteins of 8325-4, N315, Mu50, EMRSA-16, and MSSA-426 strains. As Fig. 2 shows, there are four extra amino acids (proline, serine, lysine, and aspartic acid) in the BB and EMRSA-16 (designated type I) amino acid sequences of FrpA in comparison to those of the 8325-4, RN6390-B, N315, Mu50, and MSSA-426 (designated type II) strains. There is also significant amino acid sequence variation in the C terminus between the FrpA proteins from the type I and type II isolates. In contrast, the type I and type II FrpB polypeptides have significant sequence variation, including an extra seven amino acids in the N terminus. This variation in amino acid composition may contribute to the observed size differences for FrpA and FrpB proteins observed in Fig. 1A and C.
FIG. 2.
Alignment of the amino acid sequences of the two clonal types of the FrpA protein. The cell surface anchorage motif is underlined, while boxed amino acids indicate sequence variation between the two proteins.
The 40- and 87-kDa iron-regulated cell wall proteins share sequence homology.
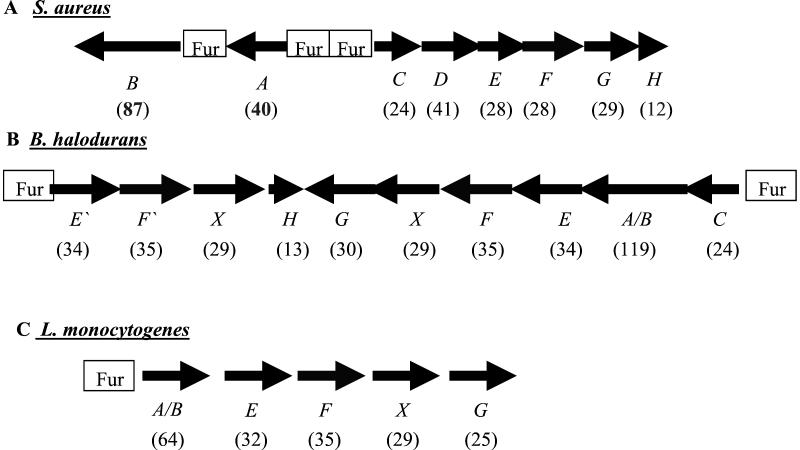
Amino acid sequence analysis showed that the FrpA N-terminal amino acid sequence ATEATNATNN is identical to a previously identified 29-kDa surface protein of unknown function in S. aureus Cowan I (GenBank accession no. AB042826). Neither FrpA nor FrpB has significant homology to any proteins of known function, although the FrpA type I C terminus has 30% identity with the Staphylococcus caprae fibronectin binding autolysin AtlC (1). However, the FrpA N-terminal region and the FrpB C-terminal region have some homology with proteins of unknown function from Listeria monocytogenes p64 (31% identity) (5) and Bacillus halodurans BH3298 (31%) and BH2299 (27%) as well as 21% identity to each other. Moreover, they have 23% identity to a putative 24-kDa membrane protein, FrpC (SA0978), encoded by an adjacent divergently transcribed gene downstream of the frpA and frpB genes (Fig. 3).
FIG. 3.
Organization of the frp operons in S. aureus (A), B. halodurans (B), and L. monocytogenes (C). The open reading frames are predicted to encode proteins as follows: A, 40-kDa cell wall protein homologue; B, 87-kDa cell wall protein homologue; C and D, membrane proteins; E, siderophore binding protein; F, transport permease protein; X, ATP binding protein; G, membrane protein; H, hypothetical protein.
Further analysis of the flanking regions of the frpABC S. aureus chromosomal region and comparison with the B. halodurans and L. monocytogenes DNA sequences surrounding the genes encoding BH3298, BH3299, and p64 revealed the surprising conclusion that there is conservation of the genes in the three regions as shown in Fig. 3. In S. aureus the frpA and frpB genes both have Fur boxes and appear not to be cotranscribed, while frpC appears to be the first gene in a divergently transcribed Fur-regulated operon containing six genes (frpC, -D, -E, -F, -G, and -H; SA0978 to SA0983). The open reading frames in the putative operon are predicted to encode a 41-kDa membrane protein with no known function (frpD), a 28-kDa protein with homology to siderophore binding proteins (frpE), a 28-kDa siderophore transport system permease protein (frpF), a 29-kDa membrane protein (frpG), and a 12-kDa hypothetical protein (frpH), the last two of which show no significant homology to proteins of known functions.
Figure 3 shows the conservation of the genes in B. halodurans (Fig. 3B), which has two convergent operons, both with Fur box consensus sequences, and L. monocytogenes, which has a single Fur-regulated operon. ATP binding proteins are found associated with the ABC transporters in B. halodurans and L. monocytogenes but not in S. aureus.
There appears to be little conservation of this region of the S. aureus chromosome in Staphylococcus epidermidis. A single open reading frame, predicted to encode a polypeptide of 84 kDa with an LPXTG motif and some homology to the frpA and frpB proteins, is found in the same region of the S. epidermidis chromosome as the frp region in S. aureus. The predicted polypeptide has no Fur box consensus sequence and shows 20% amino acid identity to a trans-sialidase from Trypanosoma cruzi.
Further sequence analysis identified a third open reading frame in S. aureus encoding a putative 100-kDa protein with 58% homology to FrpB (SA1552). This FrpB homologue (FrpJ) appears to be monocistronic and has a Fur box consensus sequence and an LPXTG motif.
FrpA and FrpB proteins are Fur regulated.
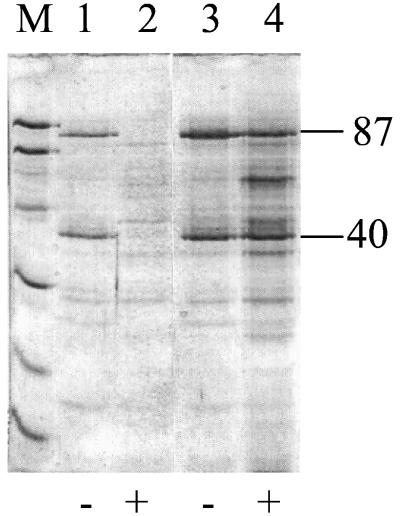
The frpA and frpB genes are preceded by Fur consensus sequences, suggesting that expression is negatively regulated by Fur in response to the presence of iron. In order to confirm the involvement of fur in the regulation of expression of these proteins, cell wall extracts prepared from a wild-type S. aureus (8325-4) and an isogenic fur mutant (MJH010) grown in CRPMI with or without 10 μM ferric citrate were compared. Figure 4 shows constitutive expression of FrpA and -B in iron-rich medium in the fur mutant, confirming the fur regulation of these proteins.
FIG. 4.
SDS-PAGE of S. aureus cell wall extracts of 8325-4 (lanes 1 and 2) and MJH010 (fur::tet) (lanes 3 and 4) grown in iron-rich medium (+) and CRPMI (−) showing Fur regulation of FrpA and FrpB.
The production of frpA, frpB, and frpAB antisense RNA downregulates frpA and frpB expression in vitro.
To determine the possible functions of the FrpA and FrpB proteins the expression of both open reading frames was disrupted using antisense RNA technology. A 606-bp fragment of the frpA gene and a 491-bp frpB fragment were cloned singly and together in the opposite orientation downstream of a constitutive promoter in an E. coli-S. aureus shuttle vector, pS10 (26). The antisense plasmids (pS10frpA, pS10frpB, and pS10frpAB) and the parental plasmid (pS10) were then transformed separately into S. aureus RN6390-B, resulting in isogenic transformants, which vary only in the production of antisense frpA, frpB, and frpAB RNA.
SDS-PAGE analysis of cell wall extracts from the staphylococcal strains grown under iron-rich or iron-restricted conditions (Fig. 5A) shows that there is a corresponding decrease in the expression of the 40-kDa FrpA protein in the presence of antisense frpA and frpAB RNA and of the 87-kDa FrpB polypeptide in the presence of frpB and frpAB antisense RNA. Thus, the production of antisense frpA and frpB RNAs significantly reduces the amount of both frpA and frpB mRNAs and proteins, respectively. Moreover, these results show that a double antisense construct can simultaneously reduce the expression of two S. aureus proteins.
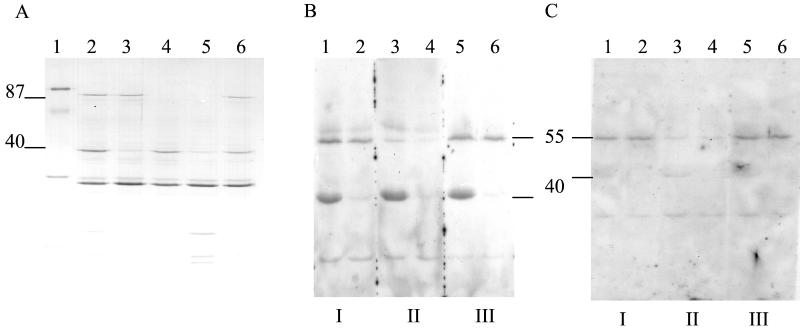
FIG. 5.
(A) SDS-PAGE of cell wall extracts from S. aureus RN6390-B grown in CRPMI showing a decrease in FrpA and FrpB proteins in the presence of antisense RNA. Lanes: 1, marker; 2 and 6, RN6390-B; 3, RN6390-B(pS10frpA); 4, RN6390-B(pS10frpB); 5, RN6390-B(pS10frpAB). (B) Affinity blots of S. aureus cell wall extracts probed with transferrin peroxidase (I), transferrin peroxidase plus 1 mg of unlabeled human holo-transferrin per ml (II), or transferrin peroxidase plus 1 mg of BSA per ml (III). (C) Affinity blots of S. aureus cell wall extracts probed with transferrin alkaline phosphatase (I), transferrin alkaline phosphatase plus 1 mg of unlabeled human holo-transferrin per ml (II), or transferrin alkaline phosphatase plus 1 mg of BSA per ml (III). Lanes: 1, 3, and 5, RN6390-B(pS10); 2, 4, and 6, RN6390-B(pS10frpA).
FrpA binds horseradish peroxidase-labeled transferrin and protein A nonspecifically.
To determine the potential function of these proteins we first analyzed the growth of the antisense disruptant strains in vitro. The growth of S. aureus in CRPMI or TSM medium in the presence or absence of ferric citrate or in rabbit serum was not affected by the downregulation of the FrpA or FrpB proteins (data not shown). Moreover, bioassays (24) showed that there were no differences in the ability of S. aureus wild-type or antisense constructs to grow on different ferric siderophores as sole iron sources (data not shown).
The presence of genes encoding iron-regulated cell surface proteins in a region of the chromosome which also encodes a putative iron transport system suggested that the FrpA and FrpB proteins may be specifically involved in the direct binding and/or removal of iron from host iron binding proteins such as hemin or transferrin.
Our initial studies showed that neither FrpA nor FrpB is involved in the binding or utilization of hemin by RN6390-B (data not shown). Although we have previously identified an S. aureus transferrin binding protein as a 42-kDa cell wall GAPDH (25), our recent studies have suggested that there may be other S. aureus transferrin binding proteins. Figure 5B shows that there are two other S. aureus RN6390-B proteins of 55 and 40 kDa capable of binding human transferrin-horseradish peroxidase conjugate (Tf-HRP) on affinity blots. Under the conditions used in these experiments, binding of the conjugate to GAPDH was not detected and no binding to FrpB was observed.
The 40-kDa binding protein detected is FrpA since in the frpA antisense strain there is a significant decrease in binding of Tf-HRP to the 40-kDa protein. However, as shown in Fig. 5C, the binding of Tf-HRP to FrpA is nonspecific since there is no competition between the Tf-HRP and unlabeled human holo-transferrin. In contrast, binding of Tf-HRP to the 55-kDa protein is significantly reduced by the presence of unlabeled holo-transferrin but not by the presence of bovine serum albumin (BSA) (Fig. 5C). This observation suggested that nonspecific binding of Tf-HRP to FrpA may be due to reaction with the horseradish peroxidase component of the conjugate. This possibility was further supported by the observation that FrpA also bound to two other peroxidase-conjugated proteins, protein A and human serum albumin, but interestingly, failed to bind horseradish peroxidase alone (data not shown).
This lack of specific binding of human transferrin to FrpA was further supported in repeat experiments using alkaline phosphatase-labeled human transferrin, which bound specifically to the 55-kDa protein but not to FrpA. Therefore, these observations suggest that the iron-regulated 55-kDa protein is a novel specific transferrin binding protein but that FrpA does not specifically bind human transferrin.
FrpA is implicated in mediating the attachment of S. aureus to plastic.
Given that both FrpA and FrpB are major surface proteins and have high pIs, they are likely to contribute significantly to the overall surface charge of S. aureus cells and their interaction with charged surfaces. The effect of reduction of expression of FrpA and -B on adherence of S. aureus to a plastic surface was therefore investigated. RN6390-B-S10 grown under iron-restricted conditions adhered to the bottom of microtiter plate wells, whereas S. aureus grown under iron-rich conditions adhered significantly less well, suggesting that attachment was in part mediated by iron-regulated surface structures (Table 1). This iron-regulated binding of S. aureus was markedly decreased in the frpA and frpAB antisense strains but not in the frpB antisense strain, implying that FrpA but not FrpB is involved in adhesion of S. aureus to plastic (Table 1).
TABLE 1.
FrpA is involved in mediating the attachment of S. aureus to plastic
| S. aureus mutant | Optical density (range) at 600 nm
|
|
|---|---|---|
| +Fe | −Fe | |
| Wild type | 0.087 (0.06-0.10) | 0.177 (0.15-0.19) |
| frpA | 0.116 (0.08-0.15) | |
| frpB | 0.155 (0.12-0.17) | |
| frpAB | 0.109 (0.07-0.15) | |
DISCUSSION
Pathogenic bacteria have adapted well to the severe iron restriction of the human body with the development of specialized iron acquisition mechanisms and the use of low iron levels as a major environmental signal for the expression of virulence determinants. A number of staphylococcal iron acquisition mechanisms have previously been identified and analysis of the recently completed S. aureus genome sequences indicates that this organism appears to have more putative iron transporters than any other pathogenic bacterium sequenced to date. This observation suggests that S. aureus is particularly well adapted to iron-restricted growth conditions and may in part explain the very low concentration of iron required to support growth of S. aureus in vitro . However, apart from two studies on capsule and slime production, to date there have been no reports of putative S. aureus virulence determinants, other than those connected with iron acquisition, being regulated by iron availability. We can now report the identification of three novel cell surface proteins, FrpA, -B, and -C, that are Fur regulated and potentially involved in functions other than iron transport.
FrpA and FrpB are cell surface proteins of 40 and 87 kDa, respectively, that are conserved among all S. aureus isolates we have tested. Our evidence suggests that these proteins are also expressed in vivo. Phenotypic analysis of bacteria recovered from rat chamber implants identified cell wall proteins of 40 and 87 kDa expressed under these growth conditions. Generation of specific antibodies to these proteins or peptide sequencing will allow us to confirm their relatedness to the proteins of similar size detected in S. aureus grown in vitro, but results of immunoblotting studies with the recombinant 40- and 87-kDa proteins and human sera provide additional if indirect evidence that they are expressed in vivo. Antibodies to proteins of 40 and 87 kDa and recombinant FrpA and FrpB are detected in sera collected from individuals with proven S. aureus septicemia and those with no known history of serious staphylococcal disease. Although we are unable to determine at this time whether these antibodies were generated in response to colonization or minor infection with S. aureus, the surface exposure, antigenicity, and conservation of FrpA and -B make these proteins good candidates as potential targets for opsonic antibodies in vivo . They may therefore be of value for the development of novel approaches to prevent or modulate S. aureus infection using specific antibodies.
Like iron-regulated proteins in other bacteria, FrpA and FrpB are regulated by Fur in response to iron; however, our studies have shown differential expression of these proteins under different iron-restricted conditions in vitro . Both FrpA and FrpB are expressed in the highly iron-restricted CRPMI, but very little FrpB is detected in the protein profiles of S. aureus incubated in low-iron TSM. Interestingly, the relative amounts of FrpA and -B detected in cells grown in vivo in rat chamber implants were similar to those observed in TSM. It is presently unclear if these in vitro and in vivo findings reflect differences in protein expression or result from modification and/or degradation of the mature protein. If the observed differences are due to alterations in protein expression, this may indicate differential levels of Fur repression of the two genes dependent on variability in iron concentration in the different growth media. Alternatively, regulation due to other as yet undetermined factors may also be involved. Our own data also indicate that FrpA and -B are degraded by proteinase K in vitro, and given published reports of degradation of other S. aureus surface proteins by proteases during batch culture (17, 22), the possibility that FrpB in particular may be degraded merits further study.
The presence of genes encoding iron-regulated cell surface proteins in a region of the chromosome which also encodes a putative iron transport system suggested that the FrpA and FrpB proteins may be specifically involved in the direct binding and/or removal of iron from host iron binding proteins. An earlier study also detected expression of a protein, Sai-1, now identified as FrpA, in S. aureus grown in serum (32) and a role for this protein as a putative virulence determinant was suggested. However, our results have shown that FrpA and FrpB are not required for growth under iron-restricted conditions, including growth in serum in vitro, and do not appear to be involved in the transport of iron from siderophores or the host iron binding proteins hemin and transferrin. Although we have shown that FrpA binds horseradish peroxidase-labeled transferrin on Western blots, this binding is nonspecific, and since FrpA also binds protein A and human serum albumin-horseradish peroxidase, binding may be due to reaction with the horseradish peroxidase component of the conjugate. It is presently unclear what properties of FrpA contribute to the nonspecific binding to these conjugates, but these findings emphasize the need to perform appropriate competition assays before assigning specific functions to putative binding proteins. Using this approach we have now identified a novel 55-kDa putative transferrin binding protein in S. aureus. Further studies will be involved in the identification of the protein and its relationship to the previously identified GAPDH transferrin binding protein.
Although we have as yet been unable to identify a role for FrpA or -B in staphylococcal iron uptake, these studies have shown that FrpA may be involved in the iron-regulated binding of S. aureus to plastic. To our knowledge, this is the first report of an iron-regulated protein being involved in staphylococcal adhesion. It seems likely that the high pI of FrpA may contribute to the overall charge of the staphylococcal surface when grown under iron restriction. Since increasing the negative charge of S. aureus cells by modifying teichoic acid structure (11) reduces adhesion to plastic, the effect of reduction of FrpA expression on adhesion observed in the present study could be explained by an increase in the overall negative charge of the cells. Changes in surface charge associated with variation in FrpA expression would primarily be expected to influence the initial nonspecific stages of adherence and we are presently investigating alternative methods for quantitating binding to look further at the role of FrpA in this process. In addition, the relevance of FrpA in staphylococcal adhesion in vivo has yet to be determined. Clearly, proteins expressed under iron restriction may contribute to adherence to biomaterials in vivo, but in tissues, biomaterials are likely to be rapidly coated with conditioning films consisting of host proteins including fibrinogen and fibronectin (31). These may modify the surface of the material and act as specific receptors to facilitate attachment via previously characterized adhesins such as FbpA and -B and ClfA (10). The role of FrpA in mediating attachment in the presence of host proteins has yet to be tested, but the studies presented here and the recent finding that slime production is also iron regulated (4) suggests that the role of iron availability in staphylococcal adherence in general merits further study.
The detection of sequence homology between specific regions of FrpA and -B and two other iron-regulated S. aureus proteins, FrpC and -J, suggests that these proteins may have had a common ancestor and function. The conservation of FrpA, -B, and -C homologues and gene organization of this region in L. monocytogenes and B. halodurans provide further support for this suggestion. For S. aureus, the predicted amino acid sequences also show that there is significant variation within the C-terminal domains of the FrpA proteins and the N-terminal domains of the FrpB proteins from different S. aureus strains, with there being at least two clonal types resulting in two differently sized FrpA and FrpB proteins as determined by SDS-PAGE. In the case of FrpA this sequence variation results in the type I but not the type II proteins having sequence homology to S. caprae AtlC, which is known to contribute to the surface hydrophobicity and potential adhesive properties of this organism (1). This divergence in amino acid sequence further supports the possibility that proteins of these different clonal types may have altered functions and/or specificities. The presence of a second FrpB homologue in S. aureus may also explain the observed lack of effect of reduction of FrpB expression on adhesion to plastic.
Taken together, these observations suggest that FrpA, -B, -C, and -J may represent a novel family of iron-regulated surface proteins which contribute to staphylococcal attachment. Conserved regions within these proteins may represent structural domains with variability in function and/or specificity encoded in the variable regions. Generation of additional antisense constructs will allow us to confirm if these iron-regulated proteins, singly or as complexes, mediate staphylococcal adherence to plastic or other substrates and the relative importance of specific or nonspecific interactions in these processes.
Acknowledgments
This work was supported by program grant G9219778 from the Medical Research Council.
We thank Malcolm Horsburgh and Simon Foster for the fur mutant (MJH010) and Andrew Sanderson for assistance with construction of pS10frpB.
Editor: E. I. Tuomanen
REFERENCES
- 1.Allignet, J., S. Aubert, K. G. H. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbuthnott, J. P., E. Arbuthnott, A. D. J. Arbuthnott, W. J. Pike, and A. Cockayne. 1992. Investigation of microbial growth in vivo: evaluation of a novel in vivo chamber implant system. FEMS Microbiol. Lett. 100:75-80. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 4.Baldassarri, L., L. Bertuccini, M. G. Ammendolia, C. R. Ariciola, and L. Montanaro. 2001. Effect of iron limitation on slime production by Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:343-345. [DOI] [PubMed] [Google Scholar]
- 5.Borezee, E., T. Msadek, L. Durant, and P. Berche. 2000. Identification in Listeria monocytogenes of MecA, a homologue of the Bacillus subtilis competence regulatory protein. J. Bacteriol. 182:5931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera, G., A. Xiong, M. Uebel, V. K. Singh, and R. K. Jayaswal. 2001. Molecular characterization of the iron-hydroxamate uptake system in Staphylococcus aureus. Appl. Environ. Microbiol. 67:1001-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockayne, A., P. J. Hill, N. B. L. Powell, K. Bishop, C. M. Sims, and P. Williams. 1998. Molecular cloning of a 32-kDa lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66:3767-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courcol, R. J., D. Trivier, M. C. Bissinger, G. R. Martin, and M. R. W. Brown. 1997. Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect. Immun. 65:1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drechsel, H., S. Freund, G. Nicholson, H. Haag, O. Jung, H. Zahner, and G. Jung. 1993. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals 6:185-192. [DOI] [PubMed] [Google Scholar]
- 10.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 11.Gross, M., S. E. Cramton, F. Götz, and A. Peschel. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69:3423-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haag, H., H. P. Fiedler, J. Meiwes, H. Drechsel, G. Jung, and H. Zahner. 1994. Isolation and biological characterisation of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol. Lett. 115:125-130. [DOI] [PubMed] [Google Scholar]
- 13.Heinrichs, J. H., L. E. Gatlin, C. Kunsch, G. H. Choi, and M. S. Hanson. 1999. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J. Bacteriol. 181:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraemer, G. R., and J. J. Iandolo. 1990. High-frequency transformation of Staphylococcus aureus by electroporation. Curr. Microbiol. 21:372-376. [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J. C., S. Takeda, P. J. Livolsi, and L. C. Paoletti. 1993. Effects of in vitro and in vivo growth conditions on expression of type 8 capsular polysaccharide by Staphylococcus aureus. Infect. Immun. 61:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay, J. A., and S. J. Foster. 1999. Interactive regulatory pathways control virulence determinant production and stability in response to environmental conditions in Staphylococcus aureus. Mol. Gen. Genet. 262:323-331. [DOI] [PubMed] [Google Scholar]
- 20.Lindsay, J. A., and S. J. Foster. 2001. zur: a Zn2+ responsive regulatory element of Staphylococcus aureus. Microbiology 147:1259-1266. [DOI] [PubMed] [Google Scholar]
- 21.Maskell, J. P. 1980. The functional interchangeability of enterobacterial and staphylococcal chelators. Antonie van Leeuwenhoek 46:343-351. [DOI] [PubMed] [Google Scholar]
- 22.McAleese, F. M., E. J. Walsh, M. Sieprawska, J. Potempa, and T. J. Foster. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276:29969-29978. [DOI] [PubMed] [Google Scholar]
- 23.Meiwes, J., H. P. Fiedler, H. Haag, H. Zahner, S. Konetschny-Rapp, and G. Jung. 1990. Isolation and characterisation of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol. Lett. 67:201-206. [DOI] [PubMed] [Google Scholar]
- 24.Modun, B., R. W. Evans, C. L. Joannou, and P. Williams. 1998. Receptor-mediated recognition and uptake of iron from human transferrin by Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 66:3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modun, B., J. A. Morrissey, and P. Williams. 2000. The staphylococcal transferrin receptor: a glycolytic enzyme with novel functions. Trends Microbiol. 8:231-237. [DOI] [PubMed] [Google Scholar]
- 26.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68:6281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pike, W. J., A. Cockayne, C. A. Webster, R. C. B. Slack, A. P. Shelton, and J. P. Arbuthnott. 1991. Development and design of a novel in vivo chamber implant for the analysis of microbial virulence and assessment of antimicrobial chemotherapy. Microb. Pathog. 10:443-450. [DOI] [PubMed] [Google Scholar]
- 28.Sebulsky, M. T. D., Hohnstein, M. D. Hunter, and D. E. Heinrichs. 2000. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182:4394-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebulsky, M. T. D., and D. E. Heinrichs. 2001. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, D. G. E., M. H. Wilcox, P. Williams, R. G. Finch, and S. P. Denyer. 1991. Characterization of the cell envelope proteins of Staphylococcus epidermidis cultures in human peritoneal dialysate. Infect. Immun. 59:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Mei, H. C., B. van de Belt-Gritter, G. Reid, H. Bialkowska-Hobrzanska, and H. J. Busscher. 1997. Adhesion of coagulase-negative staphylococci grouped according to physicochemical surface properties. Microbiology 143:3861-3870. [DOI] [PubMed] [Google Scholar]
- 32.Wiltshire, M. D., and S. J. Foster. 2001. Identification and analysis of Staphylococcus aureus components expressed by a model system of growth in serum. Infect. Immun. 69:5198-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterisation of the ferric regulator, Fur, from Staphylococcus aureus. Microbiology 146:659-668. [DOI] [PubMed] [Google Scholar]