Abstract
Salmonellae can exist in an asymptomatic carrier state in the human gallbladder. Individuals with gallstones are more likely to become typhoid carriers, and antibiotic treatments are often ineffectual against Salmonella enterica serovar Typhi in carriers with gallstones. Therefore, we hypothesized that Salmonella spp. form biofilms on the surfaces of gallstones, where the bacteria are protected from high concentrations of bile and antibiotics. A number of methods were utilized to examine biofilm formation on human gallstones and glass coverslips in vitro, including confocal, light, and scanning electron microscopy. In our assays, salmonellae formed full biofilms on the surfaces of gallstones within 14 days and appeared to excrete an exopolysaccharide layer that bound them to the surfaces and to other bacteria. Efficient biofilm formation on gallstones was dependent upon the presence of bile, as a biofilm did not form on gallstones within 14 days in Luria-Bertani broth alone. The biofilms formed by a Salmonella enterica serovar Typhi Vi antigen mutant, as well as strains with mutations in genes that eliminate production of four different fimbriae, were indistinguishable from the biofilms formed by the parents. Mutants with an incomplete O-antigen, mutants that were nonmotile, and mutants deficient in quorum sensing were unable to develop complete biofilms. In addition, there appeared to be selectivity in salmonella binding to the gallstone surface that did not depend on the topology or surface architecture. These studies should aid in the understanding of the Salmonella carrier state, an important but underresearched area of typhoid fever pathogenesis. If the basis of carrier development can be understood, it may be possible to identify effective strategies to prevent or treat this chronic infection.
Biofilms have recently been implicated as the cause of many chronic infections in humans and are frequently associated with implanted devices, such as catheters, prosthetics, and contact lenses (for reviews see references 10 and 12). A biofilm is defined as a population of one or more organisms attached to each other and a surface by means of a bacterium-initiated matrix (10, 12). This matrix provides a very stable environment and results in high levels of resistance to antimicrobial agents. Often, it is difficult to clear the infection unless the substrate to which the bacteria are attached is removed. From the sessile, matrix-bound community, planktonic cells can be continuously shed from the biofilm, which in humans can lead to a systemic infection or release of the organism into the environment.
Salmonella enterica serovar Typhi, which causes typhoid fever in humans, crosses the intestinal epithelial layer and invades macrophages. The macrophages carry the organism to the liver, pancreas, and spleen, where the bacteria are thought to replicate in both phagocytic and nonphagocytic cells. From the liver, the bacteria can be shed into the gallbladder, where either an active infection (cholecystitis) or a chronic infection (carrier state) can develop. The carrier state occurs in about 3 to 5% of the people infected and is frequently associated with gallbladder abnormalities, such as gallstones (15, 20). This infection is often asymptomatic and can last for many years with little or no deleterious effect on the host. Furthermore, it has been shown that the chronic carrier state is the single most important risk factor for development of hepatobiliary carcinomas, as evidenced by the finding that salmonella carriers with gallstones have an 8.47-fold-higher risk for developing cancer of the gallbladder (15, 33).
It is interesting that the gallbladder is the primary site of carriage, considering that it is also the storage site for bile. Bile is produced in the liver and consists of many components, including bile salts, cholesterol, and bilirubin. Bile salts are detergents that aid in degradation and dispersion of lipids and as such make bile a good antimicrobial agent. Salmonella, however, is resistant to high concentrations of bile and individual bile salts (38). In addition to being highly resistant to the effects of bile, the levels of several proteins of Salmonella spp. are affected both positively and negatively by bile (38). Bile has also been shown to be important as an environmental signal for Salmonella invasion of epithelial cells (28). The presence of bile leads to down-regulation of the transcription of genes in the type III secretion system of pathogenicity island I. This leads to a reduction in the amount of effector molecules secreted and reduced invasion of epithelial cells. Therefore, bile appears to be an important environmental signal for Salmonella pathogenesis (as well as pathogenesis with other enteric pathogens [17]), probably during both acute and chronic infections.
Antibiotic treatment can be ineffective in Salmonella carriers with gallstones, and elimination of gallbladder infection in these individuals usually requires surgery and gallstone removal (20). However, this option is less likely in several developing countries where S. enterica serovar Typhi is prevalent. Thus, it is important to determine the mechanisms by which Salmonella can establish gallbladder carriage and, possibly most importantly, to determine how it does so in the presence of gallstones. Because Salmonella spp. cause chronic infections of the gallbladder and gallstones are a major factor influencing the formation of the carrier state, in this study we addressed the hypothesis that salmonellae form biofilms on the surfaces of gallstones. Such biofilm formation may contribute to establishment of the chronic carrier state. In addition, the roles of bile and various bacterial surface structures in Salmonella biofilm formation on gallstones were examined.
MATERIALS AND METHODS
Strains and plasmids.
The Salmonella strains and plasmid used in this study are listed in Table 1. Plasmid pFPV25.1 was transformed into ATCC 14028s and Ty2 by electroporation, and Luria-Bertani (LB) agar plates containing ampicillin (50 μg/ml) were used to select for the plasmid.
TABLE 1.
Bacterial strains, plasmid, and relevant characteristics
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| S. enterica serovar Typhi- murium strains | ||
| JSG210 | ATCC 14028s | ATCCa |
| JSG224 | CS019 (Cmr) | 22 |
| JSG1149 | JSG210(pFPV25.1) | This study |
| JSG1169 | SR11x4252 | 37 |
| JSG1170 | Δ(fim-aph-11::Tn10)-391 | 37 |
| JSG1171 | Δ(aph-11::Tn10)-251 lpfC::Kan | 37 |
| JSG1172 | Δ(aph-11::Tn10)-251 pefC::Tet | 37 |
| JSG1173 | Δ(aph-11::Tn10)-251 agfB::Cam | 37 |
| JSG1174 | Δ(fim-aph-11::Tn10)-391 lpfC::Kan agfB::Cam pefC::Tet | 37 |
| JSG1225 | fliA::Tn10d-Tet | 19 |
| JSG1340 | luxS::MudJ | 34 |
| JSG1221 | zbi812::Tn10 GalE− | Karl Klose |
| S. enterica serovar Typhi strains | ||
| JSG624 | Ty2 | |
| JSG1150 | JSG624(pFPV25.1) | This study |
| JSG1213 | tviB::Kan | 2 |
| Plasmid | ||
| pFPV25.1 | GFP constitutiveb | 36 |
ATCC, American Type Culture Collection.
GFP, green fluorescent protein.
Growth conditions.
One group of uniform gallstones composed mainly of cholesterol and removed from a single patient was used in these studies. Human gallstones were incubated in test tubes with a strain of Salmonella in either LB broth or LB broth with 3% ox bile (Sigma, St. Louis, Mo.) with aeration at 37°C. Every 24 h, the medium was removed, the gallstones were washed two times in LB broth by vigorous vortexing, and fresh medium was added. Antibiotics (50 μg of ampicillin per ml, 25 μg of chloramphenicol per ml, 15 or 10 μg of tetracycline per ml, and 45 μg of kanamycin per ml) were added where appropriate. The growth periods varied from 4 to 18 days as indicated below.
Glass coverslips were placed in the bottom of a petri dish and covered with 10 ml of medium. The medium used for the coverslips was tryptic soy broth (TSB) or LB broth either with or without 3% ox bile. Strains were inoculated into the plate by using 1:100 dilutions of overnight cultures. Antibiotics were added, and the plates were rocked slowly overnight at 37°C. Every 24 h the medium was removed, each preparation was washed two times in 1× phosphate-buffered saline (PBS), and fresh medium with antibiotics was added. Enough glass slides were placed into the petri dishes so that one could be removed every day for 14 days.
Crystal violet staining of biofilms on glass slides.
Removed glass slides were washed with a continuous spray of 1× PBS and incubated at 60°C for 1 h to fix the cells. Then 0.1% crystal violet (gentian violet in isopropanol-methanol-1× PBS [1:1:18]) was placed on each glass slide, and the slides were incubated for 15 min at room temperature. The slides were then washed thoroughly with 1× PBS until the PBS ran clear. The slides were then broken, put in Eppendorf tubes, and immersed in 33% acetic acid to extract the dye. The optical density at 570 nm of the acetic acid solution was assessed to determine the amount of dye retained by the bacterial cells.
Ruthenium red staining of glass slides for detection of EPS.
Glass slides were incubated in petri dishes as described above, removed, and stained as described by Mills et al. (23). Briefly, a slide was placed in a 12-well cell culture cluster (Corning, Corning, N.Y.) with 1 ml of 0.15% ruthenium red-0.5% glutaraldehyde dissolved in 100 mM cacodylate buffer for 1 h at room temperature (Sigma). This stain was removed, 0.05% ruthenium red-5% glutaraldehyde in a 100 mM cacodylate buffer solution was added, and the preparation was incubated for 2 h at room temperature. This stain was removed, and the slides were washed five times for 10 min in 100 mM cacodylate buffer. The slides were then ready to be viewed by light microscopy for exopolysaccharide (EPS) staining.
Confocal scanning laser microscopy (CSLM).
After growth for a selected period of time, gallstones were washed two times in LB broth with vortexing and placed in a Nunc Labtek chambered coverslip for immediate viewing. Strains constitutively producing green fluorescent protein were scanned with a Fluoview Scanhead using an argon ion laser with excitation at 488 nm. The biofilm was viewed with a ×40 dry objective using a 1X70 Olympus microscope, and images were viewed by using Fluoview software, version 2.1.
Scanning electron microscopy (SEM).
After samples were grown for a specified period of time, gallstones were washed two times with vortexing and placed into 2% glutaraldehyde to fix the specimens. The specimens were then dehydrated in graded ethanol and Freon (FC-113) series and dried in an Omar SPC1500 critical point dryer. The specimens were then sputter coated with a Technics Hummer V and viewed with a JEOL JSM-840A scanning electron microscope.
RESULTS
Salmonella spp. form biofilms on the surfaces of gallstones
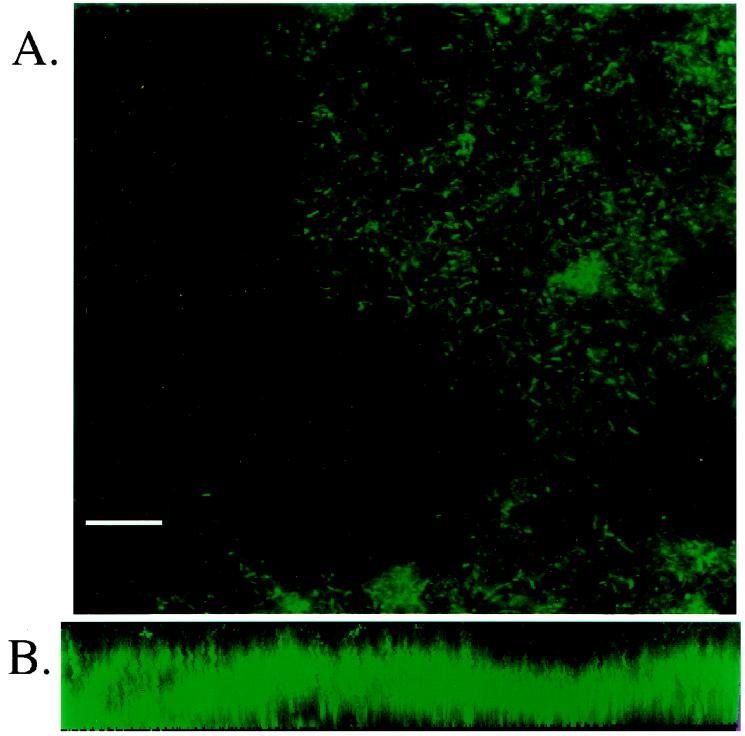
To determine if Salmonella spp. can form biofilms in vitro on gallstones, human gallstones were exposed to Salmonella spp. in liquid cultures with aeration at 37°C. The medium was replaced every 24 h in an attempt to mimic gallbladder emptying. After the specified time, the gallstones were examined for the presence of biofilms either by CSLM or by SEM. To examine gallstones for biofilm formation by CSLM, strains of S. enterica serovar Typhimurium (ATCC 14028s) and S. enterica serovar Typhi (Ty2) constitutively producing green fluorescent protein were used. Gallstones were exposed to the bacteria for 7, 11, or 18 days in LB broth containing 3% bile prior to examination by microscopy. Both S. enterica serovar Typhimurium and S. enterica serovar Typhi were bound to most of the gallstone surface after 11 days (but not after 7 days), and there was nearly full coverage of the gallstones after 18 days (Fig. 1A). Each biofilm was also examined for depth by assembly of a three-dimensional image from CSLM Z-scans. The data demonstrated that the depth of the biofilm was significant (∼31 μm), and because salmonellae are ∼2 μm in length, the cells clearly were in a multilayer community (Fig. 1B). In addition, visual inspection of the Z-scans revealed evidence of water channels, which are typical in fully formed biofilms.
FIG. 1.
CSLM micrographs of gallstones incubated with S. enterica serovar Typhi constitutively producing green fluorescent protein. (A) S. enterica serovar Typhi cells exposed to a gallstone for 18 days in the presence of 3% bile in LB broth are distributed over most of the surface of the gallstone. However, note the pockets of the stone surface where no bacteria appear to bind, suggesting that there is specificity in the Salmonella-gallstone interaction. Bar = 10 μm. (B) Images taken through the depth of the biofilm using a Z-scan were assembled into a three-dimensional form. The micrograph represents a thickness of ∼31 μm, and because salmonellae are ∼2 μm long, the results demonstrate that the cells do not exist as a monolayer.
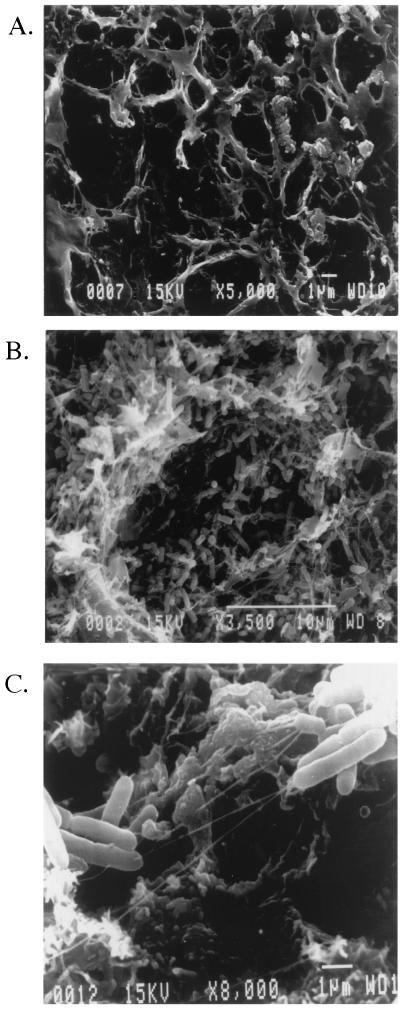
Gallstones exposed to wild-type S. enterica serovar Typhimurium and S. enterica serovar Typhi were also examined by SEM for biofilm formation, as well as for biofilm characteristics. Similar to what was observed by CSLM, both S. enterica serovar Typhimurium and S. enterica serovar Typhi were found to completely cover the surfaces of the gallstones by 14 days (Fig. 2B). The images demonstrated that the bacteria did not form densely packed monolayers but rather formed loosely assembled matrices of cells, which are typical of biofilms formed by other species on various surfaces. In addition to material that appeared to be typical of a bacterial EPS in both Salmonella serovars, a web-like network of strands that connected the bacteria to one another and to the gallstone surface was observed (Fig. 2C). Previous reports suggested that similar structures may result from the condensation of EPS that occurs during the fixation process (7). However, it is also possible that these strands are unique to Salmonella gallstone biofilms. Therefore, both CSLM and SEM demonstrated that Salmonella spp. can form biofilms on gallstone surfaces in vitro but that this process was rather slow in our culture system compared to formation of biofilms of other bacterial species, which can develop in as little as a few hours or a few days.
FIG. 2.
SEM micrographs of S. enterica serovar Typhimurium biofilm formation on the surfaces of human gallstones. (A) Example of the gallstone surface on which Salmonella are most commonly found. The gallstone was grown in the presence of bacteria in LB broth alone, and the results demonstrate that salmonellae do not form a biofilm or adhere to the surface unless bile is present in the medium. (B) Wild-type Salmonella cells exposed to a human gallstone for 14 days in the presence of LB broth containing 3% bile are loosely dispersed over the surface of a gallstone with apparent desiccated EPS. (C) Increased magnification of panel B, showing the web-like strands connecting bacteria both to other bacteria and to the gallstone surface.
Biofilm formation is bile dependent.
Once it was determined that biofilm formation occurs on gallstones, it was of interest to examine the conditions needed to form a biofilm. Because the gallbladder is the storage site for bile, it is hypothesized that bile may act as a signal for biofilm formation to occur. Therefore, gallstones were incubated in the presence of either LB broth alone or LB broth containing 3% bile. The presence of bile induced the formation of a full biofilm on a gallstone after 14 days in both S. enterica serovar Typhimurium and S. enterica serovar Typhi cultures, whereas no biofilm was observed after 14 days on the gallstones in LB broth without bile (Fig. 2). Therefore, in our model system, Salmonella biofilm formation on gallstones was dependent upon bile in the culture medium.
Because bile is necessary for biofilm formation on gallstones to occur in 14 days, it is likely that bile either signals the bacteria to form a biofilm or conditions the surfaces of the gallstones, making adhesion easier. To test this hypothesis, gallstones without bacteria were placed in either LB broth alone or LB broth with 3% bile and incubated for 7 days. Bacteria were then added, and the cultures were treated as described above for 4 days, fixed, and viewed by SEM. In addition, planktonic bacteria were grown in LB broth or LB broth with 3% bile at 37°C for 7 days with the cultures back-diluted into fresh medium every 24 h. After the 7 days, the bacteria were incubated with a gallstone for 4 days, fixed, and viewed by SEM. The results showed that neither incubating the bacteria nor conditioning the gallstones with bile before the gallstones and bacteria were incubated together induced full biofilm formation after 4 days (data not shown). These results indicate that preconditioning bacteria or gallstones with bile is not sufficient to initiate rapid biofilm formation. Therefore, it is likely that the bacteria receive signals both from bile and from interacting with the gallstone surface to initiate biofilm development.
Biofilm formation is surface specific.
Bacterial biofilms have been found on a variety of biotic and abiotic surfaces (10, 12). To test the specificity of Salmonella biofilm formation on gallstones, bacteria were incubated under identical conditions (LB broth containing 3% bile) with either a gallstone or a granite pebble that was a similar size. After 14 days, the gallstone and the pebble were examined by SEM. Although some bacteria were found to be bound to the pebble surface, the characteristics of the bound bacteria did not resemble the characteristics of the bacteria in a biofilm on the gallstone. The bacteria, which were in clumps, were tightly compacted on the surface of the pebble in scattered patches with little visible EPS (data not shown). Furthermore, the number of bacteria bound to the gallstone was much larger than the number of bacteria bound to the pebble.
The surface of an individual gallstone is not uniform, either in composition or in topology. Examination of both SEM and CSLM images showed that while salmonellae readily bound to the gallstone surface, there were patches on the surface to which the bacteria did not adhere (Fig. 1A). Based on the SEM studies, surface specificity does not appear to be due to vast differences in gallstone topology, as an unbound surface can be observed in the same plane as a bound surface, but is probably due to the materials found in the surface. Interestingly, even larger numbers of salmonellae are often observed adhering to surfaces below the newly formed outer crust of a gallstone (data not shown). Therefore, in addition to the dependence on bile for efficient Salmonella biofilm formation on gallstones, there appears to be specificity in attachment to certain gallstone surface structures or surface components.
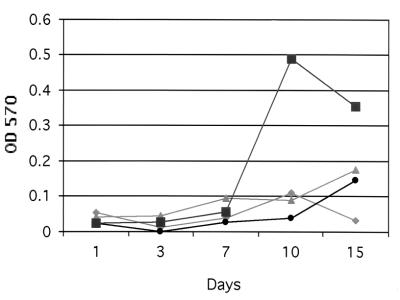
Quantification of the bacteria involved in biofilm formation on a gallstone is difficult and inaccurate due to the brittle nature of the gallstone itself, gallstone-to-gallstone variations, and the variable release of bacteria from the stone surface by chemical or mechanical means. In order to quantify Salmonella during biofilm formation, a system for growing biofilms on glass slides was developed. Biofilm density on the glass slides was determined by the crystal violet staining-destaining method described in Materials and Methods. Results comparing biofilm formation by wild-type S. enterica serovar Typhimurium in different media (LB broth, LB broth containing 3% bile, TSB, and TSB containing 3% bile) are shown in Fig. 3. The results demonstrate that biofilms are denser and form more quickly when they are grown in TSB than when they are grown in LB broth. Interestingly, Salmonella is able to form a biofilm on a glass slide in the presence of LB broth alone, something which does not occur in 14 days on a gallstone, further suggesting that the bacteria interact differently with diverse surfaces. In addition, similar to the results observed with gallstone biofilms, adding bile to either medium (TSB or LB broth) further enhanced biofilm formation on glass slides (Fig. 3).
FIG. 3.
Quantitation of biofilm density. JSG224 was grown in the presence of LB broth alone (⧫), LB broth containing 3% bile (•), TSB alone (▴), or TSB containing 3% bile (▪) and assayed quantitatively using the crystal violet staining-destaining method. OD 570, optical density at 570 nm.
Flagella are important for biofilm formation.
It has previously been demonstrated that flagella play an important role in biofilm formation for organisms such as Escherichia coli (12, 27), Vibrio cholerae (41), and Pseudomonas aeruginosa (25, 37); in these organisms nonmotile mutants have been shown to be severely defective in the ability to form a biofilm. Flagella are necessary for moving a bacterium to the surface for initial attachment and across the surface to find other bacteria with which to interact. To determine if flagella play a role in Salmonella biofilm formation on gallstones in our model system, a nonmotile strain harboring a mutation in fliA was used. fliA encodes the transcription factor σ28, which is required for production of the late flagellar genes encoding the external filament (8). As with wild-type S. enterica serovar Typhimurium, it was observed that a fliA mutant did not adhere to gallstones after 4 days. However, while this mutant was found to adhere to gallstones after 14 days (albeit in lower numbers), a full biofilm did not form as it did with the wild-type strain and did not appear to produce the strands and EPS mentioned above (Table 2). These data demonstrate that flagella, although not absolutely required, play a role in the architecture and development of Salmonella gallstone biofilms.
TABLE 2.
Summary of results from SEM visualization of gallstones
| Prepn | Appearance of bacteria on gallstones after:
|
|
|---|---|---|
| 4 Days | 14 Days | |
| S. enterica serovar Typhimurium, LB broth | No bacteria present | No bacteria present |
| S. enterica serovar Typhimurium, LB broth + 3% bile | No bacteria present | Bacteria (biofilm) present |
| S. enterica serovar Typhi, LB broth | No bacteria present | No bacteria present |
| S. enterica serovar Typhi, LB broth + 3% bile | No bacteria present | Bacteria (biofilm) present |
| S. enterica serovar Typhi, capsular mutant (ViAg−) | No bacteria present | Bacteria (biofilm) present |
| S. enterica serovar Typhimurium flagellar mutant (FliA−) | No bacteria present | Bacteria present, altered appearance |
| S. enterica serovar Typhimurium fimbrial mutants (Fim−, Pef−, Agf−, Lpf−)a | Bacteria (biofilm) present | Bacteria (biofilm) present |
| S. enterica serovar Typhimurium quorum sensing mutant (LuxS−) | No bacteria present | Few bacteria present |
| S. enterica serovar Typhimurium rough LPS mutant (GalE−) | No bacteria present | Bacteria present, altered appearance |
The background strain for these S. enterica serovar Typhimurium mutants is SR11; the background strain for all other S. enterica serovar Typhimurium mutants is ATCC 14028s. Biofilm formation on gallstones by wild-type strain SR11 is indistinguishable from biofilm formation on gallstones by the SR11 fimbrial mutants.
The capsule produced by S. enterica serovar Typhi does not play a role in biofilm formation
It has been shown that a number of organisms produce an EPS when they form a biofilm (11, 12). This polysaccharide is important for establishing a matrix within which the organisms are stabilized and for providing protection against antimicrobial agents and immune system factors. S. enterica serovar Typhi produces an EPS, Vi antigen (ViAg), which has been implicated in the pathogenesis of typhoid fever (30). This antigen is not produced by S. enterica serovar Typhimurium. Our microscopy studies described above suggested that salmonellae form an EPS when they form a biofilm. Because the type of EPS produced by Salmonella spp. within biofilms has not been defined, an S. enterica serovar Typhi ViAg-deficient strain (tviB) was examined to determine if the capsule was necessary for S. enterica serovar Typhi biofilm formation and responsible for the polysaccharide observed by SEM. Gallstones exposed to this mutant strain for 4 and 14 days showed results identical to those observed for wild-type S. enterica serovar Typhi (Table 2). Few bacteria were observed bound to the gallstone after 4 days, and a full biofilm had formed after 14 days. Furthermore, the EPS observed by SEM covering the organisms and the strands connecting bacteria to bacteria and bacteria to the gallstone surface were still present. These results suggest that the ViAg is not the major EPS produced by Salmonella spp. during biofilm development and does not play a prominent role in biofilm formation.
Salmonellae produce an EPS during biofilm formation.
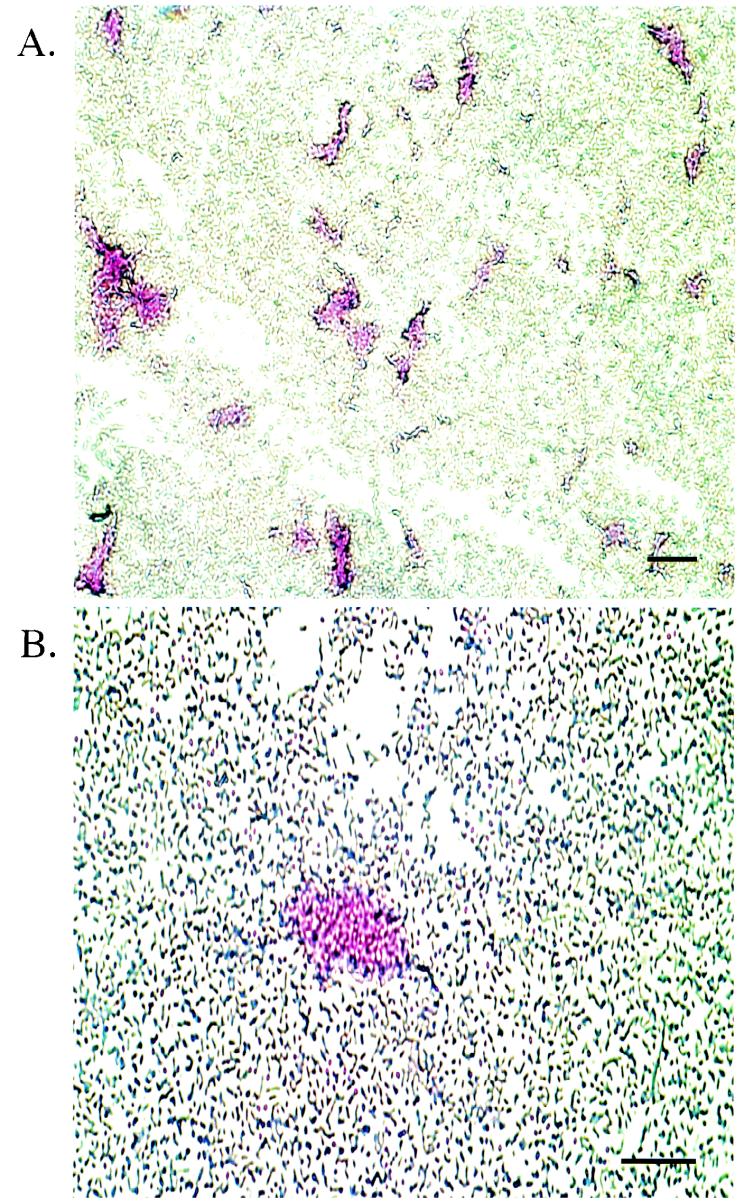
Because, as mentioned above, an EPS produced for biofilm formation has not been described previously for Salmonella spp. and ViAg of S. enterica serovar Typhi does not appear to play a role in biofilm formation, it was necessary to confirm the presence of EPS in the bacterial communities. To confirm that Salmonella does produce an extracellular matrix, ruthenium red stain, a stain specific for polysaccharides and often used in EPS detection, was used to stain biofilms formed on glass slides. The results revealed ruthenium red staining of regions containing clumps of cells likely to be in a biofilm or early microcolony formation (Fig. 4). In addition, the gallstones with biofilms were air dried for 4 days before they were placed in the fixing solution to keep the matrix from condensing during the fixing process for SEM. This drying procedure preserved the EPS, as shown in Fig. 5, in which bacteria appear to be embedded and not simply on the surface. Also, the web-like strands are not visible, suggesting that they were likely condensed EPS.
FIG. 4.
Salmonella biofilms on glass slides stained with ruthenium red for EPS. (A) Staining biofilms with ruthenium red demonstrated a specificity for groups of bacteria excreting a polysaccharide. Magnification, ×100. Bar = 10 μm. (B) Increased magnification of panel A, showing one microcolony stained with ruthenium red. Magnification, ×400. Bar = 10 μm.
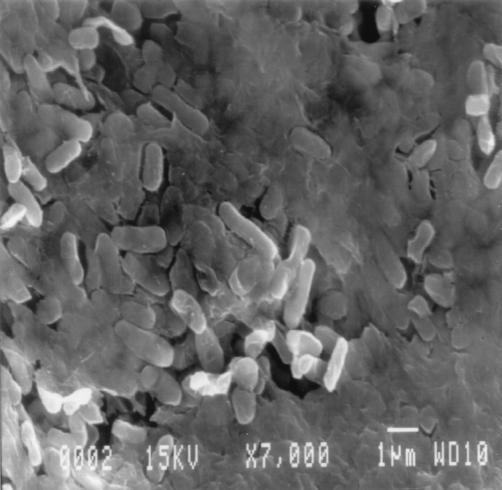
FIG. 5.
Air drying gallstones before fixation preserves the EPS. A gallstone with an S. enterica serovar Typhimurium biofilm was air dried for 3 days before fixation, and SEM revealed bacteria embedded in a putative EPS matrix without the apparent web-like strands seen in Fig. 2B. Magnification, ×7,000.
GalE is necessary for the final stages of biofilm formation.
galE, which codes for a uridine diphosphogalactose-4-epimerase, is a structural gene responsible for synthesis of galactose, which is added in both the outer core and the O-antigen. A mutation in this gene produces a lipopolysaccharide (LPS) lacking all sugars beyond the heptose region of the inner core (therefore, a rough or incomplete LPS). A strain containing a Tn10 insertion in galE was examined for its ability to form a biofilm on gallstones. The results demonstrated that the galE mutant could form a biofilm in 14 days but that there were fewer web-like strands and they were much thinner than the wild-type strands, indicating that galE is necessary for the formation of a wild-type biofilm; however, it is not clear if the biofilm deficiency was due to the lack of LPS O-antigen, EPS production (in which galE also plays a role), or both (Table 2).
Fimbriae play a negative role in biofilm formation.
It has been demonstrated that in other organisms fimbriae are necessary for biofilm formation (12, 25, 27, 32). Fimbriae play a role in attachment of the bacteria to other bacteria and to the surface, and this fimbria-mediated attachment is a signal for initiation of microcolony formation. Because fimbriae play an important role in biofilm formation in other organisms, various S. enterica serovar Typhimurium fimbrial mutants were examined in association with gallstones. Strains with individual mutations in four fimbrial operons (37), as well as a strain in which these four mutations were combined, were examined for gallstone biofilm formation. These mutations were constructed in S. enterica serovar Typhimurium strain SR11 and were not recreated in or transduced into the ATCC 14028s background, the parent of all other strains used in this study. Surprisingly, SEM examination revealed that all of the fimbrial mutants, including the quadruple knockout mutant, formed complete biofilms after 4 days (data not shown). Because an alternate wild-type S. enterica serovar Typhimurium strain was the parent of these fimbrial mutants, SR11 was tested for its ability to form a biofilm on gallstones. It was found that SR11 formed a biofilm indistinguishable from those formed by the fimbrial mutants, even after 4 days (both in the presence and in the absence of bile). Therefore, SR11 is much more efficient in the formation of gallstone biofilms than ATCC 14028s. Furthermore, in the background of rapid biofilm formation, the fimbriae examined did not appear to play a significant role in binding to the gallstone surface.
Quorum sensing is important for full biofilm formation.
Quorum sensing, which has been shown to be used in many different bacteria as a mechanism for cell signaling based on cell density, is thought to regulate a variety of processes, such as virulence, motility, conjugation, and biofilm formation (13, 21, 26, 31, 33, 34). S. enterica serovar Typhimurium has been shown to have a homologue for luxS, an autoinducer-2 gene (33, 34). The autoinducer has been implicated in helping the bacteria convert from a pathogenic state in the host to a free-living state in the environment (33). S. enterica serovar Typhimurium containing a MudJ insertion in luxS was tested for its ability to form biofilms on gallstones after 4 and 14 days in the presence of bile. The gallstones were viewed by SEM, and no adherent bacteria were found after 4 days. The gallstone incubated for 14 days had very few scattered bacteria with little apparent EPS (Table 2). These results indicate that quorum sensing is necessary for salmonellae to form a full biofilm in the in vitro system employed.
DISCUSSION
Although biofilm formation has been well described for many organisms, very little is known about the ability of Salmonella spp. to form biofilms. While it has been suggested that S. enterica serovar Typhimurium can form biofilms on stainless steel, glass, and polyvinyl chloride (6, 14, 18), the interaction of salmonellae with organic materials, such as gallstones, has not been examined. In salmonella carriers, formation of a biofilm on gallstone surfaces would offer the bacteria long-term protection from antimicrobial agents and high concentrations of bile, as well as a stable base from which to continually shed planktonic cells into the intestine and into the environment.
It has been demonstrated that individuals with gallstones who are infected with S. enterica serovar Typhi are more likely to become carriers than individuals without obvious gallbladder abnormalities (15, 20). This suggests that gallstones play a role in carrier state development. Because the gallbladder is constantly emptying, it would be advantageous for an organism to firmly attach itself to a stable surface, such as a gallstone, to avoid being washed into the environment. Currently, the best means of clearing an S. enterica serovar Typhi carrier state in an individual with gallstones is by surgical removal of the gallbladder or gallstones (20). This suggests that the organism is in a state protected from antibiotics, which further suggests the presence of a biofilm.
To determine if biofilm formation can occur on gallstones, human gallstones were grown with Salmonella spp. in the presence of LB broth containing 3% bile or LB broth alone and examined by both CSLM and electron microscopy. The results showed that salmonellae (both S. enterica serovar Typhi and S. enterica serovar Typhimurium) formed complete biofilms on gallstones within 14 days in our model system and that bile was required for efficient biofilm formation to occur. Because bile was needed in the medium for salmonellae to form a biofilm, the following question arose: does bile condition the surface of the gallstone or does it serve as a signal for the bacteria? Experimental results suggest that conditioning the bacteria or the gallstone with bile is not sufficient to initiate rapid biofilm formation, indicating that an interaction among the gallstone, the bacteria, and the bile is required for biofilm formation in our system. This could explain the length of time needed for Salmonella to form a biofilm, as time may be necessary to form the correct environment and for multiple signals to be sensed and acted upon.
Examination of several different specimens showed that salmonellae have an affinity for certain surfaces on a gallstone. Salmonella does not appear to bind as well to the outermost surface of a gallstone as to lower layers where the outer crust has not formed yet or has broken off. In addition, two very different surfaces can be in one continual plane, yet the bacteria may adhere to only one of them. This suggests that there is specificity in surface binding, possibly due to a distinct receptor(s) recognized by the bacterium. To further test the specificity of the organisms for the gallstone surface, a granite pebble of similar size was incubated under growth conditions that were optimal for biofilm formation on gallstones. The results showed that the bacteria weakly adhere to a pebble under ideal gallstone biofilm conditions, but a typical biofilm is not formed. These results further suggest that there may be a specific interaction between the gallstone and Salmonella that is necessary to initiate biofilm formation. Because cholesterol and calcium bilirubinate are the two most abundant components of gallstones, it is possible that one of these factors is the ligand bound by the bacterium.
A method for growing biofilms on glass slides was developed in order to have an accurate means of bacterial quantification. Glass slides were incubated in LB broth, LB broth containing 3% bile, TSB, and TSB containing 3% bile. The results showed that all of the media induce biofilm formation (although biofilms form faster in TSB) and that bile enhances biofilm formation even more in both LB broth and TSB. These results further confirm the role of bile in accentuating biofilm formation. In addition, it is of interest that bile is not required for biofilm formation on glass slides but is required for biofilm formation on gallstones in the system employed. These data again suggest, as mentioned above, that there is significant interplay among the bacterium, the medium, and the surface that has an impact upon the speed of formation and density of a Salmonella biofilm.
In order to further characterize the ability of salmonellae to form a biofilm on a gallstone, the roles of several bacterial surface organelles and products in this process were analyzed. Capsule, flagella, fimbriae, LPS, and quorum sensing were the initial targets of investigation. Flagella have been shown to be very important in biofilm formation, especially in the early stages when microcolonies are being formed. Flagella are needed to move the bacteria to the surface for attachment and then to propel the organisms across the surface in search of other bacteria (10, 25, 27). Nonmotile E. coli mutants show a severe defect in the ability to form a biofilm (27). To determine if flagella play a role in Salmonella biofilm formation on gallstones, a mutant that was defective in flagellar production (and therefore nonmotile) was analyzed by SEM. After 14 days of growth, a weak biofilm was present, but the phenotype was quite different from that of the S. enterica serovar Typhimurium wild type. With the flagellar mutant, there were many fewer bacteria on the surface than with the wild type, and they were widely dispersed. Furthermore, EPS was not found to be associated with the nonmotile bacteria bound to the gallstone. These results suggest that the Salmonella flagella may play a role in EPS secretion or production, as well as in both initial adherence and microcolony formation (aided by bacterial movement across the surface) on gallstones.
Fimbriae, which have also been shown to be important in biofilm development, were examined for their role in Salmonella gallstone biofilms. In E. coli, strains with mutations in type I fimbriae are severely defective in initial attachment (12, 27). P. aeruginosa strains with a mutation in type IV fimbriae do not form a biofilm as densely packed as that seen with a wild-type strain (25). The type IV fimbriae appear to play a role in full biofilm formation by stabilizing cell-to-cell interactions, and much like flagella, they give the bacteria the ability to move across the surface to form multicell aggregates (25). Based upon the genome sequence, S. enterica serovar Typhimurium contains 12 fimbrial operons, most of which have not yet been fully characterized (A. Baumler, personal communication). S. enterica serovar Typhimurium strains with mutations (either individually or all in one strain) in four of these operons (fim, agf, lpf, and pef) were examined for biofilm formation. The fim operon encodes a type I fimbria that is peritrichous (35, 37). The agf operon encodes a thin aggregative fimbria that is also peritrichous in nature (9, 37). The lpf operon encodes long polar fimbriae that are necessary for virulence and are needed for colonization of murine Peyer's patches (3, 5, 37), while the pef operon is on a plasmid and aids in adhesion to the small intestine (4, 37). The S. enterica serovar Typhimurium wild-type strain associated with these mutants is SR11, while all of the other strains examined in this study were in an ATCC 14028s background. With each of the fimbrial mutant strains, as well as the SR11 parent, a full biofilm was formed on gallstones after only 4 days. This time course can be compared to that of ATCC 14028s, which takes at least 14 days to form a complete biofilm. After several days of growth on solid medium, SR11 and the fimbrial mutants appeared to develop a rugose-like phenotype. Rugose-colony variants of V. cholerae have been shown to produce an EPS that allows biofilm formation, while smooth-colony variants are deficient for biofilm formation (39, 41). In addition, S. enterica serovar Typhimurium strain DT104 has been shown to exhibit a rugose phenotype on certain media, which may accentuate its biofilm-forming ability (1). It is possible that if SR11 can exhibit a rugose phenotype, it may overproduce an EPS that allows rapid biofilm formation. Regardless, because the fimbrial mutants formed biofilms on gallstones indistinguishable from those formed by their SR11 parent, none of the fimbriae are likely to be involved in gallstone biofilms. These data could suggest that different fimbriae are needed during stages of intestinal and gallbladder infection and that specific environmental signals, such as bile, may play a significant role in their regulation.
Strains with an incomplete O-antigen or rough LPS have been shown to be defective in biofilm formation (24). Studies with E. coli that selected for altered or defective biofilm formation revealed several deep rough mutants (16). A galE mutant, which is known to be unable to add any sugars to the O-antigen above the primary heptose in the core, was examined for its ability to form a biofilm on gallstones. The results demonstrated that while this mutant is able to adhere and form an early biofilm, it appears to be defective in full biofilm formation. Genevaux et al. suggested that in E. coli, the defective LPS has a pleiotropic effect on other extracellular structures, such as fimbriae or flagella, and that this is the reason for defective biofilm formation (16). galE has also been shown to be involved in the pathway for producing the sugars needed to make colanic acid, which is an EPS in E. coli that has been implicated in biofilm formation (11, 32). Our results demonstrate that galE plays a role in biofilm formation, but whether it is due to an incomplete LPS or to defective EPS production has not yet been determined.
Many bacteria have developed a type of communication to regulate gene expression based on cell density known as quorum sensing (for a review see reference 21). When the autoinducers, and therefore the bacteria, reach a critical density in an environment, a cascade is initiated in which genes are activated for the continued survival of the organism in that environment (13, 21, 26, 31, 33, 34). In organisms such as P. aeruginosa, quorum sensing has been shown to be required for both biofilm formation and virulence (13, 21, 26). Autoinducers have been studied in S. enterica serovar Typhimurium and are thought to provide signals that aid in modifying the bacteria for survival in the host versus survival in the environment (33). The best-studied signaling system is the acyl-homoserine lactone system (26). Autoinducers found originally in Vibrio harveyi, a bioluminescent marine organism, have been divided into two distinct groups, AI-1 and AI-2. AI-1, which is a homoserine lactone, is thought to be a unique signal used only by V. harveyi for intraspecies communication, but AI-2, produced from S-adenosylmethionine, may be used by several bacteria for interspecies relations (31, 34). The gene responsible for AI-2 production, luxS, has been shown to be highly conserved in V. harveyi, E. coli, and S. enterica serovar Typhimurium (31, 33, 34). Because these signals have been shown to be important in biofilm formation in other organisms and are present in S. enterica serovar Typhimurium, it was thought that perhaps quorum sensing also plays a role in Salmonella biofilm formation (13, 21, 26). To test if quorum sensing plays a role in Salmonella biofilm formation, a luxS::MudJ mutant was examined for its ability to form a biofilm on gallstones in our model system. The results showed that luxS was necessary for biofilm formation, since a biofilm was not formed in 14 days by the mutant, in contrast to the wild type. These data suggest that Salmonella uses quorum sensing (mediated at least in part by the Salmonella AI-2) to signal full biofilm development.
In addition to analysis of surface organelles and products associated with biofilm formation, it is also necessary to further characterize the extracellular matrix produced by salmonellae in a biofilm. Most bacteria produce an EPS within a biofilm for protection and stability, but the compositions of the matrices can differ greatly. P. aeruginosa produces an EPS, alginate, that is made up of glucose, galactose, and pyruvate (29). V. cholerae makes Vps, whose primary components are glucose, galactose, N-acetylglucosamine, mannose, and xylose (40). The EPS produced by E. coli, colanic acid, is composed of glucose, galactose, and fucose (11, 32). The EPS produced by Salmonella spp. within a biofilm remains to be characterized. Through SEM studies of air-dried and non-air-dried gallstones that had been incubated with Salmonella, evidence of EPS production was obtained. In addition, ruthenium red staining of biofilms on glass coverslips demonstrated the presence of polysaccharide associated with clumps of cells. S. enterica serovar Typhi produces an N-acetylglucosamine uronic acid capsule known as ViAg. Because EPS are known to be involved in development of the matrix of a biofilm (11, 12), the role of S. enterica serovar Typhi ViAg in gallstone biofilm formation was examined. The results of SEM of gallstones cultured with an S. enterica serovar Typhi tviB strain (for 14 days) showed that the ViAg capsule does not play a role in biofilm formation and that S. enterica serovar Typhi produces an EPS that is biofilm specific. These results suggest that S. enterica serovar Typhi (and S. enterica serovar Typhimurium as well, as it does not produce ViAg) produces another EPS in response to biofilm development. Because E. coli is thought to produce the extracellular polysaccharide colanic acid within biofilms (11), we are currently examining whether a colanic acid-like polymer may be responsible for EPS production in Salmonella biofilms.
The results presented here demonstrate that bile is required for salmonellae to form efficient biofilms on gallstones. Several proteins are both negatively and positively regulated by bile (38). In addition, it has been shown that bile can affect pathogenic properties of S. enterica serovar Typhimurium, such as invasion of epithelial cells (28). Bile resistance can also be increased by exposure to sublethal bile concentrations (38). These results suggest that salmonellae have the ability to sense bile, and the presence of bile may lead to induction of bacterial factors that promote biofilm formation. These factors could be adhesins (fimbrial or afimbrial) that promote early biofilm formation. In addition to adherence, bile sensing may also result in induction of bile resistance genes. The ability to adapt to high concentrations of bile would be important for survival within the gallbladder. Therefore, bile can act as a unique environmental signal, like classic environmental signals such as pH, osmolarity, and temperature, to alter bacterial processes. Experiments are currently being performed to determine how bile may be sensed in Salmonella, which is clearly necessary for efficient biofilm formation on gallstones in our model system.
We demonstrated that Salmonella can form biofilms on human gallstones in vitro and that biofilm formation is markedly enhanced in the presence of bile. It is likely that most, if not all, gallbladder carriage of Salmonella spp. involves biofilm formation on fully formed gallstones, newly nucleated pregallstones, or other hepatobiliary abnormalities. If the process of biofilm formation in the gallbladder is understood, it may be possible to develop therapeutic or preventative approaches to counteract this chronic infection.
Acknowledgments
We thank Daniel Guerro for technical assistance with the SEM work and Vicki Frohlich for assistance with confocal microscopy. Strains used in the study were graciously provided by Andreas Baumler, Michel Popoff, Stanley Falkow, Bonnie Bassler, and Karl Klose.
This work was supported by a grant from the San Antonio Area Foundation from the Semp Russ Foundation.
Editor: B. B. Finlay
REFERENCES
- 1.Anriany, Y. A., R. M. Weiner, J. A. Johnson, C. E. De Rezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 67:4048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arricau, N., D. Hermant, H. Waxin, C. Ecobichon, P. S. Duffey, and M. Y. Popoff. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835-850. [DOI] [PubMed] [Google Scholar]
- 3.Baumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler, A. J., R. M. Tsolis, F. A. Bowe, J. G. Kusters, S. Hoffmann, and F. Heffron. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. USA 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. Garcia-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 191:31-36. [DOI] [PubMed] [Google Scholar]
- 7.Chan, R., S. D. Acres, and J. W. Costerton. 1984. Morphological examination of cell surface structures of enterotoxigenic strains of Escherichia coli. Can. J. Microbiol. 30:451-460. [DOI] [PubMed] [Google Scholar]
- 8.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson, S. K., S. C. Clouthier, J. L. Doran, P. A. Banser, and W. W. Kay. 1996. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 11.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey, M. E., and A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhir, V. K., and C. E. Dodd. 1995. Susceptibility of suspended and surface-attached Salmonella enteritidis to biocides and elevated temperatures. Appl. Environ. Microbiol. 61:1731-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta, U., P. K. Garg, R. Kumar, and R. K. Tandon. 2000. Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gallbladder. Am. J. Gastroenterol. 95:784-787. [DOI] [PubMed] [Google Scholar]
- 16.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Gunn, J. S. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907-913. [DOI] [PubMed] [Google Scholar]
- 18.Jones, K., and S. B. Bradshaw. 1996. Biofilm formation by the Enterobacteriaceae: a comparison between Salmonella enteritidis, Escherichia coli and a nitrogen-fixing strain of Klebsiella pneumoniae. J. Appl. Bacteriol. 80:458-464. [DOI] [PubMed] [Google Scholar]
- 19.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 20.Lai, C. W., R. C. Chan, A. F. Cheng, J. Y. Sung, and J. W. Leung. 1992. Common bile duct stones: a cause of chronic salmonellosis. Am. J. Gastroenterol. 87:1198-1199. [PubMed] [Google Scholar]
- 21.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 22.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills, J., L. Pulliam, L. Dall, J. Marzouk, W. Wilson, and J. W. Costerton. 1984. Exopolysaccharide production by viridans streptococci in experimental endocarditis. Infect. Immun. 43:359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 26.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 28.Prouty, A. M., and J. S. Gunn. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68:6763-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Read, R. R., and J. W. Costerton. 1987. Purification and characterization of adhesive exopolysaccharides from Pseudomonas putida and Pseudomonas fluorescens. Can. J. Microbiol. 33:1080-1090. [DOI] [PubMed] [Google Scholar]
- 30.Robbins, J. D., and J. B. Robbins. 1984. Reexamination of the protective role of the capsular polysaccharide (Vi antigen) of Salmonella typhi. J. Infect. Dis. 150:436-449. [DOI] [PubMed] [Google Scholar]
- 31.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 34.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tinker, J. K., L. S. Hancox, and S. Clegg. 2001. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 37.van der Velden, A. W., A. J. Baumler, R. M. Tsolis, and F. Heffron. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66:2803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]