Abstract
Recombinant leptospiral outer membrane proteins (OMPs) can elicit immunity to leptospirosis in a hamster infection model. Previously characterized OMPs appear highly conserved, and thus their potential to stimulate heterologous immunity is of critical importance. In this study we undertook a global analysis of leptospiral OMPs, which were obtained by Triton X-114 extraction and phase partitioning. Outer membrane fractions were isolated from Leptospira interrogans serovar Lai grown at 20, 30, and 37°C with or without 10% fetal calf serum and, finally, in iron-depleted medium. The OMPs were separated by two-dimensional gel electrophoresis. Gel patterns from each of the five conditions were compared via image analysis, and 37 gel-purified proteins were tryptically digested and characterized by mass spectrometry (MS). Matrix-assisted laser desorption ionization-time-of-flight MS was used to rapidly identify leptospiral OMPs present in sequence databases. Proteins identified by this approach included the outer membrane lipoproteins LipL32, LipL36, LipL41, and LipL48. No known proteins from any cellular location other than the outer membrane were identified. Tandem electrospray MS was used to obtain peptide sequence information from eight novel proteins designated pL18, pL21, pL22, pL24, pL45, pL47/49, pL50, and pL55. The expression of LipL36 and pL50 was not apparent at temperatures above 30°C or under iron-depleted conditions. The expression of pL24 was also downregulated after iron depletion. The leptospiral major OMP LipL32 was observed to undergo substantial cleavage under all conditions except iron depletion. Additionally, significant downregulation of these mass forms was observed under iron limitation at 30°C, but not at 30°C alone, suggesting that LipL32 processing is dependent on iron-regulated extracellular proteases. However, separate cleavage products responded differently to changes in growth temperature and medium constituents, indicating that more than one process may be involved in LipL32 processing. Furthermore, under iron-depleted conditions there was no concomitant increase in the levels of the intact form of LipL32. The temperature- and iron-regulated expression of LipL36 and the iron-dependent cleavage of LipL32 were confirmed by immunoblotting with specific antisera. Global analysis of the cellular location and expression of leptospiral proteins will be useful in the annotation of genomic sequence data and in providing insight into the biology of Leptospira.
Leptospira is a genus of spirochetal bacteria and the causative agent of leptospirosis, a zoonotic disease of worldwide distribution. There are ca. 230 recognized serovars of pathogenic leptospires, whose distribution may be restricted geographically (17). The immense serovar diversity among pathogenic leptospires has been attributed to differences in the structure and composition of lipopolysaccharide (LPS) (5). Much work has focused on the role of leptospiral LPS in immunity (18, 33), and the genetics of LPS biosynthesis have been partly elucidated (5, 6, 13, 14). Preparations of leptospiral LPS can elicit protective immunity, but this is generally serovar specific (17). Recent work has shown that combinations of recombinant leptospiral outer membrane proteins (OMPs) (4, 28) or LPS free outer membrane preparations (46) can also elicit protective immunity in laboratory animals. Utilizing a small number of different serovars, outer membrane preparations have been shown to provide heterologous protection (46). Given the inherent difficulty in preparing multivalent LPS vaccines (36), the identification of conserved protein antigens for use in vaccination and diagnosis of leptospirosis is of critical importance.
Freeze fracture electron microscopy studies have shown Leptospira spp. to have approximately one-tenth the content of transmembrane OMPs as Escherichia coli, despite their similar genetic capacities (29). OmpL1 is the only leptospiral transmembrane OMP which has been identified to date and is thought to be a heat-modifiable porin which is present in the outer membrane in small amounts (25, 44). In contrast, the leptospiral outer membrane contains relatively abundant amounts of lipoproteins, which associate with the outer membrane via N-terminal fatty acids. The major protein of the leptospiral outer membrane is a 32-kDa lipoprotein designated LipL32 (26). In a recent evaluation of recombinant leptospiral protein antigens by enzyme-linked immunosorbent assay, LipL32 was found to be the immunodominant antigen (19). A less-abundant leptospiral OMP designated LipL36 was found to be downregulated in host-adapted leptospires, suggesting that it is not involved in pathogenesis after entry into the mammalian host (3, 27). Another highly conserved leptospiral lipoprotein, LipL41 (45), can provide 71% protection in the hamster model of leptospirosis when administered in combination with recombinant OmpL1 (28). A model of leptospiral membrane architecture has begun to be assembled (54), but preliminary investigations utilizing one-dimensional electrophoresis indicate that there are at least 16 protein constituents of the leptospiral outer membrane (29), suggesting that this model is far from complete.
Advances in the field of two-dimensional gel electrophoresis (2-DGE) have increased the ability to resolve complex protein mixtures and enhanced the reproducibility of these separations (20). Concomitant advances in mass spectrometry (MS) have decreased the amount of protein necessary for identification down to a subfemtomole level (51). These technological advances now allow high-resolution analysis of discrete cellular fractions, such as the bacterial outer membrane (37). However, the usefulness of such an approach is limited by the original ability to obtain a highly purified cellular fraction free of contaminating material from other cellular locations. By taking advantage of structural differences between spirochetes and typical gram-negative cells, relatively pure outer membrane preparations can be obtained from spirochetes. In these organisms, the cytoplasmic, rather than the outer membrane, is associated with peptidoglycan, and hence the outer membrane can be solubilized with low concentrations of Triton X-114 without disrupting the cytoplasmic membrane (12). Subsequently, the contaminating hydrophilic periplasmic proteins are removed by temperature-induced phase separation of Triton X-114. Isolation of outer membranes by Triton X-114 extraction and phase partitioning was adapted for use with leptospires by Zuerner et al. (55) and evaluated by Haake et al. (29). Electron microscopy of spirochetes subjected to this procedure shows that the cytoplasmic membrane retains its lipid bilayer structure (12). Immunoblotting shows the preparations to be free of periplasmic contaminants such as flagella and penicillin-binding proteins (29).
In this study we utilized 2-DGE and MS to separate and characterize the protein constituents of leptospiral outer membranes. Outer membranes were isolated from Leptospira interrogans serovar Lai grown at 20, 30, and 37°C in the presence of 10% fetal calf serum at 37°C and in iron-depleted medium at 30°C. Using a solution designed to solubilize hydrophobic membrane proteins, the protein components of the outer membrane preparations were separated by 2-DGE. We were able to identify all but one of the previously described leptospiral OMPs by peptide mass mapping of tryptic digest fragments. Electrospray-ionization tandem MS (ESI-MS) was then used to generate peptide sequences from eight novel leptospiral OMPs. Additionally, the expression of at least four proteins was found to vary under the conditions investigated.
MATERIALS AND METHODS
Growth of leptospires.
L. interrogans serovar Lai was grown in EMJH medium at 30°C (32) and enumerated as described previously (1). Leptospires were allowed to reach a density of 3 × 107 cells/ml. Culture conditions were then shifted to fresh media incubated at 20, 30, or 37°C in the presence of 10% fetal calf serum at 37°C and iron-depleted medium. Cultures were allowed to reach a final density of 5 × 108 cells/ml before the outer membranes were extracted. To achieve iron-limited conditions, the leptospires was pelleted at 10,000 × g and resuspended in fresh EMJH medium that had been preincubated overnight with 0.4 mM 2,2-dipyridyl (Sigma Chemical Co., St. Louis, Mo.).
Triton X-114 extraction.
L. interrogans outer membrane material was extracted by a method described previously (26). Briefly, leptospires were washed in phosphate-buffered saline-5 mM MgCl2 and then extracted in the presence of 1% protein-grade Triton X-114 (Calbiochem)-150 mM NaCl-10 mM Tris (pH 8)-1 mM EDTA at 4°C. The insoluble material was removed by centrifugation at 17,000 × g for 10 min. After centrifugation, 20 mM CaCl2 was added to the supernatant. Phase separation was performed by warming the supernatant to 37°C and subjecting it to centrifugation for 10 min at 1,000 × g. The detergent and aqueous phases were then separated and precipitated with acetone.
2-DGE.
Acetone-precipitated outer membrane material from 1.2 × 1010 leptospires was resuspended in 460 μl of membrane protein-specific sample solution [7 M urea, 2 M thiourea, 1% tetradecanoylamido-propyl-dimethyl amido-propane-sulforate (ASB-14), 2 mM tributylphosphine, and 1% carrier ampholytes] by vortexing (40). Insoluble material was removed by centrifugation at 12,000 × g for 10 min. The 460-μl samples were used to passively rehydrate pH 4 to 7 immobilized pH gradient (IPG) dry strips (Bio-Rad Laboratories, Hercules, Calif.). Isoelectric focusing was performed by using a stepwise protocol with a final voltage of 3,500 V on a Multiphor II apparatus (Amersham Pharmacia Biotech, Uppsala, Sweden) equaling a final total of 75 kVh. Focused IPG strips were reduced and alkylated for 30 min in solution containing 6 M urea, 2% sodium dodecyl sulfate (SDS), 1× Tris-HCl buffer (pH 8.8), 20% glycerol, 5 mM tributylphosphine, and 2.5% acrylamide monomer. The second dimension was performed with 8 to 18% T gradient gels (50) by using a Protean II Multi-Cell (Bio-Rad). The gels were fixed in 40% methanol-10% acetic acid with shaking for 1 h prior to overnight staining in Sypro Ruby (Molecular Probes, Eugene, Oreg.). The gels were destained in 10% methanol-7% acetic acid for a minimum of 3 h before images were acquired with a Molecular Imager Fx (Bio-Rad). All 2-DGE analyses were performed in triplicate for each of the culture conditions tested. The 2-DGE images were normalized and compared visually, prior to statistical analysis of protein spot differences by using the software package PD-Quest (Bio-Rad). Average protein spot intensities changing twofold between culture conditions and replicated across triplicate gel sets were selected as biologically significant.
Sample preparation for MS.
Protein spots were excised from gels by using a sterile scalpel and placed in a 96-well tray. Gel pieces were washed with 50 mM ammonium bicarbonate-100% acetonitrile (60:40 [vol/vol]). The gel pieces were dried in a Speed Vac (Savant Instruments, Holbrook, N.Y.) for 25 min and then rehydrated in 12 μl of 12 ng of sequencing-grade modified trypsin (Promega, Madison, Wis.)/μl for 1 h at 4°C. Excess trypsin solution was removed, and the rehydrated gel pieces were immersed in 50 mM ammonium bicarbonate and incubated overnight at 37°C. The eluted peptides were concentrated and desalted by using μ-C18 Zip-Tips (Millipore Corp., Bedford, Mass.) and washed with 10 μl of 5% formic acid. For peptide mass mapping, the bound peptides were eluted from the Zip-Tip in matrix solution (10 mg of α-cyano-4-hydroxycinnamic acid [Sigma]/ml in 70% acetonitrile) directly onto the matrix-assisted laser desorption ionization (MALDI) target plate. For ESI-MS, peptides were eluted from the Zip-Tip in 1 μl of 50% methanol-1% formic acid directly into borosilicate nanoelectrospray needles (Micromass, Manchester, United Kingdom).
MALDI-TOF MS.
MALDI-time-of-flight (TOF) mass spectra were acquired by using either a PerSeptive Biosytems Voyager DE-STR (Framingham, Mass.) or a Micromass TofSpec2E. Both instruments were equipped with 337-nm nitrogen lasers. All spectra were obtained in reflectron-delayed extraction mode, averaging 256 laser shots per sample. Two-point internal calibration of spectra was performed based upon internal porcine trypsin autolysis peptides (842.5 and 2211.10 [M+H]+ ions). A list of monoisotopic peaks corresponding to the mass of generated tryptic peptides was used to search the TrEMBL and SWISS-PROT databases via the PeptIdent program, while the Mascot program (41) was used to search the National Center for Biotechnology Information (NCBI) database. Successful identifications were based on the number of matching peptide masses and the percent sequence coverage afforded by those matches.
ESI-MS.
Tandem MS was performed with ESI on a Q-Tof hybrid quadrupole/orthogonal-acceleration TOF mass spectrometer (Micromass, Manchester, United Kingdom). Nanoelectrospray needles containing the samples were mounted in the source, and stable flow was usually obtained with a capillary voltage between 900 and 1,200 V. An initial mass/charge (m/z) scan was performed to detect peptide precursor ions. The m/z of the individual precursor ions was selected for fragmentation in the collision cell. The collision gas was argon, and collision energies and argon pressure were tuned to optimize the fragmentation pattern. Collision energies of 18 to 30 eV were usually sufficient to induce cleavage of amino acids. Data were recorded and processed by using MassLynx version 3.5 (Micromass), and amino acid sequences were deduced by the mass differences between y- or b-ion “ladder” series. The resulting sequences were searched against the NCBI nonredundant database by using the BLAST program (2).
Western blotting.
Proteins were separated by using a Bio-Rad Mini-Protean II gel electrophoresis apparatus (Bio-Rad) with a 12.5% polyacrylamide resolving gel and a 4% polyacrylamide stacking gel employing the buffer system of Laemmli (34). Proteins were then transferred to Immobilon-P membranes (Millipore) with a Trans-Blot electrophoretic transfer cell (Bio-Rad). The membranes were incubated with a 1/1,000 dilution of rabbit anti-LipL32 (26) and anti-LipL36 sera (27). Binding of rabbit antisera was detected by using horseradish peroxidase-conjugated goat-anti rabbit immunoglobulin G (Silenus Laboratories, Melbourne, Australia).
RESULTS
Characterization of leptospiral OMPs by 2-DGE and MS.
Initial separations utilizing isoelectric focusing in the pH 3 to 10 range indicated that the majority of leptospiral OMPs had isoelectric points of between four and seven (data not shown). Subsequent 2-DGE separations of leptospiral OMPs were therefore performed by using pH 4 to 7 IPGs and 2-DGE sample solubilization solution containing ASB-14 (42), which resulted in the optimal resolution of these proteins (Fig. 1). A preliminary evaluation of the purity of the outer membrane extracts was performed by comparing gel images from Triton X-114 aqueous-phase proteins with the outer membrane extract (data not shown). None of the protein spots in the Triton X-114 aqueous phases could be detected in the outer membrane extract, indicating that the outer membrane preparations were free from periplasmic contaminants.
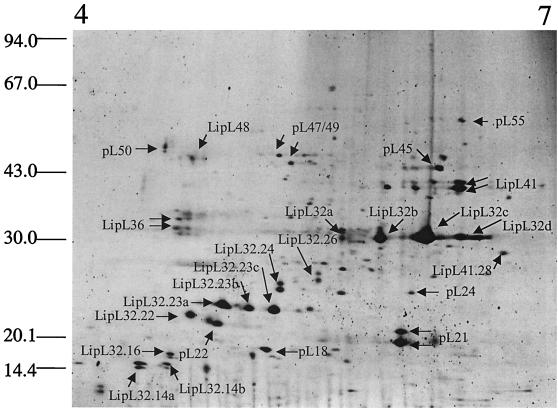
FIG. 1.
Separation of L. interrogans serovar Lai OMPs by 2-DGE utilizing a 4 to 7 IPG. Previously characterized proteins were identified by peptide mass matching and are indicated with the correct nomenclature on the gel image. Novel proteins for which peptide sequence was obtained were assigned a temporary name with the prefix pL (leptospiral protein) and a suffix indicating their approximate relative molecular masses. Cleavage products of the previously characterized proteins LipL32 and LipL41 are noted with the correct nomenclature followed by a period and the relative molecular mass of the cleavage product. When there is more than one isoform of a protein of the same molecular mass they are each arbitrarily assigned a letter. When one arrow alludes to more than one protein spot, the proteins where found to be indistinguishable, and they were considered to be an artifact of the 2-DGE. The positions of standard molecular mass markers in kilodaltons are indicated on the left.
Since L. interrogans is routinely grown in vitro at 30°C, 2-D gels derived from outer-membrane-enriched fractions from cells grown at this temperature were used as the basis for experimental comparisons of OMPs from cells grown under other conditions. To this end, we attempted to characterize proteins expressed at this temperature via 2-DGE combined with MS to generate a reference “map” of leptospiral OMPs. A total of 67 individual protein species was detected by fluorescence staining and image analysis (Fig. 1). These were given unique identifiers based on their estimated molecular mass, and 37 individual protein spots were selected for further characterization by MS.
A two-tiered strategy was employed to analyze leptospiral OMPs by MS. Initially, all protein spots were excised and subjected to in-gel tryptic digestion and analyzed by MALDI-TOF MS. The peptide masses were used to search sequence databases. Peptide mass mapping revealed significant matches to three leptospiral outer membrane lipoproteins LipL32, LipL36, and LipL41 (Table 1). Additionally, significant peptide mass matches were obtained to the unpublished sequence of the leptospiral outer membrane lipoprotein LipL48 (D. A. Haake, unpublished data). OmpL1 was the only previously characterized leptospiral OMP to which no match could be obtained. The failure to identify OmpL1 may be due to a number of factors, including low expression levels, poor solubilization in the isoelectric focusing sample buffer, or possession of a theoretical isoelectric point (pI) of 8.9, which was outside the range investigated. 2-DGE performed over the pH range 3 to 10 revealed three potential OmpL1 spots in this region with a mass of ca. 30 kDa; however, none was in sufficient abundance for further characterization.
TABLE 1.
Peptide mass matches obtained during this study to previously characterized leptospiral OMPs
| Protein | No. of peaks matcheda | % Sequence coverageb |
|---|---|---|
| LipL32.14a,b | 3 | 18.8 |
| LipL32.16 | 4 | 29.8 |
| LipL32.22 | 8 | 40.4 |
| LipL32.23a,b,c | 8 | 45.6 |
| LipL32.24 | 8 | 49.5 |
| LipL32.26 | 12 | 52.2 |
| LipL32a,b,c,d | 14 | 64.4 |
| LipL36 | 7 | 36.2 |
| LipL41 | 16 | 38.6 |
| LipL41.28 | 13 | 29.8 |
| LipL48 | 7 | 21.3 |
The number of peaks matched represents the number of peptide masses obtained by MALDI-TOF MS that match the theoretical peptide masses derived by in silico tryptic digestion of the protein.
The percent sequence coverage indicates the relative amount of protein sequence covered by the peptides with matching masses.
Several protein spots were analyzed that showed significant protein sequence coverage to segments of LipL32 but had molecular masses below that predicted or observed for the main 29-kDa form of the protein that has had the N-terminal signal sequence removed. These protein spots were designated LipL32.26, LipL32.24, LipL32.23a,b,c, LipL32.22, LipL32.16, and LipL32.14a,b, signifying the different approximate molecular masses of these forms. These cleavage products and pI isoforms of LipL32 accounted for 21 of the original 67 2-D gel-separated protein spots and ca. 75% of the visible expressed protein (volume of each protein spot with respect to the sum of all protein spot volumes). A single cleavage product was also observed for LipL41 (LipL41.28) which, together with previously uncharacterized pL21, appears to be the second major constituent of L. interrogans outer membranes. No peptide mass matches were obtained with known leptospiral proteins from cellular locations other than the outer membrane, indicating that the extracts were free from contaminating cytoplasmic or periplasmic proteins.
The second tier of the MS investigation involved analysis of several proteins that could not be matched by peptide mass mapping and may thus be previously uncharacterized members of the leptospiral OMP family. Tryptic peptides from these proteins were analyzed by tandem ESI-MS. Peptide sequences of 8 to 16 amino acids were obtained for each of eight proteins now designated pL18, pL21, pL22, pL24, pL45, pL47/49, pL50, and pL55 (Table 2). These peptide sequences were used to search the NCBI database. No significant matches were obtained, suggesting that they are all novel leptospiral proteins.
TABLE 2.
Peptide sequences, derived by ESI-MS, of the novel leptospiral OMPs identified during this study
| Protein | Amino acid sequence(s)a |
|---|---|
| pL18 | DGE[L/I]VT[F/M]GK, YE[L/I]QSH[L/I]ER |
| pL21 | TASV[L/I]VSQSQGVVK |
| pL22 | ST[L/I]QT[L/I]VYGK |
| pL24 | [L/I]SPYDEFA[L/I], PFDTNNTAE[L/I] [L/I]SNFAK |
| pL45 | [F/M]TG[L/I]NADEATK, YNTAATGTYK |
| pL47/49 | VETESVA[L/I]APHR, TVETSTPVA[L/I]TAQK |
| pL50 | PDAAVTY[L/I]FQG[L/I]GK |
| pL55 | VV[F/M]AAETVNH[L/I]R, AV[L/I][L/I]QGTR, [L/I]GDEDHET |
Both isoleucine and leucine have the same mass, leaving an ambiguity in the peptide sequence (indicated by [L/I]). Phenylalanine and oxidized methionine have the same mass, which also leaves an ambiguity in the peptide sequence (indicated by [F/M]).
Global analysis of leptospiral OMPs expressed under different environmental conditions.
Outer membranes were isolated from L. interrogans serovar Lai grown at 20, 30, and 37°C in the presence of 10% fetal calf serum at 37°C and in iron-depleted medium. 2-DGE was performed on identical amounts of starting outer membrane material, and gels were run in triplicate. To provide further statistical confidence, only protein spots showing a twofold or greater change in spot intensity or volume are reported here. Differentially expressed proteins were characterized by MS as described above.
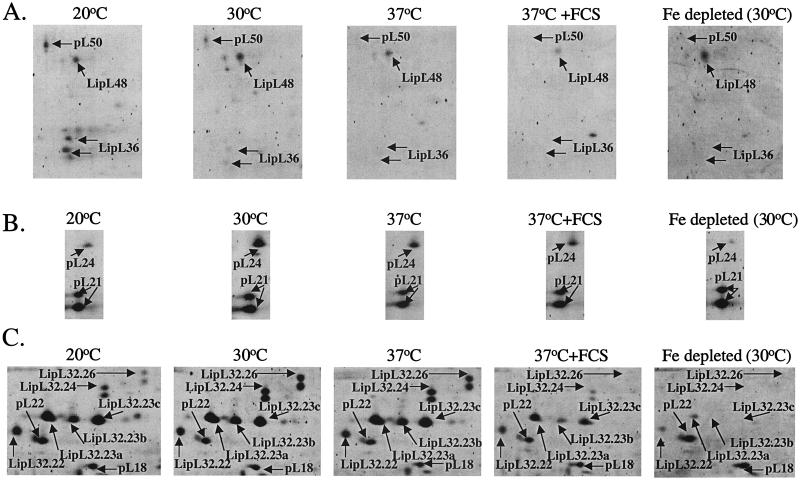
Two mass variants of LipL36 and pL50 were expressed at 20°C and significantly downregulated at 30°C but were not apparent at temperatures above 30°C or after iron depletion at 30°C (Fig. 2A). It has previously been reported that LipL36 is downregulated at temperatures above 30°C and during infection (3, 27); however, this is the first report that LipL36 is also regulated by iron availability. Under the same conditions LipL48 was unaffected (Fig. 2A). Expression of pL24 was shown to be significantly downregulated at 37°C, at 37°C in the presence of fetal calf serum, and at 20°C. Furthermore, expression of pL24 was almost entirely absent after iron starvation (Fig. 2B).
FIG. 2.
Comparisons of gel regions where differences in leptospiral OMP expression were observed. (A) Iron- and temperature-dependent expression of LipL36 and pL50. (B) Iron- and temperature-dependent expression of pL24. (C) Reduced expression and/or absence of LipL32 cleavage products under iron-depleted conditions and the reduced expression of some of the LipL32 cleavage products in the presence of fetal calf serum. For comparison, the locations of other proteins for which expression remained unchanged are indicated on the gel regions. FCS, cultures grown in the presence of 10% fetal calf serum; Fe depleted, cultures grown in iron-depleted medium.
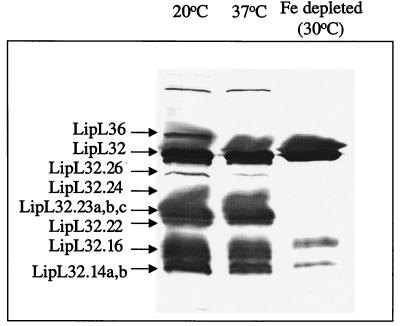
Processing of LipL32 was strikingly affected by environmental growth conditions (Fig. 2C). At 30 and 37°C all mass and pI forms of this protein could be detected at similar levels. At 20°C, LipL32.26 was significantly downregulated, whereas LipL32.24, LipL32.23, and LipL32.22 were largely unaffected. Furthermore, in the presence of fetal calf serum, LipL32.26 could not be detected, while LipL32.24, LipL32.23a,b,c, and LipL32.22 were significantly downregulated. However, significant detectable amounts of LipL32.23a and -c remained. Finally, after iron depletion LipL32.26, LipL32.24, and LipL32.23b and -c could not be detected, while only trace amounts of LipL32.23a and LipL32.22 were observed. In comparison, levels of pL18 remained constant across the five conditions (Fig. 2C). Peptide mass maps of whole LipL32 were compared with those from each of the LipL32 cleavage products. This analysis revealed an absence of peptides from the C terminus in the lower molecular mass forms (Fig. 3). Since the mass forms become smaller, more protein appears to be cleaved from the C terminus. No protein spots corresponding to the C-terminal portion of LipL32 could be detected in this analysis. Of the other major protein spots only a single additional cleavage product was observed for LipL41. This indicates that the C-terminal processing of LipL32 may be highly specific for this protein. To confirm that LipL32 processing was not an artifact of the 2-D gel process, SDS-polyacrylamide gel electrophoresis (PAGE) gels of outer membrane preparations from L. interrogans were probed with anti-LipL32 antisera (Fig. 4). The temperature dependence of LipL36 expression was also confirmed in this manner.
FIG. 3.
LipL32 protein sequence and the corresponding peptide mass matches obtained by MADLI-TOF MS on analysis of a tryptic digest of the lower-molecular-mass forms of LipL32, termed LipL32.23a,b,c (peptide mass matches are underlined). The double-underlined signal peptide does not form part of the mature protein and thus provides no peptide mass match. After cleavage of the signal peptide, the N-terminal tryptic peptide of the mature protein is lipidated and was unable to be detected by MALDI-TOF MS. Peaks corresponding to the smaller peptides were not detected in the MALDI-TOF MS spectrum since they were not within the mass gate analyzed. The absence of any mass matches to C-terminal peptides indicates that LipL32.23a,b,c is truncated at the C terminus.
FIG. 4.
Dual immunoblot of leptospiral outer membrane preparations separated by SDS-PAGE probed with anti-LipL32 and anti-LipL36 polyclonal antisera. Expression of LipL36 was observed at 20°C but not at 37°C or under iron-limiting conditions. The presence of LipL32.26, LipL32.24, and LipL32.22 and the C-terminally truncated LipL32.23a,b,c was detected at 20 and 37°C but not under iron-limiting conditions. The cleavage products of LipL36 are indicated by arrows on the left. Fe depleted, cultures grown in iron-depleted media.
DISCUSSION
Analysis of the leptospiral outer membrane fraction separated by 2-DGE resulted in the identification of four previously characterized OMPs. In addition, the amino acid sequence was obtained from several tryptic peptides of eight novel OMPs. The length of these peptide sequences is sufficient to allow identification of the corresponding genes from genomic sequence data or by design of degenerate oligonucleotide probes.
The combination of high-resolution 2-DGE and MS is a powerful approach for the analysis of complex protein mixtures but is not necessarily comprehensive. It is likely, therefore, that additional leptospiral OMPs still remain to be identified. Separation of proteins by 2-DGE is biased against very large and highly hydrophobic proteins which remain difficult to solubilize (23). In this study, advances in protein solubilization (9, 30, 42) were utilized to increase the solubility of hydrophobic OMPs and lipoproteins. Outer membrane preparations were analyzed from leptospires grown under a variety of conditions to identify proteins that may not be expressed under standard culture conditions. Although factors such as iron depletion and the presence of serum were employed in an attempt to replicate the host environment, proteins that are regulated by more subtle in vivo cues may not necessarily have been detected. Other proteins often not identified during 2-DGE are those that are extremely basic or acidic. The majority of leptospiral proteins have been found to possess isoelectric points between four and seven (21). Hence, the inability to detect proteins such as OmpL1 due to the possession of isoelectric points outside the range investigated is likely to represent an exception rather than the rule. Furthermore, commercial pH gradients are readily available for the separation of large numbers of basic proteins (10). In a 2-DGE separation the overabundance of certain cellular proteins can reduce the dynamic range necessary for the identification of lower-copy-number proteins (22). To counter this problem and increase the dynamic range, protein samples are often prefractionated (11). The preparations analyzed in this study were outer membrane fractions already free from the most abundant cellular proteins, thus obviating the need for specialized prefractionation technologies. We are therefore confident that we have successfully solubilized, separated, and identified the majority of leptospiral OMPs.
The usefulness of this kind of analysis is dependent on the ability to obtain a subcellular fraction of high purity which is free of contaminating proteins from other cellular locations. The Triton X-114 method employed in this study has been shown to be a reliable method for the isolation of the leptospiral outer membrane (24, 29, 55). The outer membrane material obtained by using Triton X-114 extraction and phase partitioning is very similar to leptospiral outer membrane vesicles isolated by sucrose density gradient ultracentrifugation (D. A. Haake and J. Matsunaga, unpublished results). There is only one published example of a spirochetal protein not located in the outer membrane being present in preparations isolated by Triton X-114. The Treponema pallidum cytoplasmic membrane lipoprotein TpN47 was found in Triton X-114 fractions; however, with more labor-intensive methods of outer membrane preparation utilizing isopycnic ultracentrifugation the fractions were not found to contain TpN47 (43). In the present study, MALDI-TOF MS analysis of all of the excised protein spots failed to identify any proteins with a known cellular location other than the outer membrane. The absence of the most abundant leptospiral cellular proteins such as elongation factor Tu, GroEL, and DnaK from the outer membrane extracts is also indicative of the high purity of these preparations. Although downstream analysis of the novel proteins identified during this study should take care to confirm the cellular location of these proteins by such methods as immunogold labeling, it is highly likely that these proteins are legitimate components of the leptospiral outer membrane.
Since many bacteria use temperature, iron availability, and undefined host factors to discern their environment and regulate protein expression, leptospiral OMPs were analyzed from cultures grown under varied conditions. Inside the host, the temperature often exceeds the environmental temperature and free iron is scarce due to the presence of host iron-binding proteins. Expression of LipL36 and the novel protein pL50 could not be detected at temperatures above 30°C or after iron depletion. This suggests that these proteins may not be expressed during mammalian infection and that they may be involved in survival outside the host. This is in agreement with previous immunohistochemical studies by Barnett et al. (3) that have shown LipL36 to be downregulated in vivo. Our results now suggest that this downregulation occurs in response to temperature and/or low iron concentration. Leptospires can be found in almost every mammal (17) and have even been isolated from amphibians (15). An alternative explanation could be that LipL36 and pL50 are expressed at low temperatures because they are required during infection of nonmammalian hosts. However, since human leptospirosis is often acquired through contact with contaminated soil or water, it seems likely that such proteins may be involved in the adaptation of leptospires to the external environment. Expression of the novel protein pL24 was also found to be downregulated after iron depletion, but its expression was still apparent at temperatures approximating that of the mammalian host. Furthermore, the protein was significantly downregulated at a lower environmental temperature. This may reflect tissue-specific expression of this protein during infections in which pL24 is expressed in organs wherein more free iron is available. Throughout this study no proteins were identified that were expressed solely under the conditions that attempted to approximate the host environment. The only protein expression changes that were observed corresponded to down regulation of certain leptospiral OMPs under simulated in vivo conditions. Downregulation of these OMPs may be important in allowing the organism to evade host defense mechanisms. Perhaps if these proteins were constitutively expressed they would potentially mediate a premature immune clearance of the leptospires before infection could be established.
A similar study by Carroll et al. (8) in which proteins were separated by 2-DGE and detected by silver staining and immunoblotting identified more than 37 changes in the Borrelia burgdorferi whole membrane proteome in response to varying pH. That study revealed that the majority of the expression changes occurred when the borreliae were shifted from a neutral pH representative of the host to a more basic pH approximating that of the tick midgut. Because there is no evidence to suggest that leptospires encounter significant pH changes during transmission from the environment to the host, we did not investigate pH-induced expression changes of leptospiral OMPs. However, it is likely that the environmental cues which regulate expression of leptospiral OMPs are multifactorial, and future studies will investigate, among other factors, the role of pH.
In the present study we confirmed that LipL32 is the major leptospiral OMP and we also identified a number of cleavage events that this protein undergoes. Cleavage of LipL32 has previously been reported and has been shown to be inhibited by the presence of E-64, a specific inhibitor of cysteine proteases (55). The presence of calcium chloride, zinc chloride, or copper chloride has also been shown to enhance the stability of LipL32 during extraction of the outer membrane (26, 55). However, since nonequimolar processing of LipL32 has been shown across some of the conditions tested here, we postulate that LipL32 cleavage may be biologically significant and that more than one process may be involved. Posttranslational processing of bacterial surface proteins that results in more than one protein being encoded by a single gene has only rarely been reported. The surface-exposed proteinaceous layer or S layer of Clostridium difficile is composed of two proteins encoded by a single gene slpA. Processing of SlpA involves the removal of a signal peptide and a second cleavage to release two mature surface layer proteins. The higher-molecular-weight cleavage product of SlpA has been shown to possess amidase activity, and it is postulated that the two cleavage products have different functions (7). LipL32 has previously been shown to enhance hemolysis mediated by sphingomyelinase SphH and has been termed the hemolysis-associated protein (35). It would be interesting to investigate whether the cleaved C-terminal portion of LipL32 is responsible for enhanced hemolytic activity, while the N terminus with its attached lipid serves some other function to the leptospiral cell. Mycoplasma hyopneumoniae possesses a 125-kDa ciliary adhesin which is thought to mediate adhesion to swine respiratory epithelium (31, 53). Variable cleavage of this surface adhesin in an avirulent M. hyopneumoniae strain has been deemed responsible for the inability of this strain to adhere to and colonize the swine respiratory epithelium (16). Leptospires that have been passaged in the laboratory lose virulence and concomitantly undergo changes to their surface that are evident by an altered colony morphology (17). It was shown that cleavage of LipL32 was absent when outer membranes were extracted from leptospires grown in the absence of iron, and it would be of interest to investigate whether there is a relationship between LipL32 cleavage and leptospiral virulence. In Porphyromonas gingivalis the surface proteins Omp40 and Omp41 undergo cleavage, and under nonreducing conditions the proteolytic fragments of these proteins have been shown to form heterodimers (49). Further investigation of LipL32 cleavage events should ascertain whether the proteolytic fragments of LipL32 associate with any other leptospiral surface structures. Divalent cations are required for charge stability of LPS, and their absence from the outer membrane extraction buffer may result in the degradation of LipL32, a finding that is consistent with, but not proof of, an association of LipL32 with LPS. The loss of LipL32 cleavage products under iron-depleted conditions did not result in a concomitant increase in levels of whole LipL32. These results, together with differential expression of several LipL32 cleavage products in the presence of serum and at 20°C, indicate that the generation of at least some of these forms occurs independently of others. Conversely, the apparent stability of whole LipL32 expression under iron-limited conditions may simply reflect a reduction in LipL32 expression that is visually compensated for by the lack of processing. LipL32 is the immunodominant protein antigen in leptospires and the most abundant protein constituent of the leptospiral outer membrane. The cleavage events identified in this study warrant further investigation.
The use of 2-DGE and MS for the analysis of the leptospiral outer membrane has yielded an impressive amount of biological data compared with previous studies. An earlier investigation of leptospiral OMPs utilizing one-dimensional electrophoresis failed to detect any iron-dependent expression of OMPs and did not elucidate any protein or nucleic acid sequence information (39). Another investigation noted the temperature-dependent expression of LipL36 and another unidentified OMP, but again no sequence information was obtained (38). The present study has shown that the L. interrogans outer membrane consists of a minimum of 67 discrete protein units, including at least 12 that are the products of different genes. The eight novel proteins for which peptide sequences were obtained in this study would be the most suitable candidates to assess for their role in immunity. The B. burgdorferi vaccinogen OspA is an OMP that is expressed only under conditions that are representative of the tick midgut, and its expression is downregulated on entry into the human host (52). Although expression of OspA is downregulated as soon as the tick begins to feed, serum antibodies to OspA can neutralize infection (47, 48). Both LipL36 and pL50 are downregulated under conditions that approximate the host environment, suggesting that they may warrant investigation as targets for neutralizing antibody on initial entry of leptospires. The new proteins identified and the protein expression information obtained during this study will provide both insight into the biology of Leptospira and a new set of proteins to assess as vaccine candidates.
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Martin R. Larsen, Ian McPherson, and Vicki Vallance. This work was facilitated by access to the Australian Proteome Analysis Facility established under the Australian government Major National Research Facility program.
S.J.C. acknowledges Bio-Rad Laboratories and Micromass for financial and instrumentation support.
Editor: J. T. Barbieri
REFERENCES
- 1.Adler, B., and S. Faine. 1977. Host immunological mechanisms in the resistance of mice to leptospiral infections. Infect. Immun. 17:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., G. Warren, W. Miller, E. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, J. K., D. Barnett, C. A. Bolin, T. A. Summers, E. A. Wagar, N. F. Cheville, R. A. Hartskeerl, and D. A. Haake. 1999. Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect. Immun. 67:853-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branger, C., C. Sonrier, B. Chatrenet, B. Klonjkowski, N. Ruvoen-Clouet, A. Aubert, G. Andre-Fontaine, and M. Eloit. 2001. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect. Immun. 69:6831-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulach, D. M., T. Kalambaheti, A. de la Pena-Moctezuma, and B. Adler. 2000. Functional analysis of genes in the rfb locus of Leptospira borgpetersenii serovar Hardjo subtype Hardjobovis. Infect. Immun. 68:3793-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulach, D. M., T. Kalambaheti, A. de la Pena-Moctezuma, and B. Adler. 2000. Lipopolysaccharide biosynthesis in Leptospira. J. Mol. Microbiol. Biotechnol. 2:375-380. [PubMed] [Google Scholar]
- 7.Calabi, E., S. Ward, B. Wren, T. Paxton, M. Panico, H. Morris, A. Dell, G. Dougan, and N. Fairweather. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187-1199. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, J. A., C. F. Garon, and T. G. Schwan. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevallet, M., V. Santoni, A. Poinas, D. Rouquie, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19:1901-1909. [DOI] [PubMed] [Google Scholar]
- 10.Cordwell, S. J., D. J. Basseal, B. Bjellqvist, D. C. Shaw, and I. Humphery-Smith. 1997. Characterization of basic proteins from Spiroplasma melliferum using novel immobilised pH gradients. Electrophoresis 18:1393-1398. [DOI] [PubMed] [Google Scholar]
- 11.Corthals, G. L., V. C. Wasinger, D. F. Hochstrasser, and J. C. Sanchez. 2000. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis 21:1104-1115. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, T. M., E. M. Walker, J. N. Miller, and M. A. Lovett. 1988. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J. Bacteriol. 170:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Pena-Moctezuma, A., D. M. Bulach, and B. Adler. 2001. Genetic differences among the LPS biosynthetic loci of serovars of Leptospira interrogans and Leptospira borgpetersenii. FEMS Immunol. Med. Microbiol. 31:73-81. [DOI] [PubMed] [Google Scholar]
- 14.de la Pena-Moctezuma, A., D. M. Bulach, T. Kalambaheti, and B. Adler. 1999. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol. Lett. 177:319-326. [DOI] [PubMed] [Google Scholar]
- 15.Diesch, S. L., W. F. McCulloch, J. L. Braun, and H. C. Ellinghausen, Jr. 1966. Leptospires isolated from frog kidneys. Nature 209:939-940. [DOI] [PubMed] [Google Scholar]
- 16.Djordjevic, S., S. Cordwell, J. Wilton, M. Walker, and M. Djordjevic. 2001. Using proteomics to unravel the complex protein cleavage events that occur on the surface of Mycoplasma hyopneumoniae. Proceedings Sixth Australian Conference on Molecular Analysis of Bacterial Pathogens, Marysville, Victoria, Australia.
- 17.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis, 2nd ed. MediSci, Melbourne, Victoria, Australia.
- 18.Farrelly, H. E., B. Adler, and S. Faine. 1987. Opsonic monoclonal antibodies against lipopolysaccharide antigens of Leptospira interrogans serovar Hardjo. J. Med. Microbiol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Flannery, B., D. Costa, F. P. Carvalho, H. Guerreiro, J. Matsunaga, E. D. Da Silva, A. G. Ferreira, L. W. Riley, M. G. Reis, D. A. Haake, and A. I. Ko. 2001. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 39:3303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1037-1053. [DOI] [PubMed] [Google Scholar]
- 21.Guerreiro, H., J. Croda, B. Flannery, M. Mazel, J. Matsunaga, M. Galvao Reis, P. N. Levett, A. I. Ko, and D. A. Haake. 2001. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 69:4958-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gygi, S. P., G. L. Corthals, Y. Zhang, Y. Rochon, and R. Aebersold. 2000. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc. Natl. Acad. Sci. USA 97:9390-9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gygi, S. P., B. Rist, S. A. Gerber, F. Turecek, M. H. Gelb, and R. Aebersold. 1999. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17:994-999. [DOI] [PubMed] [Google Scholar]
- 24.Haake, D. A. 2000. Spirochaetal lipoproteins and pathogenesis. Microbiology 146:1491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haake, D. A., C. I. Champion, C. Martinich, E. S. Shang, D. R. Blanco, J. N. Miller, and M. A. Lovett. 1993. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J. Bacteriol. 175:4225-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haake, D. A., G. Chao, R. L. Zuerner, J. K. Barnett, D. Barnett, M. Mazel, J. Matsunaga, P. N. Levett, and C. A. Bolin. 2000. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect. Immun. 68:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haake, D. A., C. Martinich, T. A. Summers, E. S. Shang, J. D. Pruetz, A. M. McCoy, M. K. Mazel, and C. A. Bolin. 1998. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect. Immun. 66:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haake, D. A., M. K. Mazel, A. M. McCoy, F. Milward, G. Chao, J. Matsunaga, and E. A. Wagar. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 67:6572-6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haake, D. A., E. M. Walker, D. R. Blanco, C. A. Bolin, M. N. Miller, and M. A. Lovett. 1991. Changes in the surface of Leptospira interrogans serovar grippotyphosa during in vitro cultivation. Infect. Immun. 59:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbert, B. 1999. Advances in protein solubilization for two-dimensional electrophoresis. Electrophoresis 20:660-663. [DOI] [PubMed] [Google Scholar]
- 31.Hsu, T., S. Artiushin, and F. C. Minion. 1997. Cloning and functional analysis of the P97 swine cilium adhesin gene of Mycoplasma hyopneumoniae. J. Bacteriol. 179:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, R. C., J. Walby, R. A. Henry, and N. E. Auran. 1973. Cultivation of parasitic leptospires: effect of pyruvate. Appl. Microbiol. 26:118-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jost, B. H., B. Adler, T. Vinh, and S. Faine. 1986. A monoclonal antibody reacting with a determinant on leptospiral lipopolysaccharide protects guinea pigs against leptospirosis. J. Med. Microbiol. 22:269-275. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structual proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. H., K. A. Kim, Y. G. Park, I. W. Seong, M. J. Kim, and Y. J. Lee. 2000. Identification and partial characterization of a novel hemolysin from Leptospira interrogans serovar Lai. Gene 254:19-28. [DOI] [PubMed] [Google Scholar]
- 36.Midwinter, A., S. Faine, and B. Adler. 1990. Vaccination of mice with lipopolysaccharide (LPS) and LPS-derived immuno-conjugates from Leptospira interrogans. J. Med. Microbiol. 33:199-204. [DOI] [PubMed] [Google Scholar]
- 37.Molloy, M. P., B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams, and A. A. Gooley. 2000. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267:2871-2881. [DOI] [PubMed] [Google Scholar]
- 38.Nally, J. E., J. F. Timoney, and B. Stevenson. 2001. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 69:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson, V. M., and J. F. Prescott. 1993. Outer membrane proteins of three pathogenic Leptospira species. Vet. Microbiol. 36:123-138. [DOI] [PubMed] [Google Scholar]
- 40.Nouwens, A. S., S. J. Cordwell, M. R. Larsen, M. P. Molloy, M. Gillings, M. D. Willcox, and B. J. Walsh. 2000. Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis 21:3797-3809. [DOI] [PubMed] [Google Scholar]
- 41.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 42.Rabilloud, T., E. Gianazza, N. Catto, and P. G. Righetti. 1990. Amidosulfobetaines, a family of detergents with improved solubilization properties: application for isoelectric focusing under denaturing conditions. Anal. Biochem. 185:94-102. [DOI] [PubMed] [Google Scholar]
- 43.Radolf, J. D., E. J. Robinson, K. W. Bourell, D. R. Akins, S. F. Porcella, L. M. Weigel, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Treponema pallidum, the syphilis spirochete. Infect. Immun. 63:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang, E. S., M. M. Exner, T. A. Summers, C. Martinich, C. I. Champion, R. E. Hancock, and D. A. Haake. 1995. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect. Immun. 63:3174-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang, E. S., T. A. Summers, and D. A. Haake. 1996. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect. Immun. 64:2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonrier, C., C. Branger, V. Michel, N. Ruvoen-Clouet, J. P. Ganiere, and G. Andre-Fontaine. 2000. Evidence of cross-protection within Leptospira interrogans in an experimental model. Vaccine 19:86-94. [DOI] [PubMed] [Google Scholar]
- 47.Steere, A. C., V. K. Sikand, F. Meurice, D. L. Parenti, E. Fikrig, R. T. Schoen, J. Nowakowski, C. H. Schmid, S. Laukamp, C. Buscarino, and D. S. Krause. 1998. Vaccination against Lyme disease with recombinant Borrelia burgdorferi outer-surface lipoprotein A with adjuvant. N. Engl. J. Med. 339:209-215. [DOI] [PubMed] [Google Scholar]
- 48.Steigbigel, R. T., and J. L. Benach. 1998. Immunization against Lyme disease: an important first step. N. Engl. J. Med. 339:263-264. [DOI] [PubMed] [Google Scholar]
- 49.Veith, P. D., G. H. Talbo, N. Slakeski, and E. C. Reynolds. 2001. Identification of a novel heterodimeric outer membrane protein of Porphyromonas gingivalis by two-dimensional gel electrophoresis and peptide mass fingerprinting. Eur. J. Biochem. 268:4748-4757. [DOI] [PubMed] [Google Scholar]
- 50.Walsh, B. J., and B. R. Herbert. 1998. Casting and running vertical slab-gel electrophoresis for 2D-PAGE, p. 245-253. In A. J. Link (ed.), Methods in molecular biology: 2-D proteome analysis protocols, vol. 112. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 51.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 52.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Q., T. F. Young, and R. F. Ross. 1995. Identification and characterization of a Mycoplasma hyopneumoniae adhesin. Infect. Immun. 63:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuerner, R., D. Haake, B. Adler, and R. Segers. 2000. Technological advances in the molecular biology of Leptospira. J. Mol. Microbiol. Biotechnol. 2:455-462. [PubMed] [Google Scholar]
- 55.Zuerner, R. L., W. Knudtson, C. A. Bolin, and G. Trueba. 1991. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar Pomona. Microb. Pathog. 10:311-322. [DOI] [PubMed] [Google Scholar]